Abstract
Missing data is a common occurrence in epidemiologic research. In this paper, 3 data sets with induced missing values from the Collaborative Perinatal Project, a multisite US study conducted from 1959 to 1974, are provided as examples of prototypical epidemiologic studies with missing data. Our goal was to estimate the association of maternal smoking behavior with spontaneous abortion while adjusting for numerous confounders. At the same time, we did not necessarily wish to evaluate the joint distribution among potentially unobserved covariates, which is seldom the subject of substantive scientific interest. The inverse probability weighting (IPW) approach preserves the semiparametric structure of the underlying model of substantive interest and clearly separates the model of substantive interest from the model used to account for the missing data. However, IPW often will not result in valid inference if the missing-data pattern is nonmonotone, even if the data are missing at random. We describe a recently proposed approach to modeling nonmonotone missing-data mechanisms under missingness at random to use in constructing the weights in IPW complete-case estimation, and we illustrate the approach using 3 data sets described in a companion article (Am J Epidemiol. 2018;187(3):568–575).
Keywords: inverse probability weighting, missing-at-random data, missing data, nonmonotone missingness
Missing data routinely occur in epidemiologic research, in patterns that are often arbitrary from one unit to another. As an illustration of a prototypical epidemiologic study with missing data, Perkins et al. (1), in a companion article in this issue of the Journal, provide a series of 3 data sets from the Collaborative Perinatal Project (CPP) (2) with induced missing values based on the “missing completely at random” (MCAR), “missing at random” (MAR), and “missing not at random” (MNAR) mechanisms. The parameter of primary scientific interest is the adjusted association between spontaneous abortion as the outcome and maternal smoking behavior as the exposure, controlling for the measured covariates body mass index (weight (kg)/height (m)2), age, and race. In these data, a person may be missing data for 1 or more confounders, the exposure, or the outcome, or a combination of all 3 in a pattern that is discernibly nonmonotone (i.e., there may be no ordering of the variables, such that observing the jth variable ensures that all variables k > j in the ordering are observed for all j) (3). In a challenge posed to us and the authors of another article (4), we were masked to the actual missing-data-generating mechanism for each of the 3 CPP data sets, as well as to the original full data. Our goal in this paper is to account for possible selection bias due to incomplete data in the estimation of the association of maternal smoking behavior with spontaneous abortion, without imposing a model for the joint distribution of variables that are prone to missingness.
Inverse probability weighting (IPW) of complete cases can achieve this goal, because the approach relies on a model for the missingness mechanism without necessarily requiring additional modeling of the full data beyond the scientific model of substantive interest (3, 5). Therefore, IPW ensures that assumptions needed to account for missing data are not conflated with assumptions encoded in the substantive model, thus avoiding issues of model compatibility and other forms of misspecification of the full-data model (6–8). In practical implementation of IPW, it is sometimes assumed that the missing-data mechanism depends only on variables fully observed in the sample, such that a simple binary regression of being a complete case (e.g., logistic or probit regression) may be used to estimate the missing-data process in order to estimate the weights. While the assumption simplifies the analysis, it is often overly restrictive, particularly in the presence of nonmonotone missing data. In fact, bias may result even if, as is commonly assumed in practice, data are MAR. Recall that the MAR assumption states that the missing-data mechanism may depend on variables observed under a given missing-data pattern but may not depend on the values of unobserved variables, while a missing-data mechanism independent of both observed and unobserved data is said to be MCAR and a missing-data mechanism that is neither MCAR nor MAR is said to be MNAR (3). The MAR assumption is commonly made in practice because, under the additional assumption that the parameters of the underlying full data distribution are distinct from those of the missing-data mechanism, the likelihood function for the missing-data mechanism factors from that of the observed data, making it possible to untangle the selection model from the model of substantive interest, even if the missing-data pattern is nonmonotone.
Untangling the 2 models under MNAR will typically require alternative, often more stringent assumptions. Under MAR, IPW exploits the fact that the likelihood factors, by explicitly incorporating assumptions about the missing-data process in a model for the missing-data mechanism, while avoiding unnecessary assumptions about the full data distribution beyond the underlying model of substantive interest. However, models for nonmonotone missing-data processes in IPW are not well developed in the statistical literature, particularly in settings where one may be willing to assume MAR but unwilling to make a further independence assumption (8). Below, we describe a recent approach for modeling nonmonotone missing-data processes which largely resolves this difficulty, and we illustrate the approach in the 3 data sets described in the companion article (1).
NOTATION AND ASSUMPTIONS
Let be a random K-vector representing the full data for a given individual. Let R be the scalar random variable encoding the different missing-data patterns. For each of n individuals, we observe an independently and identically distributed realization of . For missing-data pattern , where , we only observe . We reserve R = 1 to denote complete cases.
For identification purposes, we formalize the MAR assumption (assumption 1) for each individual as
| (1) |
with possible missing-data patterns, so that the conditional probability of having missing-data pattern m, which we denote by , depends only on the observed variables for that pattern. Throughout, we also make the following necessary positivity assumption (assumption 2) that for all individuals,
| (2) |
that is, the probability of being a complete case is bounded away from zero, for a fixed positive constant σ (i.e., we assume that every participant could have been a complete case). Assumption 2 is necessary for nonparametric identification of the full data distribution and of smooth functionals of the latter, such as means, variances, and covariances, as well as regression models involving non–fully observed variables (6), and it ensures finite asymptotic variance of IPW estimation.
A key implication of assumptions 1 and 2 is that the missing-data process is itself nonparametrically identified. This also implies that when distinct parameters are used to model the missing-data mechanism and the full data distribution, efficient estimation of the parameters of the missing-data process can be obtained by maximizing its partial likelihood, ignoring the part of the likelihood induced by the full data. Once an estimator of the probability of being a complete case is obtained, which we denote IPW then simply entails restricting the primary analysis of interest one would have performed in the absence of missing data to the complete cases, each of which is reweighted by Specifically, consider the primary scientific analysis described in the companion paper by Perkins et al. (1), which involves estimating an association between spontaneous abortion (the outcome) and maternal smoking behavior early in pregnancy (the exposure) while controlling for race, age, body mass index, and smoking status (the confounders) using a standard logistic regression model. An IPW analysis then corresponds to a CC logistic regression analysis in which each contribution is reweighted by Below, we describe the challenge of modeling and estimating the CC probability when missingness is nonmonotone, and we describe in detail our proposed approach for achieving this task.
ESTIMATING MONOTONE MISSINGNESS MECHANISMS
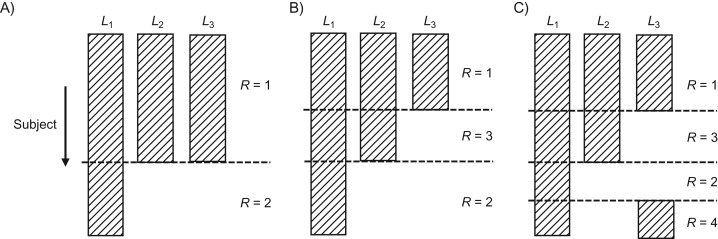
Although the missingness mechanism is in principle nonparametrically identified under assumptions 1 and 2, in practice estimation entails specifying parametric models, as dictated by Bellman’s curse of dimensionality (9), because L is typically of moderate to high dimension (e.g., hundreds) (10). To motivate our discussion of nonmonotone missing-data models, we briefly review existing strategies for modeling the missing-data mechanism. For illustration, we assume that the full data set consists of K = 3 variables. Let be the missing-data indicator for the jth variable, such that Sj takes on the value 1 or 0 when the variable Lj is observed or missing, respectively (). An example with 2 levels of missing data (i.e., ), also known as uniform missing data (11), is given in Figure 1A. In this example, the variables are either missing or observed together for each subject, so that . Since only is observed for missing-data pattern , by MAR assumption 1,
| (3) |
By the uniform missing-data assumption, and we can specify a parametric model—for instance, Note that can be estimated by fitting the logistic model among subjects with observed, that is, conditioning on the event . The weights for IPW can then be constructed as the inverse of . The uniform missing-data scenario arises in familiar settings, such as in regression analysis where only outcome data may not be fully observed.
Figure 1.
Schematic diagrams illustrating uniform (A), monotone (B), and nonmonotone (C) patterns of missing data. L1, L2, and L3 represent 3 variables in an example data set, R is the random variable encoding missing-data patterns, and the hatched rectangles denote observed portions of the data.
When there are more than 2 levels of missingness, the missing data are said to be monotone if, for some ordering of the variables in , the kth variable is observed only if the k − 1th variable was observed—that is, . This missing-data pattern occurs frequently in longitudinal studies if subjects who drop out never reenter the study. For the example at hand, a monotone missing-data pattern is shown in Figure 1B. In this case, an analyst may decide to combine missing-data patterns so that analysis can proceed in the same way as described above for uniform missing data. However, doing so will discard information from observed in subjects with for estimation of the missingness mechanism. Instead, the missing-data model can be built up sequentially for each missing-data pattern. By MAR assumption 1, is still given by equation 3, and
| (4) |
where under a monotone missing-data pattern. We may then wish to specify 2 separate logistic models for and , respectively. Note that the former can be fitted with only the variable , while the latter can be fitted with among subjects with both variables observed—that is, conditioning on the event . Therefore, remains a function of , and the weights can be constructed accordingly as the inverse of . We note that the missingness models discussed so far are guaranteed to yield which are constrained to the range , since the conditional missingness probabilities are built up sequentially at the variable level.
For settings with more than 2 levels of missing-data patterns, nonmonotone missingness is quite common in practice and building coherent models for the conditional probabilities of the various missing-data patterns is challenging even under assumptions 1 and 2 (8). An example for nonmonotone missing data is given in Figure 1C. Suppose we proceed to model the conditional probabilities for given in equations 3 and 4 as before. In both cases we need to fit a parametric model for , since it is no longer necessarily equal to 1 under nonmonotone missing data but is a function dependent on . Nonetheless, it is clear that and remain functions of and , respectively. For , by MAR assumption 1,
| (5) |
Two separate logistic models can be fitted for and ( still holds). However, it is clear that both models can only be fitted with , and therefore , which is strictly stronger than what MAR assumption 1 entails. In general, existing modeling strategies for uniform or monotone missing-data mechanisms fail to span the entire MAR model (8, 11), and the conditional probability for at least one of , , fails to incorporate all of the observed variables in its pattern—that is, , where . The set of variables is ignored, although they may be informative for the missingness mechanism.
As a remedy, Robins and Gill (12) proposed a large class of models for the missing-data mechanism, which they called the randomized monotone missingness processes, that are guaranteed to be MAR for a nonmonotone missing-data mechanism without necessarily being MCAR. This class of models does not span the space of all MAR models, and therefore it is indeed possible to test whether the proposed class of models includes the true missing-data mechanism. However, estimation of the missing-data mechanism within this class is complex and computationally demanding, even for small-to-moderate sample sizes and numbers of different missing-data patterns M, and no software is currently available with which to implement the approach, which has limited its widespread adoption. In addition, a straightforward approach to modeling nonmonotone missing-data pattern probabilities using polytomous logistic regression can at most depend on the intersection of the sets of observed variables , (i.e., the completely observed variables), where distinct patterns are observed (12, 13). This is also referred to as the “stratified MAR assumption” as discussed by Greenland and Finkle (14), and it is strictly stronger than MAR assumption 1. This suggests that standard polytomous regression is also ill-suited as a modeling strategy for a nonmonotone missing-data process under MAR. A similar problem with polytomous regression exists in the context of modeling outcome heterogeneity in epidemiologic studies (15).
NONMONOTONE MISSING-DATA MODEL
We illustrate our approach using the example described above for full data and nonmonotone missing-data patterns , , , and By MAR assumption 1, the conditional probability for each missing-data pattern can be modeled separately as
| (6) |
We have that the probability of observing complete data is given by
| (7) |
To ground ideas, we specify logistic models for each of the missing-data patterns' conditional probabilities in equation 7 directly as
| (8) |
Whereas the conditional probability is a function of only under model 5 (equation 5) as discussed above, is explicitly a function of under model 8 (equation 8). Therefore, missing-data model 8 fully incorporates the observed variables in each missing-data pattern. Nonetheless, because model 8 is not built up sequentially at the variable level, it does not naturally impose the contraints required by assumption 2—that is, for each subject,
| (9) |
As a result, we define the estimator of which maximizes the following observed data log-likelihood corresponding to missing-data model 7 (equation 7),
| (10) |
as the unconstrained maximum likelihood estimator (UMLE) , since it does not impose constraint 9 (equation 9). While straightforward to implement, it may be the case in practice that directly maximizing the oberved data log-likelihood described in equation 10 fails to converge if at least one of the fitted is near the boundary value 0 or 1. In principle, one could attempt to maximize equation 10 subject to the observable constraints
| (11) |
where is a user-specified small positive constant. However, this is potentially computationally prohibitive, as there are as many nonlinear constraints as the number of CC observations for each iteration of the optimization algorithm. As an alternative, we have previously developed a constrained Bayesian estimation (CBE) approach whereby a standard prior distribution for is combined with the log-likelihood function in equation 10 to produce a posterior distribution proportional to
| (12) |
from which samples are drawn, and only draws that fall within the constrained parameter space are retained (16). The CBE estimator is defined as the posterior median of samples drawn from the posterior distribution described in equation 12. Parameter values that fall within the resulting posterior credible intervals of are guaranteed to satisfy constraint 11 (equation 11), which is useful if one wishes to perform hypothesis testing to identify significant predictors in the missing-data regression models. It is straightforward to extend the method to handle more than the 4 levels of missing-data patterns given in the toy example above, with the missing-data model described in equation 8.
A detailed description of the CBE approach used in analyzing the CPP data sets, as well as the sample OpenBUGS code for its implementation, are provided in Web Appendix 1 (available at https://academic.oup.com/aje). For the CPP data sets, we describe the 8 levels of nonmonotone missing-data patterns in Web Table 1.
AUGMENTED IPW
Upon obtaining the UMLE (only when convergence can be achieved) or CBE of , denoted as , we may proceed to construct the CC weights . In our example with full data , let be the binary outcome of interest and be 2 continuous explanatory variables, with 2 levels of missingness: when full data are observed and when only is observed (Figure 1A). Assume the substantive model to be
| (13) |
and let , where is a vector, for instance . The IPW estimator of is the solution to the estimating equation
| (14) |
However, an important limitation of IPW is that it does not directly make use of data from incomplete observations, except in the estimation of . The efficiency of the IPW estimator can be improved by incorporating persons with missing data via augmentation of estimating equation 14 (6, 8). We emphasize that this potential efficiency gain is achieved without relying on a model for the full data beyond the substantive model of primary interest, provided that (as we assume throughout) the missingness model is correctly specified and therefore the estimator of is consistent. For a given , the class of augmented IPW (AIPW) estimators of are solutions to the estimating equations
| (15) |
for an arbitrary choice of vector . In this example with uniform missingness, the optimal choice of in terms of efficiency for fixed g(L1, L2), which we denote as , has the closed-form expression (6, 8, 17). However, the optimal function is generally no longer available in closed forms for nonmonotone missing data (6, 8). We adopt a computationally more tractable approach previously described elsewhere (8, 13) to implement AIPW. Briefly, the approach entails approximating with finite sum of basis functions. For instance, to approximate in equation 15, we may construct the random vector , where is an arbitrary constant matrix. This choice of basis functions allows for terms up to third-order terms in to be incorporated into the approximation. It is possible to estimate the unique which gives the optimal efficiency. Similarly, we can also approximate the optimal choice of in by . Together, yield an estimating equation the solution to which (the AIPW estimator) is guaranteed to be more efficient than the IPW estimator in equation 14. A detailed description of the AIPW method for the CPP data sets is included in Web Appendix 2.
EMPIRICAL ILLUSTRATION
We applied the proposed IPW approach to fit a logistic regression for the risk of spontaneous abortion as a function of maternal smoking behavior, body mass index, age, and race in the 3 data sets (1, 2, and 3) described in the companion paper (1). The UMLE for the missing-data model failed to converge in data set 1; thus, only CBE estimates for maternal smoking from the primary regression model are shown in Table 1, which also summarizes results for the complete-case (CC) logistic regression, as well as for the more efficient AIPW approach. UMLE results are also available for data sets 2 and 3 in Table 1, where convergence was attained. In the CBE approach, estimates of the parameters in the missing-data model are obtained as the median of the constrained posterior distribution with diffuse priors and . Adaptive Gibbs sampling was implemented through BRugs, the R interface to OpenBUGS software (18). We assessed convergence by visually inspecting the trace plots, as well as through the Gelman-Rubin convergence statistic (19), and allowed an adaptive phase of iterations followed by another iterations from which samples were included for estimation.
Table 1.
Parameter Estimates (Log Odds Ratios) for the 3 Data Sets in a Logistic Regression Analysis of the Risk of Spontaneous Abortion According to Maternal Smoking During Pregnancy, Collaborative Perinatal Project, 1959–1974a
| Data Set | Complete-Case Analysis | IPW Analysis | Augmented IPW Analysis | AREb | |||
|---|---|---|---|---|---|---|---|
| Estimate (SE) | 95% CI | Estimate (SE) | 95% CI | Estimate (SE) | 95% CI | ||
| 1 | −0.85 (0.40) | −1.64, −0.07 | 0.43 (0.27) | −0.10, 0.96 | 0.34 (0.17) | 0.00, 0.68 | 0.41 |
| 2 | 0.33 (0.14) | 0.06, 0.60 | 0.33 (0.14) | 0.06, 0.60 | 0.35 (0.14) | 0.09, 0.62 | 0.97 |
| 3 | −0.07 (0.26) | −0.59, 0.45 | 0.61 (0.23) | 0.16, 1.06 | 0.45 (0.18) | 0.09, 0.80 | 0.63 |
Abbreviations: ARE, asymptotic relative efficiency; CI, confidence interval; IPW, inverse-probability-weighted; SE, standard error.
a Data were obtained from Perkins et al. (1).
b Estimated ARE of augmented IPW compared with IPW estimation based on the SE.
In data set 1, the point estimate for the association of smoking with spontaneous abortion increases from −0.85 to 0.43 when comparing CC analysis with IPW analysis (corresponding to a change in the odds ratio from 0.43 to 1.54). The augmented IPW estimate for the association of smoking with spontaneous abortion is 0.34 (odds ratio = 1.40, 95% confidence interval: 1.00, 1.97) and is more efficient than IPW, as evidenced by the standard errors. In fact, the estimated asymptotic relative efficiency, defined as the ratio of estimated variances, is 0.37 for the effect of smoking behavior. The CC, IPW, and AIPW point estimates in data set 2 are similar, with asymptotic relative efficiencies ranging from 0.82 to 1.04 across the various covariates. In data set 3, the point estimate for maternal smoking behavior increases from −0.07 to 0.61 (change in odds ratio from 0.93 to 1.84) when comparing CC analysis with IPW analysis. The AIPW estimate for smoking behavior is 0.45 (odds ratio = 1.57, 95% confidence interval: 1.09, 2.23) and is somewhat smaller than the IPW estimate, with an asymptotic relative efficiency of 0.63. Thus, it appears that our IPW analysis of data set 2 provides little evidence of bias due to missing data because weighted and unweighted analyses yield similar point estimates, although IPW and AIPW estimates are substantially more efficient. This also suggests that the missing-data mechanism for data set 2 may in fact be MCAR, although we cannot rule out MNAR. Results for data sets 1 and 3 are somewhat more sensitive to IPW adjustments for missing data, suggesting that CC analyses may be subject to selection bias due to missing data. However, it is essentially impossible to know based solely on the observed data whether the adjustment leads to bias reduction without having access to the corresponding “full data” analysis, which is discussed in the companion article (1), where the mechanism used to generate missing data in each of the 3 data sets is also unveiled.
In principle, we expect IPW and AIPW analyses to be less biased than CC analysis provided that the missing-data mechanism either is MCAR or is MAR with model 8 (equation 8) correctly specified. However, IPW and AIPW can fail to be consistent when the model for the missing-data mechanism is incorrectly specified, even if the mechanism is MAR; the missing-data model is typically misspecified when the mechanism is MNAR. Table 2 summarizes results for the estimated coefficients of the missing-data model in data set 1, clearly indicating significant dependence of the model on the outcome variable “abortion.” In contrast, Web Table 2 shows results for estimation of the missing-data model in data set 2 and provides no significant evidence that the missing-data mechanism depends on the outcome. These latter results are consistent with the fact that IPW and AIPW analyses yield results similar to those of CC analysis in data set 2. Results for the fit of the missing-data model in data set 3 are provided in Web Table 3. Similar to data set 1, the results suggest significant dependence of the missing-data mechanism on the outcome variable “abortion,” which is also consistent with the fact that IPW and AIPW estimates of the regression model for spontaneous abortion differ somewhat from the unweighted CC estimates.
Table 2.
Posterior Median Values (Log Odds Ratiosa) and 95% Credible Intervals From Constrained Bayesian Estimation of Missing-Data Model Parameters in Data Set 1, Collaborative Perinatal Project, 1959–1974b
| Missing-Data Pattern | Variable | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | BMIc | Age | Abortion | Smoking | Black Race | Other Race | ||||||||
| PM | 95% CrI | PM | 95% CrI | PM | 95% CrI | PM | 95% CrI | PM | 95% CrI | PM | 95% CrI | PM | 95% CrI | |
| 1 | −3.73 | −4.08, −3.39d | 1.64 | 0.34, 2.77d | 1.07 | 0.81, 1.32d | 1.06 | 0.91, 1.22d | 0.08 | −0.10, 0.27 | −0.41 | −0.90, 0.03 | ||
| 2 | −6.41 | −7.01, −5.57d | 3.50 | 0.32, 6.25d | 3.08 | 0.90, 4.91d | 0.98 | 0.63, 1.32d | −0.15 | −0.57, 0.25 | 0.41 | −0.32, 1.02 | ||
| 3 | −2.76 | −2.95, −2.52d | 1.21 | 0.34, 1.89d | 1.11 | 0.99, 1.22d | −0.05 | −0.18, 0.09 | 0.12 | −0.14, 0.37 | ||||
| 4 | −3.45 | −3.92, −3.14d | 1.24 | −0.43, 2.46 | 0.98 | 0.31, 2.54d | 1.73 | 1.49, 1.96d | −0.07 | −0.25, 0.11 | −0.46 | −0.89, −0.08d | ||
| 5 | −3.43 | −3.65, −3.13d | 1.61 | 0.63, 2.53d | 1.41 | 1.14, 1.67d | 0.05 | −0.13, 0.22 | 0.07 | −0.29, 0.39 | ||||
| 6 | −2.94 | −3.35, −2.64d | 1.73 | 0.47, 3.09d | 1.35 | 0.18, 2.41d | 0.09 | −0.05, 0.22 | 0.07 | −0.19, 0.33 | ||||
| 7 | −4.15 | −4.80, −3.68d | −2.12 | −4.17, 0.19 | −0.06 | −0.50, 0.38 | −1.39 | −3.24, −0.14d | ||||||
Abbreviations: BMI, body mass index; CrI, credible interval; PM, posterior median.
a Log odds of observing the specific missing-data pattern per unit change in the relevant covariate.
b Data were obtained from Perkins et al. (1).
c Weight (kg)/height (m)2.
d 95% CrI excludes 0.
DISCUSSION
In this paper, we analyzed 3 CPP data sets using recently proposed IPW methods for handling nonmonotone missing data. After accounting for the missing values, the estimated associations of maternal smoking with spontaneous abortion were substantially different from those obtained in CC analyses for data sets 1 and 3. We provided an overview of methods for estimating a missing-data mechanism for use in IPW estimation under MAR, in 3 common incomplete-data situations encountered in practice: a simple 2-level missing-data pattern, a more general monotone missing-data pattern, and a nonmonotone missing-data pattern. The first two settings can be handled using fairly standard statistical models for the missing-data process, but the third requires some care to ensure that estimation is conducted without inadvertently imposing a more stringent assumption than intended about the nature of the missing data. To achieve this latter goal, we described in considerable detail 2 methods for modeling a nonmonotone missing-data pattern, UMLE and CBE. While the former is appealing in its simplicity, it may fail to converge in practice due to violation of certain model restrictions, for which the latter constrained Bayesian approach is proposed as a remedy (13). The performance of the described methods in analysis of the 3 CPP data sets with induced missing values is discussed further in the companion paper (1), where Perkins et al. describe the nature of missingness in each of the 3 CPP data sets, revealing which if any of the results are consistent with the full data analysis, up to sampling variability.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Division of Intramural Population Health Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Rockville, Maryland (Neil J. Perkins, Enrique F. Schisterman); Department of Statistics, College of Liberal Arts and Sciences, University of Connecticut, Storrs, Connecticut (Ofer Harel); Centers for Financing, Access and Cost Trends, Agency for Healthcare Research and Quality, Rockville, Maryland (Emily M. Mitchell); Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, Massachusetts (Eric J. Tchetgen Tchetgen, BaoLuo Sun); and Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Stephen R. Cole).
This research was partially supported by the Long-Range Research Initiative of the American Chemistry Council (Washington, DC) and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. This work was also partially supported by award K01MH087219 from the National Institute of Mental Health. E.T.T.’s work was funded by National Institutes of Health grant R01 AI127271.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
Conflict of interest: none declared.
Abbreviations
- AIPW
augmented inverse probability weighted/weighting
- CBE
constrained Bayesian estimation
- CC
complete-case
- CPP
Collaborative Perinatal Project
- IPW
inverse probability weighted/weighting
- MAR
missing at random
- MCAR
missing completely at random
- MNAR
missing not at random
- UMLE
unconstrained maximum likelihood estimator
REFERENCES
- 1. Perkins NJ, Cole SR, Harel O, et al. Principled approaches to missing data in epidemiologic studies. Am J Epidemiol. 2018;187(3):568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardy JB. The Collaborative Perinatal Project: lessons and legacy. Ann Epidemiol. 2003;13(5):303–311. [DOI] [PubMed] [Google Scholar]
- 3. Little RJ, Rubin DB. Statistical Analysis with Missing Data. 2nd ed Hoboken, NJ: John Wiley & Sons, Inc.; 2002. [Google Scholar]
- 4. Harel O, Mitchell EM, Perkins NJ, et al. Multiple imputation for incomplete data in epidemiologic studies. Am J Epidemiol. 2018;187(3):576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Horvitz DG, Thompson DJ. A generalization of sampling without replacement from a finite universe. J Am Stat Assoc. 1952;47(260):663–685. [Google Scholar]
- 6. Robins JM, Rotnitzky A, Zhao LP. Estimation of regression coefficients when some regressors are not always observed. J Am Stat Assoc. 1994;89(427):846–866. [Google Scholar]
- 7. van der Laan MJ, Robins JM. Unified Methods for Censored Longitudinal Data and Causality. New York, NY: Springer-Verlag New York; 2003. [Google Scholar]
- 8. Tsiatis A. Semiparametric Theory and Missing Data. New York, NY: Springer Publishing Company; 2006. [Google Scholar]
- 9. Bellman RE. Adaptive Control Processes: A Guided Tour. Princeton, NJ: Princeton University Press; 1961. [Google Scholar]
- 10. Robins JM, Ritov Y. Toward a curse of dimensionality appropriate (CODA) asymptotic theory for semi-parametric models. Stat Med. 1997;16(1-3):285–319. [DOI] [PubMed] [Google Scholar]
- 11. Li L, Shen C, Li X, et al. On weighting approaches for missing data. Stat Methods Med Res. 2013; 22(1):14–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robins JM, Gill RD. Non-response models for the analysis of non-monotone ignorable missing data. Stat Med. 1997;16(1–3):39–56. [DOI] [PubMed] [Google Scholar]
- 13. Sun BL, Tchetgen Tchetgen EJ. On inverse probability weighting for nonmonotone missing at random data [published online ahead of print December 16, 2016]. J Am Stat Assoc. (doi:10.1080/01621459.2016.1256814). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greenland S, Finkle WD. A critical look at methods for handling missing covariates in epidemiologic regression analyses. Am J Epidemiol. 1995;142(12):1255–1264. [DOI] [PubMed] [Google Scholar]
- 15. Sun B, VanderWeele T, Tchetgen Tchetgen EJ. A multinomial regression approach to model outcome heterogeneity. Am J Epidemiol. 2017;186(9):1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gelfand AE, Smith AFM, Lee TM. Bayesian analysis of constrained parameter and truncated data problems using Gibbs sampling. J Am Stat Assoc. 1992;87(418):523–532. [Google Scholar]
- 17. Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22(3):278–295. [DOI] [PubMed] [Google Scholar]
- 18. Lunn DJ, Spiegelhalter D, Thomas A, et al. The BUGS Project: evolution, critique and future directions. Stat Med. 2009;28(25):3049–3067. [DOI] [PubMed] [Google Scholar]
- 19. Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7(4):457–511. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.