Abstract
The association between cigarette smoking and inflammation is well known. However, the biological mechanisms behind the association are not fully understood, particularly the role of DNA methylation, which is known to be affected by smoking. Using 2-step epigenetic Mendelian randomization, we investigated the role of DNA methylation in the association between cigarette smoking and inflammation. In 822 African Americans from the Genetic Epidemiology Network of Arteriopathy, phase 2 (Jackson, Mississippi; 2000–2005), study population, we examined the association of cigarette smoking with DNA methylation using single nucleotide polymorphisms identified in previous genome-wide association studies of cigarette smoking. We then investigated the association of DNA methylation with levels of inflammatory markers using cis-methylation quantitative trait loci single nucleotide polymorphisms. We found that current smoking status was associated with the DNA methylation levels (M values) of cg03636183 in the coagulation factor II (thrombin) receptor-like 3 gene (F2RL3) (M = −0.64, 95% confidence interval (CI): −0.84, −0.45) and of cg19859270 in the G protein-coupled receptor 15 gene (GPR15) (M = −0.21, 95% CI: −0.27, −0.15). The DNA methylation levels of cg03636183 in F2RL3 were associated with interleukin-18 concentration (−0.11 pg/mL, 95% CI: −0.19, −0.04). These combined negative effects suggest that cigarette smoking increases interleukin-18 levels through the decrease in DNA methylation levels of cg03636183 in F2RL3.
Keywords: cigarette smoking, DNA methylation, inflammation, Mendelian randomization
Cigarette smoking is a risk factor for many diseases, including cancer, pulmonary diseases, and cardiovascular diseases (1, 2). Inflammatory responses have been suggested as one of the mechanisms behind smoking-induced disease risks (3). Serum C-reactive protein (CRP) and fibrinogen concentrations are markers of inflammation. The production of serum CRP is stimulated by the proinflammatory cytokine interleukin-6 (IL-6) (4). The synthesis of IL-6 is promoted by interleukin-18 (IL-18) (5). Cigarette smoking has been associated with elevated levels of inflammatory markers (6), including serum CRP and fibrinogen (7, 8).
Recently, epigenetics has received more attention in environmental epidemiology because of the plasticity and responsiveness of epigenetic modifications to environmental changes (9). DNA methylation is one of the most studied epigenetic mechanisms that regulate levels of gene expression (10). DNA methylation mostly occurs at cytosine-phosphate-guanine (CpG) sites of DNA, and levels of DNA methylation in gene promoter regions are often inversely associated with gene expression levels (11). DNA methylation plays an important role in embryonic development (12), cell differentiation (13), and X chromosome inactivation (14).
Several epigenome-wide association studies have identified CpG sites at which DNA methylation levels are associated with cigarette smoking (15–20). In particular, Dogan et al. (18) showed that long-term cigarette smoking is associated with changes in DNA methylation levels in the promoter regions of genes involved in inflammation, immune function, and coagulation. For example, methylation levels of the CpG sites in the coagulation factor II (thrombin) receptor-like 3 gene (F2RL3) have been consistently associated with cigarette smoking (15, 17, 20). Among African Americans in the Genetic Epidemiology Network of Arteriopathy (GENOA) Study, Sun et al. (20) identified 15 CpG sites that were associated with current smoking status, and findings for 5 of those 15 CpG sites were replicated in an independent study of 262 African Americans in the Grady Trauma Project. However, researchers in the previous study did not provide information on the direction of the associations and did not investigate whether DNA methylation plays a role in cigarette smoking-induced inflammatory responses (20).
The Mendelian randomization (MR) approach is an instrumental-variable analysis that uses a genetic marker as an instrument (21). This approach enables us to examine the causal relationship in observational studies and is less vulnerable to confounding and reverse causation (22, 23). In general, a genetic allele would not be changed by other factors (except somatic mutations in cancer cells). Hence, use of a genetic marker as an instrument is less likely to be affected by confounding. In addition, we can evaluate the direction of the association from one variable to the other (24). The 2-step epigenetic MR approach is an extension of MR that allows us to study the causal role of DNA methylation in the association between an environmental exposure and a health-related outcome (25).
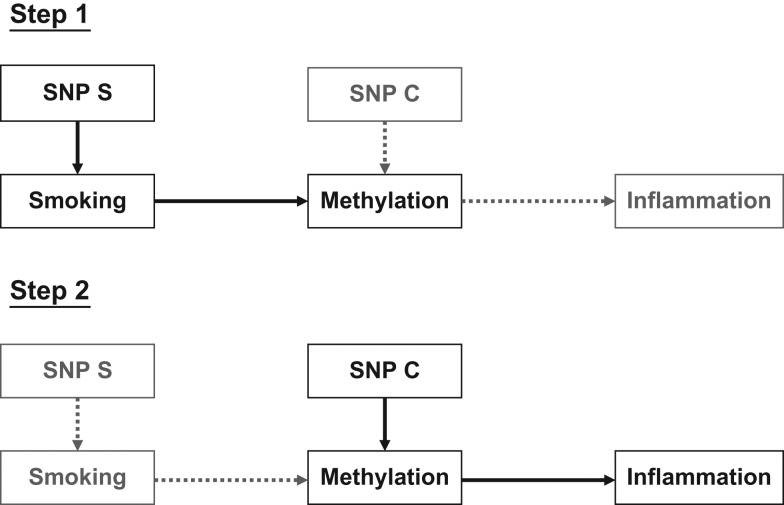
In this study, we hypothesized that cigarette smoking induces changes in DNA methylation levels that result in changes in inflammatory responses. We utilized the 2-step epigenetic MR approach to examine the association of cigarette smoking with DNA methylation using single nucleotide polymorphisms (SNPs) identified in previous genome-wide association studies (GWAS) of cigarette smoking and a genetic risk score (GRS) integrating those SNPs (26). We then investigated the association of DNA methylation with levels of inflammatory markers using cis-methylation quantitative trait loci SNPs and a GRS (Figure 1).
Figure 1.
A 2-step epigenetic Mendelian randomization approach to the study of relationships between cigarette smoking, methylation, and inflammation. Rectangular dark boxes represent variables of interest at each step, and light boxes represent variables of interest at the other step. Solid dark arrows indicate the directed association of interest at each step, and dashed gray arrows indicate the directed association of interest at the other step. Single nucleotide polymorphism (SNP) S is an instrument SNP or a genetic risk score for smoking, and SNP C is an instrument SNP or a genetic risk score for methylation levels.
METHODS
Study population
The GENOA Study was established by the National Heart, Lung, and Blood Institute in 1995 as part of the Family Blood Pressure Program (27). The African-American cohort of GENOA is from Jackson, Mississippi (28). In phase 1 of the GENOA Study (1995–2000), 1,854 subjects who were members of 683 sibships with at least 2 individuals diagnosed with essential hypertension before age 60 years were recruited. In phase 2 (2000–2005), 1,482 of the initial subjects returned. Study visits were made in the morning after an overnight fast. Data on demographic factors, medical history, clinical characteristics, and lifestyle factors were collected, and blood samples were taken. Written informed consent was obtained from all subjects, and approval was granted by participating institutional review boards.
Genotypes
Using blood samples collected in phase 1 of the GENOA Study, genotype data were obtained by means of the Affymetrix Genome-Wide Human SNP Array 6.0 platform (Affymetrix, Inc., Santa Clara, California). Samples that failed to provide genotype data from the platform were genotyped with the Illumina Human 1M-Duo, 660-Quad, or 610-Quad BeadChips array (Illumina, Inc., San Diego, California). Quality control was performed based on the following exclusion criteria: <95% SNP call rate, <1% minor allele frequency, and <95% sample call rate. Since GENOA is a family-based study, Hardy-Weinberg equilibrium was not used for quality control.
The genotype data were prephased using the Segmented HAPlotype Estimation and Imputation Tool (SHAPEIT), version 2 (29). Imputation was performed using IMPUTE, version 2 (30, 31), and reference panels from the 1000 Genomes Project phase 1 integrated variant set release (version 3) in National Center for Biotechnology Information build 37 (hg19; Genome Reference Consortium GRCh37) coordinates (32, 33). Outlier samples from principal component analyses (>6 standard deviations) and monomorphic markers were excluded. After quality control, there were 30,022,375 imputed SNPs available for 1,599 study subjects.
DNA methylation
DNA methylation levels were measured from peripheral blood leukocytes isolated from blood samples collected in phase 2 of the GENOA Study. The EZ DNA Methylation Gold Kit (Zymo Research, Orange, California) was used for bisulfite conversion. The methylation assay was performed at the Mayo Clinic Advanced Genomics Technology Center using Illumina Infinium HumanMethylation27 BeadChips (Illumina, Inc.) and the Illumina BeadXpress reader (Illumina, Inc.). Seven samples with poor bisulfite conversion efficiency (intensity <4,000) and 28 samples with poor background signals were excluded. The R package “lumi” (R Foundation for Statistical Computing, Vienna, Austria) (34) was used for background adjustment, color balance adjustment, and quantile normalization. The methylated and unmethylated intensities for each CpG site were used to calculate a β value,
and an M value,
where max() is a function that returns the larger value (35). We used the M value of DNA methylation levels because the β value has a bounded range from 0 to 1 that violates the Gaussian distribution assumption (35). Thirty samples were removed because more than 5% of probes had a detection P value greater than 0.01. The detection P value for CpG locus j is given by pj = 1 − Φ[(Ij − μneg)/σneg], where Ij is the sum of intensities from the cyanine (Cy) dyes Cy3 and Cy5 (or bead A and bead B for Infinium I), whereas μneg and σneg are the mean value and standard deviation of signals of internal negative controls and Φ(.) is the normal cumulative probability distribution function (36). A total of 1,130 probes were removed due to a detection P value greater than 0.01 in more than 5% of samples. After quality control, there were 26,448 CpG sites measured for 943 subjects.
DNA methylation is a mechanism that differentiates cells (13). Therefore, the proportion of different cell types could affect measurements of DNA methylation. Cell proportions for CD8 T lymphocytes, CD4 T lymphocytes, natural killer cells, B cells, monocytes, and granulocytes were estimated using the method of Houseman et al. (37) and added as covariates.
For DNA methylation, 96 samples were assayed in a single plate. To reduce technical batch effects, the plate information was added as a covariate.
Our study had potential for confounding due to population stratification. To minimize this confounding effect, we computed 4 SNP-based principal components and adjusted for them in the analysis (38).
Smoking variables
Information on cigarette smoking was collected in both phase 1 and phase 2 of the GENOA Study. Participants were asked the following smoking-related questions: 1) “Do you now smoke cigarettes?”; 2) “Have you smoked more than 100 cigarettes in your entire life?”; 3) “On average, how many cigarettes per day do/did you usually smoke?”; 4) “In what year or how old were you when you first started smoking?”; and 5) “In what year or how old were you when you last quit smoking?”.
To be consistent with the previous smoking epigenome-wide association study in GENOA (20), a current smoker was defined as an individual who had smoked within the past year, based on the answers to questions 1, 2, and 5. For example, a person who was not currently smoking (“no” to question 1) but had smoked more than 100 cigarettes in their lifetime (“yes” to question 2) and had quit smoking within the past year (question 5) was classified as a current smoker. An “ever smoker” was defined as anyone who had ever smoked more than 100 cigarettes, based on the answers to question 2.
Inflammatory markers
Levels of inflammatory markers were measured from fasting blood samples obtained during phase 2 of the GENOA Study. CRP levels were measured by means of a highly sensitive immunoturbidimetric assay (39). IL-6 and IL-18 were measured with a 6-plex enzyme-linked immunosorbent assay using a contracted service with SearchLight Technologies (Boston, Massachusetts). Fibrinogen level was measured from citrated plasma using the clotting time-based Clauss method (40). More details on the inflammatory markers have been given previously (41). Data for CRP, IL-6, and IL-18 were log-transformed to reduce skewness.
2-step epigenetic MR
Previously in GENOA, 15 CpG sites were associated with cigarette smoking, findings for 5 of which were replicated in an independent study (20): cg03636183 in F2RL3, cg19859270 in the G protein-coupled receptor 15 gene (GPR15), cg13668129 in the heterogeneous nuclear ribonucleoprotein U-like 1 gene (HNRPUL1), cg01500140 in the lens intrinsic membrane protein 2 gene (LIM2), and cg11314684 in the AKT serine/threonine kinase 3 gene (AKT3). We reevaluated the associations to identify CpG sites that are causally associated with cigarette smoking. Further, we investigated the role of DNA methylation of the identified CpG sites in the association between cigarette smoking and inflammation using the 2-step epigenetic MR approach (Figure 1) (25).
Step 1 MR
In step 1, we used previously identified GWAS SNPs associated with cigarette smoking as an instrument for the MR approach to reevaluate the associations between cigarette smoking and the DNA methylation levels of the 5 CpG sites (step 1 in Figure 1). The 210 SNPs that had been identified in the 3 previous GWAS of cigarette smoking in African Americans or in multiethnic populations (42–44) (see Web Table 1, available at https://academic.oup.com/aje) were tested for the assumptions of the MR approach (Web Appendices 1 and 2) (21). We constructed a GRS composed of the SNPs that satisfied the assumptions and were not in linkage disequilibrium with one another (r2 < 0.3). Using each of the SNPs or the GRS as an instrument, we implemented the step 1 MR approach by conducting 2-stage least-squares regression with adjustment for age, sex, plate, 4 principal components, 5 cell proportions, and random intercepts for family (Web Appendices 1 and 2).
In addition to current smoking status, we evaluated the effect of ever smoking status.
Step 2 MR
In step 2, we examined the association of DNA methylation with levels of inflammatory markers using the MR approach (step 2 in Figure 1). The best candidates for the instrument SNP associated with DNA methylation levels may be SNPs found in methylation quantitative trait loci studies, which identify SNPs associated with DNA methylation levels. To identify appropriate candidates for instrument SNPs, we reviewed studies that identified SNPs associated with the DNA methylation levels of the CpG sites and investigated cis-methylation quantitative trait loci within 10 kilobases of the genes where the CpG sites were located. The candidate SNPs were tested for the assumptions of the MR approach (SNP C in Figure 1; Web Appendices 1 and 2). We also constructed a GRS using the instrument SNPs in step 2. Using each of the identified SNPs or the GRS as an instrument, we implemented the MR approach by running 2-stage least-squares regression with adjustment for age, sex, plate, 4 principal components, 5 cell proportions, and random intercepts for family. To conduct the analysis, we used SAS 9.4 (SAS Institute, Inc., Cary, North Carolina) and the R package “nlme.”
RESULTS
Descriptive statistics
Out of 943 subjects with methylation data, 121 did not have available SNP information and were excluded, resulting in the inclusion of 822 GENOA African Americans in the analysis. The mean age of participants was 66.6 years (standard deviation (SD), 7.5), and 72% (n = 592) of the population were women. In this study population, 13% (n = 104) were current smokers and 42% (29% + 13%) were ever smokers (Table 1). Among ever smokers, the average number of cigarettes smoked per day was 15.1 (SD, 11.8), and the average number of pack-years of smoking was 25.1 (SD, 21.7). Current cigarette smoking status was significantly associated with log(CRP), log(IL-6), and fibrinogen levels and was marginally associated with log(IL-18) levels after adjustment for age and sex (Web Table 2).
Table 1.
Characteristics of African-American Participants in the GENOA Study at the Phase 2 Examination (n = 822), Jackson, Mississippi, 2000–2005
| Variable | No. of Persons | % | Mean (SD) |
|---|---|---|---|
| Continuous variables | |||
| Age at examination, years | 822 | 66.6 (7.5) | |
| Cigarette smoking | |||
| Cigarettes/day | 342 | 15.1 (11.8) | |
| Pack-years | 342 | 25.1 (21.7) | |
| C-reactive protein, mg/L | 795 | 6.1 (6.8) | |
| Interleukin-6, pg/mL | 717 | 10.0 (7.0) | |
| Interleukin-18, pg/mL | 712 | 71.9 (44.8) | |
| Fibrinogen, mg/dL | 797 | 369.2 (80.6) | |
| CD8 T lymphocytes, proportion | 822 | 0.16 (0.08) | |
| CD4 T lymphocytes, proportion | 822 | 0.18 (0.07) | |
| Natural killer cells, proportion | 822 | 0.03 (0.02) | |
| B cells, proportion | 822 | 0.07 (0.03) | |
| Monocytes, proportion | 822 | 0.07 (0.03) | |
| Granulocytes, proportion | 822 | 0.49 (0.11) | |
| Categorical variables | |||
| Sex | |||
| Female | 592 | 72 | |
| Male | 230 | 28 | |
| Cigarette smoking status | |||
| Current smoker | 104 | 13 | |
| Former smoker | 238 | 29 | |
| Never smoker | 480 | 58 |
Abbreviations: CD4, cluster of differentiation 4; CD8, cluster of differentiation 8; GENOA, Genetic Epidemiology Network of Arteriopathy; SD, standard deviation.
Step 1 MR: smoking→DNA methylation
In the previous epigenome-wide association study in GENOA, the DNA methylation levels of 5 CpG sites were associated with current cigarette smoking status and replicated (20). In the current study, we reevaluated the associations using previously identified GWAS SNPs associated with cigarette smoking as an instrument for the MR approach (step 1 in Figure 1; Web Table 1) (42–44). The previous GWAS SNPs were checked for the assumptions of the MR approach, and the SNPs satisfying the assumptions were used for the analyses (Web Appendices 1 and 2).
Current smoking status was associated with decreases in the DNA methylation levels (M values) of cg03636183 in the F2RL3 gene (M = −0.64, 95% confidence interval (CI): −0.84, −0.45) and of cg19859270 in the GPR15 gene (M = −0.21, 95% CI: −0.27, −0.15), using rs4074134 as an instrument (Table 2). Each additional coded allele (T) of rs4074134 was associated with a 0.40-unit increase in the log odds of current smoking (Web Table 3), which decreased the DNA methylation level of cg03636183 by 0.26 (0.40 × (−0.64) = 0.26) and decreased the DNA methylation level of cg19859270 by 0.08 (0.40 × (−0.21) = 0.08). Using different GWAS SNPs and a GRS as an instrument each time, we observed consistent results with variations in the effect sizes (Table 2). The GRS used is defined in Table 2 and Web Appendix 2. The other 3 of the 5 CpGs investigated were not significantly associated with current smoking status (Web Table 4).
Table 2.
Step 1 Mendelian Randomization Results for the Association Between Current Smoking Status and the DNA Methylation Levels of cg03636183 in the F2RL3 Gene and cg19859270 in the GPR15 Gene, GENOA Study, Jackson, Mississippi, 2000–2005
| Gene and Instrument SNPa |
Cytosine-Phosphate-Guanine Site | |||||
|---|---|---|---|---|---|---|
| cg03636183 in F2RL3 | cg19859270 in GPR15 | |||||
| βb | 95% CI | P Value | βb | 95% CI | P Value | |
| BDNF-AS | ||||||
| rs4074134 | −0.64 | −0.84, −0.45 | 3.9 × 10−10 | −0.21 | −0.27, −0.15 | 1.7 × 10−10 |
| rs4923457 | −0.64 | −0.84, −0.45 | 4.0 × 10−10 | −0.21 | −0.27, −0.15 | 1.7 × 10−10 |
| rs4923460 | −0.65 | −0.85, −0.45 | 4.7 × 10−10 | −0.21 | −0.27, −0.15 | 3.0 × 10−10 |
| IREB2 | ||||||
| rs1964678 | −0.47 | −0.65, −0.29 | 6.5 × 10−7 | −0.20 | −0.26, −0.14 | 9.4 × 10−11 |
| HYKK | ||||||
| rs952216 | −0.47 | −0.65, −0.29 | 6.2 × 10−7 | −0.20 | −0.26, −0.15 | 1.7 × 10−11 |
| rs11636131 | −0.48 | −0.67, −0.29 | 7.9 × 10−7 | −0.21 | −0.27, −0.15 | 1.4 × 10−11 |
| rs11632604 | −0.48 | −0.67, −0.29 | 8.1 × 10−7 | −0.21 | −0.27, −0.15 | 1.4 × 10−11 |
| rs12910289 | −0.49 | −0.68, −0.30 | 6.6 × 10−7 | −0.22 | −0.28, −0.16 | 1.1 × 10−11 |
| rs1504546 | −0.48 | −0.67, −0.29 | 8.8 × 10−7 | −0.21 | −0.27, −0.15 | 1.5 × 10−11 |
| rs12916999 | −0.48 | −0.67, −0.29 | 9.1 × 10−7 | −0.21 | −0.27, −0.15 | 1.5 × 10−11 |
| PSMA4 | ||||||
| rs12915366 | −0.48 | −0.66, −0.29 | 9.6 × 10−7 | −0.21 | −0.27, −0.15 | 1.5 × 10−11 |
| rs12916483 | −0.48 | −0.66, −0.29 | 9.7 × 10−7 | −0.21 | −0.27, −0.15 | 1.5 × 10−11 |
| rs3813571 | −0.46 | −0.64, −0.27 | 1.8 × 10−6 | −0.21 | −0.27, −0.15 | 2.2 × 10−11 |
| CHRNA3 | ||||||
| rs1317286 | —c | — | — | −0.18 | −0.25, −0.11 | 5.2 × 10−7 |
| GRSd | −0.18 | −0.30, −0.07 | 2.4 × 10−3 | −0.11 | −0.15, −0.07 | 1.5 × 10−7 |
Abbreviations: BDNF-AS, brain-derived neurotrophic factor antisense RNA; CHRNA3, cholinergic receptor, nicotinic α 3; CI, confidence interval; F2RL3, coagulation factor II (thrombin) receptor-like 3; GENOA, Genetic Epidemiology Network of Arteriopathy; GPR15, G protein-coupled receptor 15; GRS, genetic risk score; HYKK, hydroxylysine kinase; IREB2, iron-responsive element binding protein 2; PSMA4, proteasome subunit α 4; SNP, single nucleotide polymorphism.
a The instrument SNPs were identified in previous genome-wide association studies of cigarette smoking in an independent cohort and satisfied the assumptions of Mendelian randomization in GENOA.
b β coefficient for the exposure (current smoking status) in the second stage of 2-stage least-squares regression analysis. In all models, results were adjusted for age, sex, plate, 4 principal components, 5 cell proportions, and random intercepts for family.
c Missing data for the Mendelian randomization results because of the violation of assumption 2.
d GRS = [rs4074134 + (2 − rs1964678) + (2 − rs952216) + (2 − rs12915366) + (2 − rs1317286)]/5. When the coded allele was negatively associated with current smoking status, we coded for the other allele by subtracting the allele dosage from 2.
Using rs9920506 as an instrument, we found that ever smoking was also associated with decreases in the DNA methylation levels of cg03636183 in F2RL3 (M = −0.62, 95% CI: −0.82, −0.41) and of cg19859270 in GPR15 (M = −0.22, 95% CI: −0.29, −0.16) (Table 3), as well as a GRS. The GRS is defined in Table 3 and Web Appendix 2. As a sensitivity analysis, we ran the MR analyses using different GWAS SNPs each time. We observed consistent results with variations in effect sizes.
Table 3.
Step 1 Mendelian Randomization Results for the Association Between Ever Smoking Status and the DNA Methylation Levels of cg03636183 in the F2RL3 Gene and cg19859270 in the GPR15 Gene, GENOA Study, Jackson, Mississippi, 2000–2005
| Gene and Instrument SNPa |
Cytosine-Phosphate-Guanine Site | |||||
|---|---|---|---|---|---|---|
| cg03636183 in F2RL3 | cg19859270 in GPR15 | |||||
| βb | 95% CI | P Value | βb | 95% CI | P Value | |
| CHRNB4 | ||||||
| rs9920506 | −0.62 | −0.82, −0.41 | 9.1 × 10−9 | −0.22 | −0.29, −0.16 | 3.1 × 10−11 |
| rs8023822 | −0.57 | −0.76, −0.38 | 1.1 × 10−8 | −0.20 | −0.26, −0.14 | 4.3 × 10−10 |
| LOC105370913 | ||||||
| rs4887077 | −0.54 | −0.73, −0.34 | 1.8 × 10−7 | −0.20 | −0.26, −0.14 | 6.8 × 10−10 |
| rs11638372 | −0.55 | −0.75, −0.35 | 9.0 × 10−8 | −0.20 | −0.26, −0.14 | 4.2 × 10−10 |
| rs922692 | −0.57 | −0.77, −0.37 | 4.8 × 10−8 | −0.20 | −0.27, −0.14 | 3.7 × 10−10 |
| rs11072791 | −0.57 | −0.77, −0.37 | 2.8 × 10−8 | −0.21 | −0.27, −0.15 | 5.3 × 10−11 |
| ADAMTS7 | ||||||
| rs12286 | −0.47 | −0.65, −0.29 | 5.0 × 10−7 | −0.19 | −0.25, −0.14 | 1.7 × 10−10 |
| rs1809420 | −0.41 | −0.59, −0.24 | 3.8 × 10−6 | −0.18 | −0.23, −0.12 | 1.2 × 10−9 |
| rs7174367 | −0.45 | −0.63, −0.28 | 5.9 × 10−7 | −0.18 | −0.24, −0.13 | 5.0 × 10−10 |
| rs3825807 | —c | — | — | −0.21 | −0.27, −0.15 | 7.8 × 10−11 |
| rs7177699 | — | — | — | −0.21 | −0.27, −0.15 | 1.1 × 10−10 |
| GRSd | −0.41 | −0.59, −0.24 | 3.0 × 10−6 | −0.17 | −0.22, −0.11 | 5.6 × 10−9 |
Abbreviations: ADAM, a disintegrin and metalloproteinase; ADAMTS7, ADAM metallopeptidase with thrombospondin type 1 motif 7; CHRNB4, cholinergic receptor, nicotinic β 4; CI, confidence interval; F2RL3, coagulation factor II (thrombin) receptor-like 3; GENOA, Genetic Epidemiology Network of Arteriopathy; GPR15, G protein-coupled receptor 15; GRS, genetic risk score; LOC105370913, uncharacterized LOC105370913; SNP, single nucleotide polymorphism.
a The instrument SNPs were identified in previous genome-wide association studies of cigarette smoking in an independent cohort and satisfied the assumptions of Mendelian randomization in GENOA.
b β coefficient for the exposure (ever smoking status) in the second stage of 2-stage least-squares regression analysis. In all models, results were adjusted for age, sex, plate, 4 principal components, 5 cell proportions, and random intercepts for family.
c Missing data for the Mendelian randomization results because of the violation of assumption 2.
d GRS = [rs9920506 + rs4887077 + rs12286]/3. When the coded allele was negatively associated with ever smoking status, we coded for the other allele by subtracting the allele dosage from 2.
Step 2 MR: DNA methylation→inflammation
In step 1 MR, we observed the associations of current and ever smoking with the DNA methylation levels of cg03636183 in F2RL3 and cg19859270 in GPR15. We further investigated the associations of DNA methylation levels of cg03636183 and cg19859270 with the inflammatory markers CRP, IL-6, IL-18, and fibrinogen (step 2 in Figure 1). We did not find an appropriate instrument SNP for cg19859270; hence, we conducted step 2 MR only for cg03636183 (Web Appendices 1 and 2).
In a study carried out in the HapMap Yoruba (Africans) samples, Bell et al. (45) found that rs2227341 was associated with the DNA methylation levels of cg03636183 (http://eqtl.uchicago.edu/Methylation/cis-meQTL.results). This SNP also satisfied the assumptions of the MR approach (F = 480) (Web Table 5). Using rs2227341 as an instrument, we found that the DNA methylation level of cg03636183 in F2RL3 was associated with log(IL-18) levels (−0.11 pg/mL, 95% CI: −0.19, −0.04) (Table 4). Combining step 1 and step 2 MR results, each additional coded allele of rs4074134 for current smokers was associated with a 0.26-unit decrease in the DNA methylation level (M value) of cg03636183 (0.40 × (−0.64) = 0.26) (Web Table 3, Table 2), which resulted in a 3% (e−0.11×(−0.26) = 1.03, 95% CI: 1, 5) increase in serum IL-18 levels (Table 4). In other words, as the log odds of current smoking increased by 1, the DNA methylation level (M value) of cg03636183 decreased by 0.64, which resulted in a 7% (e−0.11×(−0.64) = 1.07, 95% CI: 3, 13) increase in serum IL-18 levels.
Table 4.
Step 2 Mendelian Randomization Results for the Association Between the DNA Methylation Levels of cg03636183 in the F2RL3 Gene and the Inflammatory Markers log(CRP), log(IL-6), log(IL-18), and Fibrinogen, GENOA Study, Jackson, Mississippi, 2000–2005a
| Gene and Instrument SNPb | Inflammatory Marker | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| log(CRP) | log(IL-6) | log(IL-18) | Fibrinogen | ||||||
| βc | 95% CI | βc | 95% CI | βc | 95% CI | P Value | βc | 95% CI | |
| F2RL3 | |||||||||
| rs2227341d | 0.04 | −0.08, 0.16 | 0.002 | −0.07, 0.07 | −0.11 | −0.19, −0.04 | 4.1 × 10−3 | 1.09 | −8.30, 10.49 |
| rs2227353 | 0.04 | −0.08, 0.17 | −0.01 | −0.08, 0.07 | −0.12 | −0.20, −0.05 | 1.8 × 10−3 | 2.55 | −7.10, 12.20 |
| rs773905 | −0.05 | −0.24, 0.15 | −0.05 | −0.16, 0.07 | −0.21 | −0.33, −0.10 | 3.4 × 10−4 | −8.91 | −23.90, 6.08 |
| CPAMD8 | |||||||||
| rs2227368 | 0.01 | −0.15, 0.18 | 0.01 | −0.08, 0.11 | −0.17 | −0.26, −0.07 | 1.2 × 10−3 | −2.26 | −15.03, 10.50 |
| rs10418195 | 0.03 | −0.14, 0.20 | 0.004 | −0.10, 0.10 | −0.18 | −0.28, −0.08 | 6.9 × 10−4 | −0.44 | −13.59, 12.71 |
| rs773901 | −0.05 | −0.25, 0.15 | −0.05 | −0.16, 0.06 | −0.22 | −0.34, −0.11 | 1.8 × 10−4 | −9.93 | −24.99, 5.13 |
| rs7245967 | −0.05 | −0.26, 0.15 | −0.04 | −0.16, 0.07 | −0.20 | −0.32, −0.09 | 8.3 × 10−4 | −10.05 | −25.47, 5.37 |
| rs2981474 | −0.04 | −0.24, 0.16 | −0.04 | −0.15, 0.08 | −0.22 | −0.33, −0.10 | 2.8 × 10−4 | −10.34 | −25.60, 4.92 |
| rs2608732 | −0.06 | −0.26, 0.14 | −0.05 | −0.17, 0.06 | −0.22 | −0.34, −0.11 | 2.3 × 10−4 | −10.68 | −25.92, 4.55 |
| SIN3B | |||||||||
| rs773895 | 0.01 | −0.19, 0.20 | —e | — | −0.20 | −0.32, −0.09 | 6.3 × 10−4 | — | — |
| rs773899 | 0.00 | −0.20, 0.20 | — | — | −0.20 | −0.31, −0.08 | 8.0 × 10−4 | — | — |
| rs1044773 | 0.00 | −0.20, 0.20 | — | — | −0.20 | −0.31, −0.08 | 8.0 × 10−4 | — | — |
| GRSf | 0.06 | −0.10, 0.21 | 0.01 | −0.08, 0.10 | −0.15 | −0.24, −0.06 | 1.4 × 10−3 | 1.39 | −10.34, 13.11 |
Abbreviations: CI, confidence interval; CPAMD8, C3 and PZP-like, α-2-macroglobulin domain containing 8; CRP, C-reactive protein; F2RL3, coagulation factor II (thrombin) receptor-like 3; GENOA, Genetic Epidemiology Network of Arteriopathy; GRS, genetic risk score; IL-6, interleukin-6; IL-18, interleukin-18; PZP, pregnancy zone protein; SIN3B, SIN3 transcription regulator family member B; SNP, single nucleotide polymorphism.
a Because results for log(CRP), log(IL-6), and fibrinogen were not statistically significant, P values are not presented.
b All of the instrument SNPs were located within 10 kilobases of the F2RL3 gene and satisfied the assumptions of Mendelian randomization in GENOA.
c β coefficient for the exposure (cg03636183) in the second stage of 2-stage least-squares regression analysis. In all models, results were adjusted for age, sex, plate, 4 principal components, 5 cell proportions, and random intercepts for family.
d A SNP identified as a methylation quantitative trait locus for cg03636183 in an independent cohort and in GENOA.
e Missing data for the Mendelian randomization results because of the violation of assumption 2.
f GRS = [(2 − rs2227341) + (2 − rs10418195) + (2 − rs773895) + rs7245967 + (2 − rs2981474)]/5. When the coded allele was negatively associated with the DNA methylation levels of cg03636183, we coded for the other allele by subtracting the allele dosage from 2.
For sensitivity analyses, we sought additional instrument SNPs, investigating nearby SNPs that are strongly associated with the DNA methylation levels of cg03636183, and checked the SNPs for the assumptions of the MR approach (Web Appendices 1 and 2). We identified nearby SNPs that satisfied the assumption of MR for cg03636183 (Web Table 5). The SNP rs2227341 was also identified in the Yoruba study (45). In addition, we constructed a GRS, which is defined in Table 4 and Web Appendix 2. In sensitivity analyses using other nearby SNPs and the GRS as an instrument, we observed consistent results with variations in effect sizes. In sensitivity analyses using β values of DNA methylation levels instead of M values, we obtained a similar finding (Web Tables 6–8).
Additionally, we checked associations between step 1 and step 2 instruments and the instrumented exposures and outcomes (Web Table 9), and we compared descriptive statistics between compliers and noncompliers (Web Table 10). Given lifelong tendencies in smoking habits and the cross-sectional nature of our study design, we cannot fully rule out the possibility of reverse causation. However, differences in step 1 SNP genotypes between former smokers and current smokers (Web Table 11) suggested that it is less likely that there is a path from instrument SNPs for current smoking to the outcome through former smoking. Our data suggested that the effect of cigarette smoking due to nicotine-related genes on inflammation and mediation by cg03636183 may be causal.
DISCUSSION
Cigarette smoking is known to induce inflammation (46); however, the biological mechanisms behind the association are not fully understood. DNA methylation is known to be affected by cigarette smoking (47). To our knowledge, however, no study to date has investigated the role of DNA methylation in smoking-induced inflammation. We investigated the role of DNA methylation in the association between cigarette smoking and inflammation using the 2-step epigenetic MR approach (25) in GENOA African Americans. Our findings suggest that cigarette smoking may increase serum IL-18 levels through a decrease in the DNA methylation levels of cg03636183 in the F2RL3 gene.
In the present study, each additional coded allele of rs4074134, which was used as an instrument for current smokers, was associated with a 3% increase in IL-18 concentration that is mediated by the DNA methylation level of cg03636183. Homozygous coded allele carriers, as compared with those without a coded allele, would be expected to have 6% higher IL-18 levels through cg03636183. Using 1-step MR investigating the association of cigarette smoking with serum IL-18 concentration, each additional coded allele of rs4074134 used as an instrument for current smokers was associated with a 3.5% increase in IL-18 level. Therefore, approximately 3% of the 3.5% difference in IL-18 concentration between current smokers and former/never smokers is explained by the DNA methylation levels of cg03636183. Using a conventional mediation analysis, serum IL-18 concentration was 13% higher in current smokers than in former/never smokers in GENOA, which is similar to the results of another study (48). This 13% difference drops to 2% with adjustment for cg03636183 (data not shown). The effect size (13 − 2 = 11%) from conventional analysis may not be directly comparable to the 3% from the 2-step MR, which is based on rs4074134 and is free of confounding and reverse causation.
We found an association of cg03636183 in the F2RL3 gene with IL-18 but not with other inflammatory markers. This may be explained by the unique characteristics of IL-18 as a cytokine that enhances cell-mediated cytotoxicity and both T helper 1 (proinflammatory) and T helper 2 (antiinflammatory) immune responses (49). As an upstream biomarker of inflammation, IL-18 promotes the synthesis of IL-6 (5), which stimulates the production of CRP (4). In GENOA, the correlations between IL-18 and levels of other inflammatory markers were substantially lower than the correlations between other inflammatory markers (, , ). In addition, a previous study in mice and humans demonstrated that IL-18 plays an important role in the pathogenesis of cigarette smoking-induced pulmonary emphysema and inflammation (50). Even though we did not find an association of cg03636183 with the other inflammatory markers, other CpG sites could have an association with other inflammatory markers. Thus, extended study of the associations of other CpGs with inflammation is warranted.
The 2-step epigenetic MR approach enabled us to identify the CpG sites involved in the association between an environmental exposure and an outcome. However, this approach relies on the assumption that the instrument SNP affects the outcome only through the exposure. Yet often a gene or a SNP has associations with different phenotypes (51). Additional paths from the instrument SNP to an outcome can potentially introduce biases into the estimates. We reduced the possibility of violation of the assumption by excluding SNPs that were significantly associated with potential confounders between the exposure and the outcome and SNPs with a significant effect on the outcome through other paths (Web Appendices 1 and 2). We investigated the exclusion restriction assumption in several ways; however, the exclusion restriction assumption cannot be exhaustively tested empirically and must be justified conceptually. Hence, the assumption might not be satisfied. The MR approach also assumes a linear relationship between variables. However, nonlinear relationships are commonly found in epidemiologic studies (52); hence, we might have missed nonlinear relationships. In spite of these limitations, when applied carefully, the 2-step epigenetic MR approach may expand our understanding of the role of DNA methylation in disease development and provide information on the biological mechanisms behind it.
One limitation of our study could be the small number of current smokers. Smoking status is likely to be affected by disease conditions; hence, there were relatively few current smokers in this elderly population with a high prevalence of hypertension. In addition, differential survivorship may also have influenced our findings, as selection of older individuals into the study may have been influenced by genetic factors and/or smoking behaviors. To avoid false findings, we applied the MR approach using all of the smoking GWAS SNPs that satisfied the assumptions and demonstrated consistent results across the use of diverse instrument SNPs. We also investigated ever smoking status and obtained similar results. These consistent findings across diverse instrument SNPs and the GRS and smoking measures increase the reliability of our results. Given the cross-sectional nature of the present study, reverse causation cannot be ruled out. For example, current DNA methylation levels could be influenced by previous IL-18 levels, which could be a result of past smoking and/or past DNA methylation levels. In addition, our study may not have fully eliminated potential ancestry effects, despite adjustment for 4 principal components.
The gene of interest in this study, F2RL3, has been associated with multiple other adverse health outcomes, including platelet activation and perioperative myocardial injury, postinfectious irritable bowel syndrome, and gastric cancer (53–55). IL-18 has been found to play a role in autoimmune disease, diabetes, atherosclerosis, myocardial infarction, coronary heart disease, heart failure, and cardiovascular death (5, 56–61). Elevated serum IL-18 level has been associated with the development of type 2 diabetes (62) and with atherosclerosis and renal dysfunction in type 2 diabetes patients (63, 64). In addition, Mallat et al. (58) found that serum IL-18 levels were substantially higher in patients with ischemic or nonischemic cardiomyopathy than in those without chronic heart failure. Median levels of serum IL-18 were also substantially higher in patients who died than in survivors (58). Thus, further study of the health-related effects of DNA methylation levels in the F2RL3 gene is warranted.
In conclusion, we investigated the role of DNA methylation in the association between smoking and inflammation to shed light on biological processes that occur through DNA methylation. Our findings suggest that DNA methylation levels play a role in cigarette smoking-induced inflammation.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan (Min A Jhun, Jennifer A. Smith, Sharon L. R. Kardia, Patricia A. Peyser, Sung Kyun Park); Research Center for Group Dynamics, Institute for Social Research, University of Michigan, Ann Arbor, Michigan (Erin B. Ware); Department of Medicine and Neurology, University of Mississippi Medical Center, Jackson, Mississippi (Thomas H. Mosley, Jr.); and Division of Nephrology and Hypertension, Mayo Clinic, Rochester, Minnesota (Stephen T. Turner).
This work was supported by the National Heart, Lung, and Blood Institute (grants HL054457, HL087660, HL100185, HL119443, HL081331, and HL133221). S.K.P. was supported by grants from the National Institute of Environmental Health Sciences (grant P30-ES017885) and the Centers for Disease Control and Prevention/National Institute for Occupational Safety and Health (grant T42-OH008455).
We thank Jodie L. Van de Rostyne, Pamela I. Hammond, Dr. Julie M. Cunningham, and the Mayo Clinic Advanced Genomics Technology Center for providing technical assistance. We thank Dr. Kirsten Herold of the University of Michigan School of Public Health Writing Lab for her assistance with manuscript preparation.
Conflict of interest: none declared.
REFERENCES
- 1. Ockene IS, Miller NH. Cigarette smoking, cardiovascular disease, and stroke: a statement for healthcare professionals from the American Heart Association. American Heart Association Task Force on Risk Reduction. Circulation. 1997;96(9):3243–3247. [DOI] [PubMed] [Google Scholar]
- 2. Office on Smoking and Health, National Center for Chronic Disease Prevention and Health Promotion, Centers for Disease Control and Prevention The Health Consequences of Smoking: 50 Years of Progress. A Report of the Surgeon General Atlanta, GA: Centers for Disease Control and Prevention; 2014.
- 3. Wannamethee SG, Lowe GD, Shaper AG, et al. . Associations between cigarette smoking, pipe/cigar smoking, and smoking cessation, and haemostatic and inflammatory markers for cardiovascular disease. Eur Heart J. 2005;26(17):1765–1773. [DOI] [PubMed] [Google Scholar]
- 4. Jones SA, Novick D, Horiuchi S, et al. . C-reactive protein: a physiological activator of interleukin 6 receptor shedding. J Exp Med. 1999;189(3):599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerdes N, Sukhova GK, Libby P, et al. . Expression of interleukin (IL)-18 and functional IL-18 receptor on human vascular endothelial cells, smooth muscle cells, and macrophages: implications for atherogenesis. J Exp Med. 2002;195(2):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Vaart H, Postma DS, Timens W, et al. . Acute effects of cigarette smoke on inflammation and oxidative stress: a review. Thorax. 2004;59(8):713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietrich T, Garcia RI, de Pablo P, et al. . The effects of cigarette smoking on C-reactive protein concentrations in men and women and its modification by exogenous oral hormones in women. Eur J Cardiovasc Prev Rehabil. 2007;14(5):694–700. [DOI] [PubMed] [Google Scholar]
- 8. Sinha S, Luben RN, Welch A, et al. . Fibrinogen and cigarette smoking in men and women in the European Prospective Investigation into Cancer in Norfolk (EPIC-Norfolk) population. Eur J Cardiovasc Prev Rehabil. 2005;12(2):144–150. [DOI] [PubMed] [Google Scholar]
- 9. Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13(2):97–109. [DOI] [PubMed] [Google Scholar]
- 10. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. [DOI] [PubMed] [Google Scholar]
- 11. Weber M, Hellmann I, Stadler MB, et al. . Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–466. [DOI] [PubMed] [Google Scholar]
- 12. Geiman TM, Muegge K. DNA methylation in early development. Mol Reprod Dev. 2010;77(2):105–113. [DOI] [PubMed] [Google Scholar]
- 13. Nagae G, Isagawa T, Shiraki N, et al. . Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum Mol Genet. 2011;20(14):2710–2721. [DOI] [PubMed] [Google Scholar]
- 14. Goto T, Monk M. Regulation of X-chromosome inactivation in development in mice and humans. Microbiol Mol Biol Rev. 1998;62(2):362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Breitling LP, Yang R, Korn B, et al. . Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88(4):450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeilinger S, Kühnel B, Klopp N, et al. . Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8(5):e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shenker NS, Polidoro S, van Veldhoven K, et al. . Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. [DOI] [PubMed] [Google Scholar]
- 18. Dogan MV, Shields B, Cutrona C, et al. . The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics. 2014;15:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harlid S, Xu Z, Panduri V, et al. . CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the Sister Study. Environ Health Perspect. 2014;122(7):673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun YV, Smith AK, Conneely KN, et al. . Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum Genet. 2013;132(9):1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greenland S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol. 2000;29(4):722–729. [DOI] [PubMed] [Google Scholar]
- 22. Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33(1):30–42. [DOI] [PubMed] [Google Scholar]
- 23. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timpson NJ, Nordestgaard BG, Harbord RM, et al. . C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int J Obes (Lond). 2011;35(2):300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41(1):161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horne BD, Anderson JL, Carlquist JF, et al. . Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Ann Hum Genet. 2005;69(2):176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. FBPP Investigators Multi-center genetic study of hypertension: the Family Blood Pressure Program (FBPP). Hypertension. 2002;39(1):3–9. [DOI] [PubMed] [Google Scholar]
- 28. Daniels PR, Kardia SL, Hanis CL, et al. . Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) Study. Am J Med. 2004;116(10):676–681. [DOI] [PubMed] [Google Scholar]
- 29. Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. [DOI] [PubMed] [Google Scholar]
- 30. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howie BN, Marchini J IMPUTE2. http://mathgen.stats.ox.ac.uk/impute/impute_v2.html. Updated December 23, 2014. Accessed September 25, 2017.
- 32. 1000 Genomes Project Consortium; Abecasis GR, Auton A, et al. . An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. University of California, Santa Cruz Frequently asked questions: assembly releases and versions. List of UCSC genome releases. https://genome.ucsc.edu/FAQ/FAQreleases.html. Accessed September 25, 2017.
- 34. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. [DOI] [PubMed] [Google Scholar]
- 35. Du P, Zhang X, Huang CC, et al. . Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuan PF, Wang S, Zhou X, et al. . A statistical framework for Illumina DNA methylation arrays. Bioinformatics. 2010;26(22):2849–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Houseman EA, Accomando WP, Koestler DC, et al. . DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barfield RT, Almli LM, Kilaru V, et al. . Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38(3):231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kullo IJ, Seward JB, Bailey KR, et al. . C-reactive protein is related to arterial wave reflection and stiffness in asymptomatic subjects from the community. Am J Hypertens. 2005;18(8):1123–1129. [DOI] [PubMed] [Google Scholar]
- 40. Khaleghi M, Singletary LA, Kondragunta V, et al. . Haemostatic markers are associated with measures of vascular disease in adults with hypertension. J Hum Hypertens. 2009;23(8):530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim CX, Bailey KR, Klee GG, et al. . Sex and ethnic differences in 47 candidate proteomic markers of cardiovascular disease: the Mayo Clinic Proteomic Markers of Arteriosclerosis Study. PLoS One. 2010;5(2):e9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hamidovic A, Kasberger JL, Young TR, et al. . Genetic variability of smoking persistence in African Americans. Cancer Prev Res (Phila). 2011;4(5):729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. David SP, Hamidovic A, Chen GK, et al. . Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bell JT, Pai AA, Pickrell JK, et al. . DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shiels MS, Katki HA, Freedman ND, et al. . Cigarette smoking and variations in systemic immune and inflammation markers. J Natl Cancer Inst. 2014;106(11):dju294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamagami H, Kitagawa K, Hoshi T, et al. . Associations of serum IL-18 levels with carotid intima-media thickness. Arterioscler Thromb Vasc Biol. 2005;25(7):1458–1462. [DOI] [PubMed] [Google Scholar]
- 49. Nakanishi K, Yoshimoto T, Tsutsui H, et al. . Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Factor Rev. 2001;12(1):53–72. [DOI] [PubMed] [Google Scholar]
- 50. Kang MJ, Homer RJ, Gallo A, et al. . IL-18 is induced and IL-18 receptor alpha plays a critical role in the pathogenesis of cigarette smoke-induced pulmonary emphysema and inflammation. J Immunol. 2007;178(3):1948–1959. [DOI] [PubMed] [Google Scholar]
- 51. Sivakumaran S, Agakov F, Theodoratou E, et al. . Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. May S, Bigelow C. Modeling nonlinear dose-response relationships in epidemiologic studies: statistical approaches and practical challenges. Dose Response. 2005;3(4):474–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y, Yu G, Jiang P, et al. . Decreased expression of protease-activated receptor 4 in human gastric cancer. Int J Biochem Cell Biol. 2011;43(9):1277–1283. [DOI] [PubMed] [Google Scholar]
- 54. Muehlschlegel JD, Perry TE, Liu KY, et al. . Polymorphism in the protease-activated receptor-4 gene region associates with platelet activation and perioperative myocardial injury. Am J Hematol. 2012;87(2):161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han W, Wang Z, Lu X, et al. . Protease activated receptor 4 status of mast cells in post infectious irritable bowel syndrome. Neurogastroenterol Motil. 2012;24(2):113–119, e82. [DOI] [PubMed] [Google Scholar]
- 56. Wei XQ, Leung BP, Arthur HM, et al. . Reduced incidence and severity of collagen-induced arthritis in mice lacking IL-18. J Immunol. 2001;166(1):517–521. [DOI] [PubMed] [Google Scholar]
- 57. Raeburn CD, Dinarello CA, Zimmerman MA, et al. . Neutralization of IL-18 attenuates lipopolysaccharide-induced myocardial dysfunction. Am J Physiol Heart Circ Physiol. 2002;283(2):H650–H657. [DOI] [PubMed] [Google Scholar]
- 58. Mallat Z, Heymes C, Corbaz A, et al. . Evidence for altered interleukin 18 (IL)-18 pathway in human heart failure. FASEB J. 2004;18(14):1752–1754. [DOI] [PubMed] [Google Scholar]
- 59. Suchanek H, Myśliwska J, Siebert J, et al. . High serum interleukin-18 concentrations in patients with coronary artery disease and type 2 diabetes mellitus. Eur Cytokine Netw. 2005;16(3):177–185. [PubMed] [Google Scholar]
- 60. Blankenberg S, Luc G, Ducimetière P, et al. . Interleukin-18 and the risk of coronary heart disease in European men: the Prospective Epidemiological Study of Myocardial Infarction (PRIME). Circulation. 2003;108(20):2453–2459. [DOI] [PubMed] [Google Scholar]
- 61. Blankenberg S, Tiret L, Bickel C, et al. . Interleukin-18 is a strong predictor of cardiovascular death in stable and unstable angina. Circulation. 2002;106(1):24–30. [DOI] [PubMed] [Google Scholar]
- 62. Thorand B, Kolb H, Baumert J, et al. . Elevated levels of interleukin-18 predict the development of type 2 diabetes: results from the MONICA/KORA Augsburg Study, 1984–2002. Diabetes. 2005;54(10):2932–2938. [DOI] [PubMed] [Google Scholar]
- 63. Nakamura A, Shikata K, Hiramatsu M, et al. . Serum interleukin-18 levels are associated with nephropathy and atherosclerosis in Japanese patients with type 2 diabetes. Diabetes Care. 2005;28(12):2890–2895. [DOI] [PubMed] [Google Scholar]
- 64. Araki S, Haneda M, Koya D, et al. . Predictive impact of elevated serum level of IL-18 for early renal dysfunction in type 2 diabetes: an observational follow-up study. Diabetologia. 2007;50(4):867–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.