Abstract
Sequencing pathogen samples during a communicable disease outbreak is becoming an increasingly common procedure in epidemiologic investigations. Identifying who infected whom sheds considerable light on transmission patterns, high-risk settings and subpopulations, and the effectiveness of infection control. Genomic data shed new light on transmission dynamics and can be used to identify clusters of individuals likely to be linked by direct transmission. However, identification of individual routes of infection via single genome samples typically remains uncertain. We investigated the potential of deep sequence data to provide greater resolution on transmission routes, via the identification of shared genomic variants. We assessed several easily implemented methods to identify transmission routes using both shared variants and genetic distance, demonstrating that shared variants can provide considerable additional information in most scenarios. While shared-variant approaches identify relatively few links in the presence of a small transmission bottleneck, these links are highly accurate. Furthermore, we propose a hybrid approach that also incorporates phylogenetic distance to provide greater resolution. We applied our methods to data collected during the 2014 Ebola outbreak, identifying several likely routes of transmission. Our study highlights the power of data from deep sequencing of pathogens as a component of outbreak investigation and epidemiologic analyses.
Keywords: Ebola virus, epidemics, genomics, infection control, infectious disease outbreaks, molecular epidemiology
Genomic data offer new insights into epidemiologic and evolutionary dynamics, and sequencing pathogen samples is becoming increasingly routine, providing new insights for a range of issues of public health importance (1). Pathogen genomic data allow us to determine the phylogeny of isolates, which in turn sheds light on the potential transmission networks between the hosts from whom they were collected. Identifying routes of infection enables the estimation of risk factors for disease transmission, which in turn can inform the implementation of infection control strategies. As such, inference of transmission trees using genomic data is an increasingly well-studied field (2–8). Although low-resolution pathogen typing has been used for some time to discriminate between independent outbreaks (9–11), whole-genome sequencing provides additional resolution with which genetic distance between identical phenotypes may be ascertained (12–14). However, this too has limits. Studies have shown that while transmission clusters may be identified with genomic data, individual-level transmission routes can rarely be identified with a great degree of certainty (3, 4). Characterizing an infected host by a single pathogen genome (isolation and purification of a single colony for bacteria, or using the consensus sequence for viral pathogens) is common practice yet neglects within-host diversity. The variation in sampled genetic distances can be large relative to the expected number of mutations between hosts, rendering the number of single nucleotide polymorphisms a rather crude measure of relatedness on an individual level (15). As such, particularly for rapidly evolving pathogens, or those whose mode of transmission is associated with a large and potentially diverse inoculum (“transmission bottleneck”—the number of pathogens transmitted in an infection event), single-genome sampling can cause hosts to appear misleadingly similar or dissimilar.
Deep sequencing can potentially provide new insights into within-host diversity. Currently, sequencing a mixed population sample to sufficient depth to identify minor nucleotide variants has mostly been limited to viral samples. While consensus sequences may appear identical for 2 samples, comparing minor variants can offer additional resolution. For instance, if the same nucleotide variation is observed at the same locus in pathogen samples from 2 individuals (henceforth referred to as a “shared variant” (SV)), this could be considered strong evidence for direct transmission, particularly if the variant is not observed in any other host. This naturally relies on the possibility that a pathogen population of size greater than 1 survives the transmission bottleneck; otherwise, each infection must initially be monoclonal, implying that any variation found within distinct hosts must have arisen independently. There is evidence for larger bottlenecks occurring in viral pathogens such as influenza (16, 17) and Ebola (18), and it is plausible that this is also the case for bacterial pathogens (19).
The connection between SV presence and direct transmission has previously been suggested. Gire et al. (20) noted the presence of SVs in Ebola virus samples from individuals who were potentially linked by transmission. Data collected from 2 influenza A animal transmission studies were used to explore the presence of SVs between hosts, and it was shown that such data were consistent with known contact patterns (21). This study used known contact patterns to identify characteristics of SVs that were more likely to be associated with transmission, allowing variants to be split into those consistent and inconsistent with transmission, minimizing false connections. Poon et al. (22) identified routes of influenza transmission occurring during a household contact study using both consensus whole-genome sequence data and the presence of SVs. In the case of bacterial pathogens, the diversity in Staphylococcus aureus infections, which can be considerable, has been linked to transmission in a veterinary hospital (23).
Pathogens vary considerably in their bottleneck size, mutation rates, and transmission dynamics. It remains unclear how methods based on SVs are expected to perform in different regions of this parameter space. Establishing this is a crucial component of the interpretation of SVs and the value of the approach.
In the present study, we investigated the predictive power of SVs for identifying transmission routes. In addition to pathogen genomes, many other sources of data may contribute information towards inference of transmission routes, including temporal and spatial data, contact patterns, and expression of symptoms. However, here we aimed to examine the information contributed by genomic data alone and, in particular, the additional benefit offered by considering SVs.
METHODS
We generated infectious disease outbreaks with within-host pathogen evolution under various mutation rates and bottleneck sizes by simulation, using the R (R Foundation for Statistical Computing, Vienna, Austria) package Seedy (24). In contrast to other simulation approaches, this method explicitly simulates within-host evolution and allows sampling of mixed pathogen populations across time. We expanded upon methods previously used to infer transmission routes using deep sequence data (20–22), comparing their performance with analogous genetic distance–based approaches. We additionally proposed hybrid approaches that combine SV information with phylogenetic distance data. We considered the following approaches:
Weighted variant tree: For each host, we weighted potential sources by the number of observed SVs, such that the host sharing the largest number of variants is attributed the largest weight. Hosts sharing no variants with any other were not assigned a source. Weighting edges provided an extension to previous approaches (20–22).
Maximum variant tree: For each host, we defined the source to be the individual with whom the largest number of SVs was observed. Hosts sharing no variants with any other host were not assigned a source.
Weighted distance tree: Using consensus sequences, the genetic distance (number of single nucleotide polymorphisms) between isolates was calculated, and potential sources were weighted inversely by this metric. This approach has been described previously (3).
Minimum distance tree: Using consensus sequences, the source of a given host was defined to be the carrier of the genetically closest isolate to that of the host. This approach, with the incorporation of sampling times to provide directionality, has been described previously (7).
Hybrid weighted tree: First, the weighted variant tree was constructed. Hosts with no source were then assigned potential sources based upon weighted genetic distance.
Hybrid maximum tree: First, the maximum variant tree was constructed. Hosts with no source were then assigned potential sources based upon minimum genetic distance.
These 6 simple heuristics by no means comprise an exhaustive list of approaches to identify routes of transmission but are instead a range of readily implemented, distance-based approaches that require neither knowledge of evolutionary dynamics nor infection or sampling times. As has previously been demonstrated, simple methods based on genomic data alone can provide powerful insights into transmission dynamics (25). Further details of the approaches used here, as well as the metrics used to assess the accuracy of tree reconstruction and reliability of estimated transmission routes, are provided in Web Appendix 1 (available at https://academic.oup.com/aje). We additionally applied SV methods to previously published data collected during the Ebola virus outbreak in West Africa in 2014 (20, 26).
RESULTS
Simulation studies
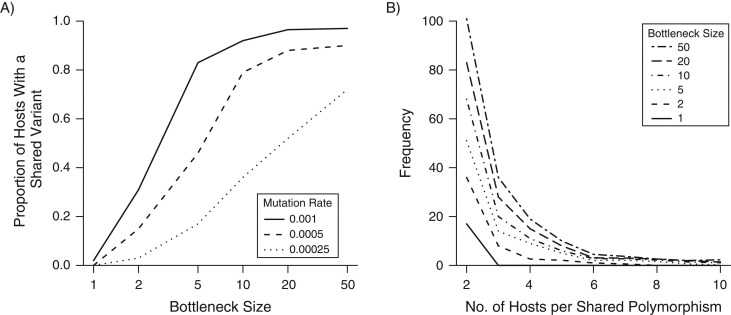
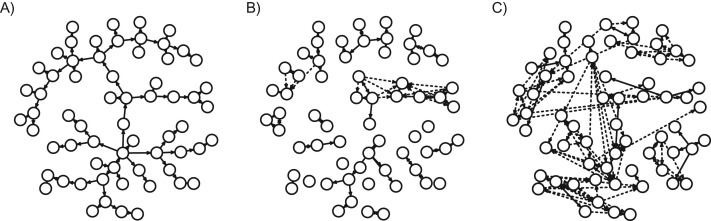
As expected, the proportion of cases in which a SV was observed in at least 1 other host increased rapidly with mutation rate and bottleneck size (Figure 1A). The majority of SVs were observed in exactly 2 individuals, with the proportion shared among larger groups declining rapidly as the size of the group increased (Figure 1B). For each simulation, we constructed a weighted transmission tree according to the 6 methods outlined previously. An example simulated outbreak of a pathogen with characteristics similar to S. aureus (see Web Appendix 1) is shown in Figure 2, along with reconstructions based upon two of these methods. While many edges are bidirectional and symmetric, asymmetry can occur under most methods due to the lack of commutativity (i.e., even if B is the closest host to A, A may not be the closest host to B).
Figure 1.
Summary of genetic variant frequency across simulated outbreaks. We simulated 10 outbreaks for each combination of 6 bottleneck sizes and 3 mutation rates (180 in total). A) Mean proportion of cases in the outbreak who shared at least 1 variant with another host. B) Distribution of shared-variant group size for different bottleneck sizes, with a mutation rate of 5 × 10−4 per genome per generation.
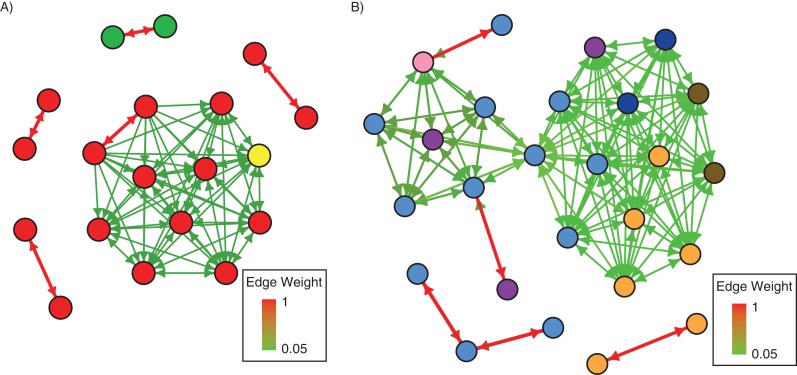
Figure 2.
Simulated and reconstructed transmission trees. A simulated tree (A) was generated with bottleneck size 10 and mutation rate 0.001 per genome per generation. Cases are represented as numbered circles linked by arrows representing transmission routes. Based on the simulated genetic data from this outbreak, trees were reconstructed according to the maximum variant approach (B) and the minimum genetic distance approach (C) described in “Methods.” Arrows denoting unambiguous routes (weight = 1) are solid, while dashed arrows denote edges with weight <1. Networks were plotted with the igraph package in R (Foundation for Statistical Computing) (38).
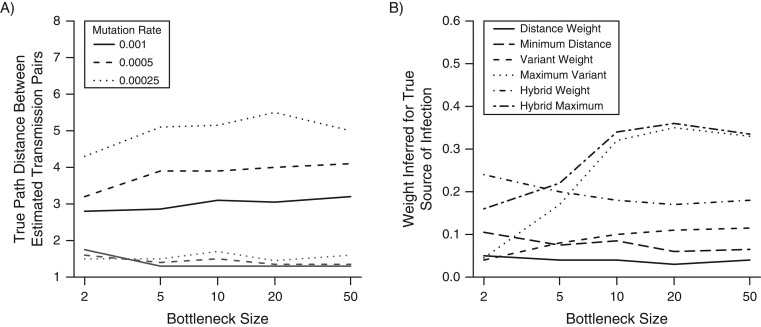
We used two metrics to assess the reliability of individual estimated transmission routes and compared the different methods described above. First, we considered the true path distance between inferred transmission pairs. We found that under the maximum variant tree, the mean path distance was typically less than 2, outperforming the minimum distance approach (Figure 3A). Second, we examined the mean weight assigned to the true source of each host. In the case of small (<5) bottleneck sizes, methods based on SVs perform poorly because the likelihood of a monoclonal infection is high, resulting in most true links being assigned a weight of zero. Furthermore, those links that are inferred by SVs in small-bottleneck settings identify direct transmission with high confidence. The hybrid approaches perform best for small bottlenecks, incorporating SV information when available but not relying upon it. For larger bottlenecks, the distance-based approaches were markedly outperformed by the variant-based approaches (Figure 3B). As the rate of mutation increases, SV approaches outperform those based on distance alone to an increasing degree as mutation generates increasing diversity in the infecting population (Web Figure 1A–1C).
Figure 3.
Reliability of estimated transmission routes. A) A comparison of the minimum distance (black) and the maximum variant (gray) approaches. The true path distance between estimated transmission pairs gives insight into the extent to which transmission links are misspecified. A perfect reconstruction would have mean path length of 1; greater values indicate increasing misspecification. Maximum variant path lengths are averaged over identified transmission pairs—that is, excluding hosts with no shared variants. B) The mean weight assigned to transmission links for each tree reconstruction method, under a range of scenarios and methodologies. Results are shown for a mutation rate of 5 × 10−4 per genome per generation and were averaged across 10 simulated outbreaks for each scenario.
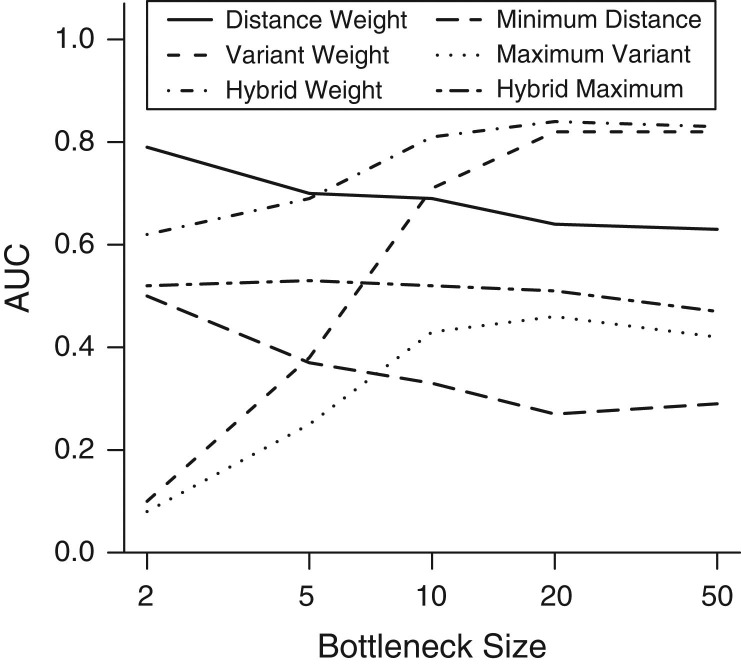
In addition to the reliability of individual links, we also considered the overall accuracy of a transmission tree reconstruction. This was measured by the area under the receiver operating characteristic curve (AUC) statistic. For small bottlenecks, variant-based methods provide a poor tree reconstruction by this metric (Figure 4). We note that here we regard assigning no source (e.g., where no SVs exist) as an incorrect classification, leading to values below 0.5, the value expected under random source assignment. A tight bottleneck leads to little diversity persisting across transmission events, and as such, SVs are rarely observed, leading to a sparsity of informed links across the network. However, larger bottlenecks led to a considerably better performance under this measure for the variant-based approaches, which exceeded even the weighted distance approach with a sufficiently large bottleneck size and mutation rate (Figure 4, Web Figure 1D–1F). In contrast, distance-based approaches typically declined in accuracy as the bottleneck size increases, for reasons that are well understood (3, 27)
Figure 4.
Transmission tree reconstruction accuracy. The area under the receiver operating characteristic curve (AUC) metric provides an overall measure of network accuracy: Values closer to 1 indicate better overall reconstruction. Results for a mutation rate of 5 × 10−4 per genome per generation are shown here. The mean AUC across 10 simulations was measured for each scenario.
We additionally investigated the effect of “mutational hotspots,” which can generate potentially confounding homoplasy. We found that while variant approaches performed less well, they generally continued to outperform distance-based approaches for larger (>10) bottleneck sizes (see Web Appendix 1, Web Figures 2 and 3 for further details).
Ebola virus data
We next examined previously published data sets for Ebola in Sierra Leone, for which sequence data are available, and we can determine the presence and properties of intrahost variants. In order to reduce the risk of counting variant calling errors as true intrahost variants, we identified only variants in which the minor within-host allele frequency was at least 5% (routes estimated under a 1% threshold are shown in Web Figure 4). Figure 5 shows the transmission trees reconstructed for each data set under the weighted variant approach, using no epidemiologic information. In the first data set (Figure 5A, (26)) 19 of 78 hosts were found to share a variant with at least 1 other individual. Four pairs of patients shared more than 1 variant (3 pairs with 2 SVs and 1 pair with 4), while 1 additional pair shared 1 unique variant. Consistent with transmission, each of these pairs originated from the same geographic location, and permutation testing revealed that this geographic similarity was significantly higher than would be expected via random selection (P = 0.0075). Pairs were also temporally clustered; 3 of these links were sampled 2 or fewer days apart, while the remaining 2 were sampled 12 and 22 days apart, which is plausible given serial interval estimates for Ebola virus infection of 15.3 ± 9.3 days (28). Under the minimum distance tree, 2 of these pairs were reproduced, 2 pairs belonged to a much larger group of samples with identical consensus sequences, and 1 pair, differing by a single nucleotide according to consensus sequences, remained unconnected due to the presence of other identical sequences (Web Figure 5).
Figure 5.
Estimated Ebola transmission routes, Sierra Leone, 2014. Transmission links between sampled hosts during an Ebola outbreak under the maximum variant approach. Node colors denote distinct geographic regions to which hosts belong, while arrow color and thickness correspond to the relative weight attributed to each potential transmission event. Variant detection threshold was 5%. Cases are shown from the first (20) (A) and the second (26) (B) data sets. Further details are provided in Web Appendix 1.
While a consistent result of our simulations was the sharing of variants among small numbers of hosts, rarely more than 2, in the Ebola data collected by Gire et al. (20), 1 variant was shared by 11 hosts. Samples with this variant are highly clustered geographically (10 of 11 in the same chiefdom, P = 0.022) and temporally (observed within an 18-day period), as well as phylogenetically, lending support to this group representing a transmission cluster.
In the second data set (Park et al. (26)) 26 of 150 (for which replicate sequencing and variant calling was performed) shared a variant with at least 1 other host (Figure 5B). There were 5 pairs of individuals sharing a unique variant. As before, 1 variant was shared by multiple hosts, but unlike the previous data set these were not geographically or temporally clustered, coming from different regions and spanning several weeks. Furthermore, while some of these samples clustered on the phylogenetic tree, many fell in different clades (26), suggesting the group is unlikely to represent a single transmission cluster but rather multiple transmission events in combination with homoplasy.
DISCUSSION
We have described some simple methods for reconstructing transmission trees using SVs, testing how well this approach performs for a range of parameters governing the rates of diversification within and between hosts. We have then applied the methods to data from the 2014 Ebola outbreak to identify links, using the genomic data alone, that are likely to be consistent with transmission given time and location.
For the great majority of parameter space, excluding only very low mutation rates and tight bottlenecks, these methods outperform genetic-distance comparison methods, which have increasingly been used to identify potential transmission events (7, 29, 30). The limitations of distance-based methods that characterize a single genome are well appreciated. We note that although, for the purpose of comparison, additional data sources were not included in our inference of transmission routes, incorporating these independently would be a relatively straightforward step with these methods. Most simply, sampling dates could be used to provide directionality to inferred connections.
The additional information we derive from SVs can inform the transmission tree in two distinct ways, depending on the region of parameter space. First, small but nonsingular bottlenecks (e.g., for airborne influenza transmission (31) or sexually transmitted HIV (32)) produce few inferred transmission pairs, but these are highly accurate. The small bottleneck means that the probability of observing a SV between individuals who are in the same transmission cluster, but not directly linked, is negligible. Second, SV data for pathogens with larger transmission bottlenecks (e.g., Ebola (18), influenza transmitted via contact (31), or intravenous drug–associated HIV transmission (33)) provide good information on the overall tree structure and transmission clusters, but individual links may be more uncertain. In all cases, higher mutation rates make the observation of SVs more probable, which typically results in better inference of transmission routes.
A hybrid approach that combines SVs and the sequence of either an individual, sequenced genome, or the consensus offers substantial benefit in the case of small bottleneck sizes (<5), where we predict a method based on SV alone would struggle. Because transmission routes are assessed independently of one another, estimated transmission trees frequently comprise several unconnected nodes or clusters. Such unconnected clusters could be linked to one another if further structure is required, using the weighted distance approach on pooled within-cluster samples. Here we have simply used genetic distance, which is predicted to be efficient and reliable under the relatively short time scale of an outbreak, but more sophisticated models of sequence evolution could be applied.
We applied these methods to Ebola data collected from Sierra Leone in 2014. While the first data set is thought to represent relatively dense coverage of the initial stages of the epidemic in the country, with around 70% of cases sampled (20, 34), the later data set comprised a sparser sample. While this reduces the expected number of linked cases in the data set, the reliability of transmission routes identified via SVs remains largely unaffected (Web Figures 6 and 7). As such, while only relatively few transmission routes were identified in the data sets, this is likely a function of both the proportion of missing data and the relatively low mutation rate of Ebola virus (20, 35). Confidence in the transmission pairs identified was reinforced by investigating temporal and geographic clustering, which proved to be significant, and while the aim of our study was to assess the accuracy of transmission-route identification via genomic data alone, methodology combining spatial and temporal data sources will naturally provide further insight. Identifying even a small proportion of direct-transmission pairs can be of great interest in studying pathogen-level transmission dynamics as well as conducting outbreak investigations.
Studying the Ebola data showed that both data sets contained a large group of hosts sharing the same variant, which was rare in all our simulations. The observation can be explained in at least two ways: recurrent mutation (as might arise through selection) or an anomalously large number of contacts with large bottleneck size (such as might be associated with a funeral-based exposure). Park et al. (26) suggest that the large group in the second data set likely arose through a combination of patient-to-patient transmission and recurrent mutation. Subsets of this group do cluster on the phylogenetic tree, and having identified clusters of hosts with SVs, we might partition these groups by genetic background (e.g., ruling out transmission between hosts with >1-nucleotide difference present across nonvariant sites).
Sample contamination may be an additional source of error. Cross-contamination may potentially lead to SVs observed between unlinked hosts. However, in many settings we do not believe this would present a major concern. If no minor variants are contained in the contaminating sample, SVs would not link this to the contaminated sample. Still, it remains important to verify that observed SVs are consistent with transmission and to minimize the risk of contamination as much as possible. The second Ebola data set highlights the potential for false-positive connections to be identified between spatiotemporally inconsistent pairs. SVs observed across several samples with differing genetic backgrounds may be indicative of contamination. While such rules can eliminate obvious sources of error, further work is required to formally evaluate the risk of contamination based on deep sequence data.
Another deliberate simplification in the present work is the assumption of neutral evolution. While this is plainly faulty over longer time scales, over the relatively short timescale of an outbreak it is a first approximation, and this is supported by real data from outbreaks (20, 26) and even longer periods (36) showing evidence of incomplete purifying selection. Selection may not, however, have as severe an effect on these methods as we might assume. If a specific variant is maintained through balancing selection, it is likely to be found in multiple hosts and, as a result, will be less informative as to specific transmission links; if several hosts are connected by the same SV, this will be misleading only if no additional variants are observed. In contrast, diversifying selection by the host immune system is expected to produce the mutational hotspots we have studied here, which again have little impact. A similar argument can be made that sequencing errors will be less important than expected, because they are likely to be found in just one sample and hence be uninformative as to links to other samples. A more formal approach to this problem would be to test for selection and down-weight the identified loci from the analysis.
As yet, there are still few studies in which adequate data have been collected in order to use SVs as a feature to identify transmission routes. Deep and high-quality sequencing is required to reliably call minor variants, as well as dense sampling of the outbreak population such that the majority of infection sources are included in the study population. It is likely that such data will become more commonly collected in the near future, for both viral and bacterial pathogens, as the associated sequencing costs fall and the benefits become more evident. This work demonstrates that deep sequence data can be informative in outbreak investigations and epidemiologic studies, and it should motivate both the wider collection of such data and the further development of methodologies that might accommodate scenarios that stray further from neutral evolutionary dynamics. It is noticeable that bottleneck size in nature, as opposed to minimal infectious dose, has not received the attention it deserves. The importance of this parameter for these methods, as well as other factors such as the evolution of virulence (37), should motivate further study.
We have demonstrated the power of deep sequencing data to identify transmission routes with greater resolution than analogous methods using the genome of a single isolate. While homoplasy and contamination might generate false-positive results and, in some settings, might be relatively common, consideration of additional data sources (temporal, spatial, and similarity of genetic background) can sometimes identify and rule out such cases. Rigorous collection of epidemiologic data remains a crucial component of outbreak investigation, and combining this with deep sequencing and SV analysis can provide unprecedented insight into individual-level transmission dynamics. The development of models that suitably incorporate all data sources remains an important goal.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, Massachusetts (Colin J. Worby, Marc Lipsitch, William P. Hanage). C.J.W. is currently at the Department of Ecology and Evolutionary Biology, Princeton University, Princeton, New Jersey.
Research reported in this paper was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant U54GM088558).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest: none declared.
REFERENCES
- 1. Grad YH, Lipstich M. Epidemiologic data and pathogen genome sequences: a powerful synergy for public health. Genome Biol. 2014;15(11):538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cottam EM, Thébaud G, Wadsworth J, et al. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc Biol Sci. 2008;275(1637):887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Worby CJ, Lipsitch M, Hanage WP. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput Biol. 2014;10(3):e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Didelot X, Gardy J, Colijn C. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol. 2014;31(7):1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ypma RJ, Bataille AM, Stegeman A, et al. Unravelling transmission trees of infectious diseases by combining genetic and epidemiological data. Proc Biol Sci. 2012;279(1728):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ypma RJF, van Ballegooijen WM, Wallinga J. Relating phylogenetic trees to transmission trees of infectious disease outbreaks. Genetics. 2013;195(3):1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jombart T, Eggo RM, Dodd PJ, et al. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity (Edinb). 2011;106(2):383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jombart T, Cori A, Didelot X, et al. Bayesian reconstruction of disease outbreaks by combining epidemiologic and genomic data. PLoS Comput Biol. 2014;10(1):e1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Struelens MJ, Deplano A, Godard C, et al. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30(10):2599–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strommenger B, Braulke C, Heuck D, et al. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koreen L, Ramaswamy SV, Graviss EA, et al. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42(2):792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gardy JL, Johnston JC, Ho Sui SJ, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364(8):730–739. [DOI] [PubMed] [Google Scholar]
- 13. Bryant JM, Schürch AC, van Deutekom H, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013;13:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walker TM, Lalor MK, Broda A, et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med. 2014;2(4):285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Worby CJ, Chang HH, Hanage WP, et al. The distribution of pairwise genetic distances: a tool for investigating disease transmission. Genetics. 2014;198(4):1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes J, Allen RC, Baguelin M, et al. Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks. PLoS Pathog. 2012;8(12):e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murcia PR, Hughes J, Battista P, et al. Evolution of an Eurasian avian-like influenza virus in naive and vaccinated pigs. PLoS Pathog. 2012;8(5):e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emmett KJ, Lee A, Khiabanian H, et al. High-resolution genomic surveillance of 2014 ebolavirus using shared subclonal variants. PLoS Curr. 2015;7 (doi:10.1371/currents.outbreaks.c7fd7946ba606c982668a96bcba43c90). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balloux F. Demographic influences on bacterial population structure In: Robinson DA, Falush D, Feil EJ, eds. Bacterial Population Genetics in Infectious Diseases. Hoboken, NJ: John Wiley & Sons Inc.; 2010:103–120. [Google Scholar]
- 20. Gire SK, Goba A, Andersen KG, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345(6202):1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stack JC, Murcia PR, Grenfell BT, et al. Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation. Proc Biol Sci. 2013;280(1750):20122173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poon LL, Song T, Rosenfeld R, et al. Quantifying influenza virus diversity and transmission in humans. Nat Genet. 2016;48(2):195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paterson GK, Harrison EM, Murray GG, et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun. 2015;6:6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Worby CJ, Read TD. “SEEDY” (Simulation of Evolutionary and Epidemiological Dynamics): an R package to follow accumulation of within-host mutation in pathogens. PLoS One. 2015;10(6):e0129745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wertheim JO, Leigh Brown AJ, Hepler NL, et al. The global transmission network of HIV-1. J Infect Dis. 2014;209(2):304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park DJ, Dudas G, Wohl S, et al. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161(7):1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10(8):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. WHO Ebola Response Team ; Aylward B, Barboza P, et al. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371(16):1481–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snitkin ES, Zelazny AM, Thomas PJ, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4(148):148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spada E, Sagliocca L, Sourdis J, et al. Use of the minimum spanning tree model for molecular epidemiological investigation of a nosocomial outbreak of hepatitis C virus infection. J Clin Microbiol. 2004;42(9):4230–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varble A, Albrecht RA, Backes S, et al. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe. 2014;16(5):691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joseph SB, Swanstrom R, Kashuba AD, et al. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol. 2015;13(7):414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84(12):6241–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stadler T, Kühnert D, Rasmussen DA, et al. Insights into the early epidemic spread of Ebola in Sierra Leone provided by viral sequencing. PLoS Curr. 2014;6 (doi:10.1371/currents.outbreaks.02bc6d927ecee7bbd33532ec8ba6a25f). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoenen T, Safronetz D, Groseth A, et al. Virology. Mutation rate and genotype variation of Ebola virus from Mali case sequences. Science. 2015;348(6230):117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rocha EP, Smith JM, Hurst LD, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239(2):226–235. [DOI] [PubMed] [Google Scholar]
- 37. Leggett HC, Cornwallis CK, West SA. Mechanisms of pathogenesis, infective dose and virulence in human parasites. PLoS Pathog. 2012;8(2):e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. The igraph core team igraph—the network analysis package. 2015. www.igraph.org. Accessed December 12, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.