Abstract
Pseudomonas aeruginosa secretes numerous toxins and destructive enzymes that play distinct roles in pathogenesis. The Type III secretion system (T3SS) of Pseudomonas is a system that delivers a subset of toxins directly into the cytoplasm of eukaryotic cells. The secreted effectors include ExoS, ExoT, ExoU, and ExoY. In this chapter, we describe methods to induce T3S expression and measure the enzymatic activities of each effector in in vitro assays. ExoU is a phospholipase and its activity can be measured in a fluorescence-based assay monitoring the cleavage of the fluorogenic substrate, PED6. ExoS and ExoT both possess ADP-ribosyltransferase (ADPRT) and GTPase-activating protein (GAP) activity. ADPRT activity can be assessed by using radiolabeled nicotinamide adenine dinucleotide (NAD+) and measuring the covalent incorporation of ADP-ribose into a target protein. GAP activity is measured by the release of radiolabeled phosphate from [γ-32P]GTP-bound target proteins. In accordance with recent trends towards reducing the use of radioactivity in the laboratory, alternative assays using fluorescent or biotin-labeled reagents are described. ExoY is a nucleotidyl cyclase; cAMP production stimulated by ExoY can be monitored using reverse-phase HPLC or with commercially available immunological assays.
Keywords: Pseudomonas aeruginosa, Type III secretion, ExoU, ExoT, ExoS, ExoY, Bacterial toxins, ADP-ribosyltransferase, GTPase-activating protein, Phospholipase, Nucleotidyl cyclase
1 Introduction
The T3SS encodes a protein machinery that allows P. aeruginosa to directly inject effectors into the cytoplasm of mammalian host cells. Cells of the innate immune system are thought to be the central targets for T3SS intoxication. Intoxication of immune cells such as neutrophils, monocytes, and macrophages allows the bacteria to evade clearance mechanisms and establish an initial infection [1]. T3S is stimulated by specific environmental cues, including contact with mammalian cells [2, 3]. Interestingly each of the identified effectors possesses a domain whose enzymatic activity requires a eukaryotic cofactor, suggesting that these enzymes evolved to specifically target eukaryotic competitors. Intoxication of cells by the P. aeruginosa T3SS is thought to contribute to severe human disease [4–6] which is recapitulated in various animal models of acute infection [7–10]. The role of T3SS enzymes in the unique setting of Pseudomonas chronic infection remains controversial. While most environmental isolates secrete T3S proteins, the percentage of Pseudomonas isolates cultured from CF patients that are able to secrete T3S effectors decreases relative to increased duration of infection [11].
The P. aeruginosa T3SS is regulated by a member of the AraC/XylS family of transcriptional activators, ExsA, which is involved in controlling the ten promoters that drive expression of operons encoding the secretion machinery, chaperones and effectors [12, 13]. Inducing signals for ExsA expression include growth in a calcium-limited medium and bacterial contact with host cells [3, 14, 15]. To measure the extracellular accumulation of effector enzymes, simple mediums have been devised which contain chelators such as EGTA or nitrilotriacetic acid (NTA). The concentration of chelators varies between investigators but ranges between 2 and 10 mM [14–18]. A minimal medium has also been reported that supplements with a calcium chelator and magnesium salts [19, 20].
ExoU is an A2 phospholipase [21–23] which targets cellular membranes, resulting in cytotoxicity in cellular models and pathology, such as lung damage, disseminated infection, and mortality in animal models [9, 20, 24–28]. In humans, infection with an ExoU-expressing strain is associated with fatal pneumonia [4, 5, 29]. ExoU activity can be measured by a fluorescence-based assay in which PED6, a lipid analog with a BODIPY dye-labeled sn-2 acyl chain and a dinitrophenol quencher-labeled head group [30], is utilized as a fluorogenic substrate for ExoU phospholipase activity. In intact PED6, the quencher group prevents fluorescence emission. Cleavage of the dye-labeled acyl chain of PED6 by an A2 phospholipase eliminates the quenching effect and results in a fluorescent signal [31]. A eukaryotic cofactor is required for the in vitro activity of ExoU. This cofactor is present in yeast and mammalian cellular extracts [21, 22, 30, 32] and was initially identified as superoxide dismutase (SOD1) [30]. Later ubiquitin was shown to be the essential cofactor for ExoU activity. Ubiquitylated proteins such as SOD1 also activate ExoU [33]. Interestingly, ExoU itself can be ubiquitylated at a specific lysine residue [32]; however, ubiquitylation is not required for ExoU activity either in vitro [33] or in vivo [32].
ExoS and ExoT have 75 % amino acid identity [34] and are bifunctional enzymes with both GAP and ADPRT activity [34–39]. These activities target cellular signaling and the host cell cytoskeleton. Specifically, the GAP domain of ExoS and ExoT targets small GTPases such as Rho, Rac, and Cdc42 [36–38, 40]. GTPases cycle between an active GTP-bound state and an inactive GDP-bound state. Overexpression of a GAP, such as in a cell injected with ExoS or ExoT, results in the inactive GDP-bound form [36, 41]. GTPase activity can be measured by loading target proteins such as RhoA with [γ-32P]GTP and determining the release of [32Pi] from the target protein. P. aeruginosa inhibits phagocytosis by macrophages in vitro via the GAP activity of ExoS [42]. ExoS GAP activity disrupts the host cell’s cytoskeleton and inhibits bacterial uptake [41, 43, 44]. ExoT GAP activity targets the cellular actin cytoskeleton and prevents bacterial internalization [45].
ADPRT activity involves transfer of an ADP-ribose moiety from NAD to a target protein. This covalent modification interferes with the normal function of the target protein. ADPRT activity can be measured by monitoring the incorporation of [32P]adenylate phosphate-NAD+ onto target protein arginine residues. ExoS and ExoT require a eukaryotic cofactor initially identified as the Factor-Activating Exoenzyme S (FAS) and later verified as members of the 14-3-3 protein family [39, 46–49]. Unlike most bacterial enzymes with ADPRT activity, ExoS has many targets, including small GTPases and members of the ezrin, radixin, and moesin family of proteins [35, 50–55]. ExoS ADPRT activity interferes with host cell endocytosis [54] and vesicular trafficking [44], is cytotoxic [56] and induces apoptosis in cellular models [57, 58]. ExoS ADPRT activity may also play a role in immune evasion through inhibiting secretion of the pro-inflammatory mediator IL-1β [59]. The targets of ExoT ADPRT activity, CrkI and CrkII adaptor proteins [39], are more limited than that of ExoS. ExoT ADPRT activity contributes to disruption of the host cell actin cytoskeleton, impairment of wound healing, and inhibits bacterial internalization [60]. Although not cytotoxic, ExoT has been shown to contribute to disease in animal models [27, 28, 45, 61].
ExoY was initially identified as an adenylyl cyclase. More recent analyses suggest its substrate specificity may be broader than initially thought, acting upon UTP, GTP, and CTP. In the future, these data may lead to the reclassification of ExoY as a nucleotidyl cyclase ([62], R. Seifert, personal communication). The full activity of ExoY also requires an unidentified eukaryotic cofactor, although basal adenylyl cyclase activity can be detected in the absence of a cofactor [63]. ExoY disrupts the actin cytoskeleton [63, 64], inhibits bacterial uptake [65] and causes junctional gaps in endothelial cells [66, 67]. The mechanism mediating endothelial gap formation appears to be associated with microtubule disruption as a result of Tau phosphorylation [66, 67]. Ultimately ExoY activity is postulated to increase lung permeability [66]. Recently, ExoY has also been shown to possess guanylyl cyclase activity that impacts endothelial gap formation and Tau phosphorylation, although not to as great an extent as adenylyl cyclase activity [62]. The specific role of ExoY in animal models is uncertain [28, 61]. Nucleotidyl cyclase activity can be measured by incubating toxin with its ATP substrate and monitoring cAMP formation by reverse-phase high-performance liquid chromatography (HPLC) [63], immunological assays measuring cAMP [62, 66], and mass spectrometry-based assays [68].
2 Materials
2.1 Induction of Type III Secretion and Harvest of Secreted Proteins from the Supernatant
10× Vogel Bonner Minimal (VBM) medium: 900 ml dH2O, 2 g MgSO4·7H2O, 20 g citric acid (free acid), 100 g K2HPO4, 35 g NaNH4HPO4·4H2O. Adjust the pH to 7.0. Adjust final volume to 1 l. Autoclave. Store at room temperature (RT).
1× VBM Agar medium: 450 ml water, 7.5 g Bacto Agar (BD). Autoclave. Place melted agar and 10× VBM in a 50 °C water bath and add 50 ml of sterile 10× VBM salts to the 450 ml agar/water. Swirl to mix. Pour into 10 cm bacteriological petri dishes.
10× Trypticase Soy Broth Dialyzate (10× TSBD): Add 150 g of tryptic soy broth (Difco) to 450 ml of filtered water. Add 50 g Chelex 100 resin (Bio-Rad). Stir at RT for approximately 5.5 h or overnight (4 °C). To remove the Chelex resin subject the mixture to centrifugation at 10,000 × g, 4 °C for 20 min. Pour off the supernatant and filter it through Whatman #1 filter paper. Soak an Amicon PM30 76 mm membrane (30K MW cutoff) in water to remove glycine. Assemble a 400-ml capacity Amicon stirred ultrafiltration cell (EMD Millipore): place the filter shiny-side up on the bottom of the concentrator. Place the unit on the magnetic stir plate and start stirring. Connect the nitrogen gas line to the Amicon unit. Set the inlet pressure of the nitrogen cylinder to 40–70 psi. Rinse the membrane several times with filtered water. Pour the filtered medium into the concentrator. Collect the filtrate in a covered vessel. Filtration will take several hours at 4 °C. Continue filtering until ~75 ml medium is left in the Amicon unit. Save the filtered medium (filtrate) and freeze at −20 °C in 40 ml aliquots.
1× TSBD: Thaw 10× filtered medium, dilute 1:10 with high-quality cation-deficient water (18 mΩ) and autoclave.
1× TSBD++: Add glycerol to 1 % final concentration and mono-sodium glutamate (MSG) to 100 mM final concentration.
2–10 mM NTA or EGTA.
Saturated ammonium sulfate.
SDS-PAGE loading buffer.
2.2 ExoU: Phospholipase Assay
Pseudomonas culture supernatant containing ExoU, unconcentrated.
Phospholipase Assay Buffer (pH 6.3): 50 mM MOPS, 50 mM NaCl, 250 mM MSG, 30 μM N-((6-(2,4-Dinitrophenyl) amino)hexanoyl)-2-(4,4-Difluoro-5,7-Dimethyl-4-Bora-3a, 4a-Diaza-s-Indacene-3-Pentanoyl)-1-Hexadecanoyl-sn-Glycero-3-Phosphoethanolamine, Triethylammonium Salt (PED6) (Molecular Probes).
K48-linked polyubiquitin (stock solution in 10 mM KPO4, pH 6.3) (Enzo Life Sciences).
96-well black microplate.
Spectramax M5 microplate reader (Molecular Devices).
2.3 ExoS/T Bifunctional enzymes: GAP Assay
2.4 ExoS/T Bifunctional Enzymes: ADPRT Assay
Secreted ExoS or ExoT: Supernatants from cultures grown as described in TSBD low-calcium medium [39].
Soybean trypsin inhibitor.
Purified Crk-I [39].
ADPRT Assay Buffer: 50 mM Tris–HCl (pH 7.4), 0.1 mM NAD+ ([32P]adenylate phosphate-NAD+), 500 nM FAS [70], and 0.2 μg/μl bovine serum albumin.
SDS sample buffer.
Scintillation counter.
2.5 ExoY: Adenylyl Cyclase Assay
1 μM recombinant ExoY [63].
ExoY Assay Buffer: 10 mM Tris–HCl (pH 8), 6 mM MgCl2, 0.2 mM CaCl2, 2 mM ATP, and 2 mM DTT.
Postnuclear extract (PNE) from CHO cells [71].
100 % ethanol.
Reversed-phase HPLC C18 column (0.46 cm × 15 cm).
HPLC instrument.
Buffer A: 0.1 M potassium phosphate (pH 6.0).
Buffer B: 0.1 M potassium phosphate (pH 6.0) in 10 % CH3OH.
Purified ATP, AMP, and cAMP for use as standards for HPLC.
3 Methods
3.1 Induction of Type III Secretion and Harvest of Secreted Proteins from the Supernatant
Culture Pseudomonas strain on VBM.
Incubate plates at 37 °C for 24 h, then store at RT for 1–2 days (do not store plates at 4 °C).
Emulsify a large loopful of bacteria in 1 ml of TSBD++ containing 0.01 M NTA. Obtain an optical density reading at 540 nm (OD540).
Dilute the suspension to a final OD540 = 0.02 in 10 ml TSBD++, +/− antibiotic, +/− chelator: use either 2.5–10 mM NTA or 2–5 mM EGTA (see Note 1).
Grow 10–18 h at 32 °C with shaking to an OD540 ≈ 4.0.
Harvest the supernatant in a microcentrifuge (13,500 rpm or 16,000×g) for 10 min at 4 °C (or 8,000 × g in a preparative centrifuge).
For enzyme activity assays, unconcentrated supernatant material may be titrated into the assay immediately or frozen at −80 °C until the day the assay is to be performed (see Notes 2–5).
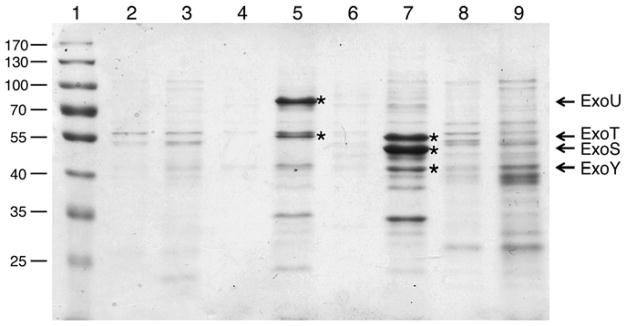
For SDS-PAGE analysis, concentrated supernatant material should be prepared. Continue from step 6 as follows: Remove 0.64 ml of the supernatant to a microcentrifuge tube containing 0.8 ml saturated NH4 sulfate (final concentration 55 %). Keep on ice 2 h or −20 °C overnight. Centrifuge for 10 min in the microcentrifuge (13,500 rpm or 16,000 × g) at 4 °C. Discard supernatant without disturbing the pellet. Repeat centrifugation. Remove remaining supernatant and suspend the pellet in 32 μl of 1× SDS-PAGE loading buffer. Final concentration of supernatant is 20× (see Note 6). See Fig. 1 for a sample gel showing concentrated supernatants from 4 strains of P. aeruginosa.
Fig. 1.
Expression and secretion of the Pseudomonas aeruginosa Type III effectors. Four stains of P. aeruginosa were grown in Chelex-treated trypticase soy broth with monosodium glutamate, glycerol and in the absence (lanes 2, 4, 6, and 8) or in the presence of 10 mM nitrilotriacetic acid (lanes 3, 5, 7, and 9). Supernatant fractions were prepared and concentrated 20-fold as described in Subheading 3 . Samples were normalized to the OD540 at the time of harvest and separated by SDS-PAGE (10 % acrylamide gel). The gel was stained with Coomassie blue. Lane 1 shows a prestained molecular weight standard. Lanes 2 and 3 , PA01; lanes 4 and 5, PA103; lanes 6 and 7 , PAK and lanes 8 and 9 , the concentrated culture supernatants from strain PA14. Type III toxins secreted from PA103 and PAK are marked with an asterisk (*) in lanes 5 and 7 . Concentrated supernatants from strains with low protease expression (PA103, lanes 4 and 5) and strains that overproduce type III effectors (PAK, lanes 6 and 7 ) exhibit a clearly inducible pattern of protein bands in the extracellular supernatant fraction. In contrast, it can be difficult to detect the effectors in strains that produce proteases (PA01, lanes 2 and 3) or strains that do not grow as well under these conditions (PA14, lanes 8 and 9) (see Note 3). Methods that have a high level of sensitivity and specificity such as Western blot analysis with specific antibodies, silver stained gels, enzymatic activity, or mass spectrometry can be used as alternative approaches to identify the complement of secreted effectors
3.2 ExoU: Phospholipase Assay
Carry out all procedures at RT.
In a 50 μl total reaction volume, add 20 μl culture supernatant to Phospholipase Assay Buffer (see Notes 7 and 8). Add K48-linked polyubiquitin stock solution to a final amount of 0.4–3.0 μg (see Notes 9 and 10).
Vortex briefly.
Add to a 96-well black microplate.
Measure fluorescence intensity (relative fluorescence units, RFU) at 15-min increments for up to 120 min at RT at an excitation wavelength of 488 nm and an emission wavelength of 511 nm (495 nm cutoff filter) (see Note 11).
Normalize RFU values relative to growth of the bacterial culture by dividing RFU value by the OD540 value at time of harvest (see Notes 12 and 13).
Alternatives to the described assay are described in Subheading 4 (see Notes 14 and 15).
3.3 ExoS/T Bifunctional enzymes: GAP Assay
Incubate recombinant Rho proteins at 2 μM final concentration (see Note 16) with 10 μM [γ-32P]GTP for 5 min at 37 °C in GAP Assay Buffer.
For initiation of intrinsic GTPase activity, add 12 mM MgCl2 and 2 mM unlabeled GTP to the reaction.
For GAP stimulation, add 100 nM ExoS or ExoT (see Notes 17–19) and incubate at 37 °C for 4 min.
Analyze GTPase activity by filter binding (see Note 20): Spot reaction mixtures on 0.4 μm nitrocellulose, wash filters with GAP Assay Buffer to remove free radiolabel, dry and then measure remaining radioactivity by scintillation counting.
Report results as percentage of GTP bound initially (100 %) (see Note 21).
3.4 ExoS/T Bifunctional Enzymes: ADPRT Activity Assay
Incubate 5 nM secreted ExoS or ExoT (see Note 22) with 3 μM soybean trypsin inhibitor (SBTI) (see Note 23) or purified Crk-I in ADPRT Assay Buffer (see Notes 24–26).
Stop reactions at 2, 4, 8, and 16 min by adding SDS sample buffer.
Fractionate samples by SDS-PAGE, followed by Coomassie staining.
Excise radioactive bands and subject to scintillation counting to measure incorporation of radiolabel (see Note 27).
Determine specific activity as moles of NAD+ incorporation per mole of enzyme per minute (see Note 28).
Alternatives to the described assay are described in Subheading 4 (see Notes 29 and 30).
3.5 ExoY: Adenylyl Cyclase Assay (see Note 31)
In a 100 μl final reaction volume, incubate 1 μM ExoY (see Note 32) in ExoY Assay Buffer. 30 μg PNE from CHO cells may also be added to further stimulate ExoY activity (see Note 33).
Incubate at 30 °C for 4 h (30 min if +PNE).
Stop reaction with the addition of 186 μl of ethanol (65 %, final concentration).
Incubate at RT 5 min to precipitate protein.
Centrifuge at 14,000 × g for 5 min.
Harvest supernatants, lyophilize, and suspend in 50 μl distilled water.
Perform reverse-phase HPLC to resolve cAMP from ATP and AMP (see Notes 34 and 35): Inject samples into a C-18 column and resolve by washing for 5 min at 100 % Buffer A, followed by an 8-min gradient to 100 % Buffer B, hold for 5 min at 100 % Buffer B, and return to 100 % Buffer A for 5 min.
Monitor absorbance at 259 nm.
Calculate formation of cAMP as a percentage of original ATP substrate present in the reaction.
Calculate retention times using purified ATP, AMP, and cAMP as standards (see Note 36).
Footnotes
Alternate mediums for the expression of T3S are discussed in Subheading 1. The authors include the original medium developed by the Iglewski Laboratory [14] since it can be used in the absence of chelators to assay for extracellular exotoxin A activity [72].
Pseudomonas supernatant material has routinely been used as a source of toxin for enzymatic assays [39, 73].
The methods described for the induction of type III secretion have been optimized for strains PA103 and PAK. Depending on the Pseudomonas strain used, the expression of proteases may degrade secreted proteins and affect the yield of protein from secreted supernatants (as shown in Fig. 1). Importantly, some clinical isolates, particularly those from cystic fibrosis patients, are difficult to grow in the presence of a chelator. Therefore several induction parameters may need optimization depending on the strain used for a model system. These parameters include the type of medium, the concentration of chelator, the type of chelator, growth temperature, or length of time that the culture is induced. For strains that express and secrete alkaline protease or elastase, harvesting the culture at a lower optical density enhances the extracellular yield of ExoS and ExoT.
An alternate approach to using secreted supernatants as a source of Type III toxins is to express and purify recombinant toxins. This approach is particularly useful for assessing the activity of genetically modified toxins and determining structure–activity relationships.
Total time required for the assay: 1 day is required for growth on plates followed by 10–18 h growth in liquid culture. Harvest of supernatant will take an additional 10 min. Total estimated time (minimum) of 2 days. Optional concentration of supernatant requires an additional 2–3 h.
This assay is sufficient to detect activity from unconcentrated Pseudomonas culture supernatants [73].
Recombinant ExoU may also be used successfully in this assay at a final concentration of approximately 34 nM [33].
Although other isoforms of ubiquitin, such as mono-, di-, tetra-, or octa-ubiquitin will activate ExoU, the affinity of ubiquitin for ExoU increases with increasing ubiquitin chain length. Polyubiquitin was chosen for this assay because of its high affinity for ExoU.
Other isoforms of ubiquitin may also be used in this assay with the concentration dependent upon the isoform used [33].
Determine normal background fluorescence intensity associated with PED6 by performing the assay in the absence of ubiquitin. Subtract this value from the sample value.
The described assay is highly sensitive and can provide kinetic data of ExoU activation. It is the first example in the literature of an ExoU activity assay with enough sensitivity to detect catalytically active ExoU from an injected eukaryotic cell [31].
Total time required for the assay: This assay runs for 120 min, plus additional time to prepare reagents and the reaction mixture.
The activity of ExoU has also been measured with 14C-labeled lipid substrates [23, 75]. Recombinant ExoU was incubated with labeled liposomes and eukaryotic cell extract (as a source for the cofactor). Hydrolysis of the substrate was measured by thin layer chromatography, or alternately by using a scintillation counter.
The activity of ExoU has also been indirectly measured with Ellman’s reagent [76]. Briefly, ExoU-expressing yeast cells were lysed and incubated with a cleavable phospholipid substrate and then Ellman’s reagent. Cleavage of the substrate yields a free thiol group that reacts with Ellman’s reagent to form a colored product.
Recombinant Rac and Cdc42 may be substituted for Rho in this assay [36].
This assay has only been demonstrated for recombinant ExoS or ExoT expressed from E. coli [36, 37] and ExoS expressed from P. aeruginosa and subsequently purified by gel filtration and ion exchange chromatography [36, 69]. The methods described here apply to recombinant ExoS. Slight variations to the materials and methods (including concentration of RhoA, toxin, unlabeled GTP, and MgCl2 and temperature of the assay) may be necessary for assaying activity of ExoT [37] or ExoS purified from P. aeruginosa [36].
Varying concentrations of ExoS or ExoT (5–100 nM) may be used in this assay. A linear rate of activity can be found at concentrations of ExoT/ExoS that stimulate hydrolysis of less than 25 % of the available Rho-GTP [37].
GAP activity of ExoT and ExoS does not require the presence of a eukaryotic cofactor.
An alternate approach to the filter-binding assay described would be to measure GDP:GTP:P(i) ratios by HPLC [77]. A similar reaction mixture could be used and radiolabeled nucleotides would not be necessary.
Total time required for the assay: This assay runs for approximately 10 min. Additional time for the filter-binding assay and scintillation counting (estimated less than one h) is required.
This assay is sufficient to detect activity from concentrated Pseudomonas culture supernatants.
For analysis of ExoS activity with a more physiologically relevant target than SBTI, a similar reaction can also be performed in the presence of 500 nM ezrin, radixin, moesin, or Ras [55].
ADPRT activity of ExoS and ExoT requires the presence of the cofactor FAS, which is present in the ADPRT Assay Buffer.
Control reactions should include a reaction without FAS. No activity should be detected in this reaction.
Rather than incubation with FAS and a specific target protein, enzyme and labeled NAD may be incubated with wheat germ extract or CHO/HeLa cell lysate, which are sources of both FAS and target proteins for ADP-ribosylation [39, 69].
As an alternative to band excision and scintillation counting, the reaction mixture can be fractionated by SDS-PAGE, subjected to autoradiography, and the incorporation of radiolabel determined by densitometry [78].
This assay runs for approximately 16 min. Additional time for SDS-PAGE and scintillation counting will be required.
A fluorescent assay for ExoS ADPRT activity has also been developed [79]. In this assay, ExoS is added to a reaction mixture of FAS, recombinant Ras, and fluorescent ε-NAD+. Time-dependent change in fluorescence can then be monitored.
A biotin-based assay for ExoS ADPRT activity has also been developed [80]. Briefly, biotinylated-NAD is added to enzyme and target protein and incubated for 1 h. ADP-ribosylated proteins are then separated by SDS-PAGE, blotted onto nitro-cellulose, detected with a streptavidin-HRP conjugate, and finally visualized by chemiluminescence. This assay avoids the use of radioactivity.
ExoY, initially identified as an adenylyl cyclase, has recently been demonstrated to have broader substrate specificity, acting upon UTP, GTP, and CTP, and may be reclassified as a nucleotidyl cyclase ([62], R. Seifert, unpublished).
Previously published ExoY activity assays have only been performed with recombinant ExoY or in ExoY-expressing eukaryotic cells.
Control reactions should include reactions without enzyme and with a post-nuclear extract of eukaryotic cells alone.
Formation of cAMP, cGMP, cCMP, and cUMP can also be detected via HPLC-MS/MS [68].
cAMP and cGMP formation can also be detected using commercially available immunological assays (Biomedical Technologies [62, 66]).
Total time required for the assay: The enzyme assay runs for approximately 16 min, with additional time required for SDS-PAGE and scintillation counting.
References
- 1.Diaz MH, Hauser AR. Pseudomonas aeruginosa cytotoxin ExoU is injected into phagocytic cells during acute pneumonia. Infect Immun. 2010;78:1447–1456. doi: 10.1128/IAI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank DW. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 3.Vallis AJ, et al. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy-Burman A, et al. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 5.Hauser AR, et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 6.EL-Solh AA. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med. 2012;40:1157–1163. doi: 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawa T, et al. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med. 1999;5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 8.Kurahashi K, et al. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J Clin Invest. 1999;104:743–750. doi: 10.1172/JCI7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allewelt M, et al. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun. 2000;68:3998–4004. doi: 10.1128/iai.68.7.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holder IA, Neely AN, Frank DW. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns. 2001;27:129–130. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 11.Jain M, et al. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 13.King JM, et al. Orientation of Pseudomonas aeruginosa ExsA monomers bound to promoter DNA and base-specific contacts with the P(exoT) promoter. J Bacteriol. 2012;194:2573–2585. doi: 10.1128/JB.00107-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iglewski BH, et al. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson MR, et al. Exoenzyme S: an ADP-ribosyltransferase produced by Pseudomonas aeruginosa. In: Sugimura T, Smulson M, editors. Novel ADP-ribosylation of regulatory enzymes and proteins. Elsevier/North-Holland; Amsterdam: 1980. pp. 425–433. [Google Scholar]
- 16.Dacheux D, et al. Cell death of human polymorphonuclear neutrophils induced by a Pseudomonas aeruginosa cystic fibrosis isolate requires a functional type III secretion system. Infect Immun. 1999;67:6164–6167. doi: 10.1128/iai.67.11.6164-6167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCaw ML, et al. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol. 2002;46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, et al. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology. 2005;151:3575–3587. doi: 10.1099/mic.0.28277-0. [DOI] [PubMed] [Google Scholar]
- 19.Nicas TI, Iglewski BH. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauser AR, Kang PJ, Engel JN. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 21.Sato H, et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips RM, et al. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J Biol Chem. 2003;278:41326–41332. doi: 10.1074/jbc.M302472200. [DOI] [PubMed] [Google Scholar]
- 23.Tamura M, et al. Lysophospholipase A activity of Pseudomonas aeruginosa type III secretory toxin ExoU. Biochem Biophys Res Commun. 2004;316:323–331. doi: 10.1016/j.bbrc.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 24.Finck-Barbançon V, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 25.Finck-Barbançon V, Frank DW. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol. 2001;183:4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankhaniya RR, et al. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit Care Med. 2004;32:2293–2299. doi: 10.1097/01.ccm.0000145588.79063.07. [DOI] [PubMed] [Google Scholar]
- 27.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee VT, et al. Activities of Pseudomonas aeruginosa effectors secreted by the Type III secretion system in vitro and during infection. Infect Immun. 2005;73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulert GS, et al. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J Infect Dis. 2003;188:1695–1706. doi: 10.1086/379372. [DOI] [PubMed] [Google Scholar]
- 30.Sato H, Feix JB, Frank DW. Identification of superoxide dismutase as a cofactor for the pseudomonas type III toxin, ExoU. Biochemistry. 2006;45:10368–10375. doi: 10.1021/bi060788j. [DOI] [PubMed] [Google Scholar]
- 31.Benson MA, Schmalzer KM, Frank DW. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA2 activity. Clin Chim Acta. 2010;411:190–197. doi: 10.1016/j.cca.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stirling FR, et al. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol. 2006;8:1294–1309. doi: 10.1111/j.1462-5822.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 33.Anderson DM, et al. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol. 2011;82:1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahr TL, Barbieri JT, Frank DW. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganesan AK, et al. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 36.Goehring UM, et al. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J Biol Chem. 1999;274:36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 37.Krall R, et al. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect Immun. 2000;68:6066–6068. doi: 10.1128/iai.68.10.6066-6068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazmierczak BI, Engel JN. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase-activating protein for RhoA, Rac1, and Cdc42. Infect Immun. 2002;70:2198–2205. doi: 10.1128/IAI.70.4.2198-2205.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J Biol Chem. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 40.Henriksson ML, et al. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem J. 2002;367:617–628. doi: 10.1042/BJ20020714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pederson KJ, et al. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol Microbiol. 1999;32:393–401. doi: 10.1046/j.1365-2958.1999.01359.x. [DOI] [PubMed] [Google Scholar]
- 42.Rocha CL, et al. Characterization of Pseudomonas aeruginosa exoenzyme S as a bifunctional enzyme in J774A.1 macrophages. Infect Immun. 2003;71:5296–5305. doi: 10.1128/IAI.71.9.5296-5305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krall R, et al. In vivo Rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect Immun. 2002;70:360–367. doi: 10.1128/IAI.70.1.360-367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Q, Barbieri JT. Modulation of host cell endocytosis by the type III cytotoxin, Pseudomonas ExoS. Traffic. 2008;9:1948–1957. doi: 10.1111/j.1600-0854.2008.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrity-Ryan L, et al. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect Immun. 2000;68:7100–7113. doi: 10.1128/iai.68.12.7100-7113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coburn J, et al. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 47.Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, et al. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J Bacteriol. 1997;179:1609–1613. doi: 10.1128/jb.179.5.1609-1613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, et al. Residues of 14-3-3 zeta required for activation of exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:12159–12164. doi: 10.1021/bi991019l. [DOI] [PubMed] [Google Scholar]
- 50.Coburn J, et al. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coburn J, et al. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–9008. [PubMed] [Google Scholar]
- 52.Coburn J, Gill DM. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riese MJ, Wittinghofer A, Barbieri JT. ADP ribosylation of Arg41 of Rap by ExoS inhibits the ability of Rap to interact with its guanine nucleotide exchange factor, C3G. Biochemistry. 2001;40:3289–3294. doi: 10.1021/bi002729q. [DOI] [PubMed] [Google Scholar]
- 54.Barbieri AM, et al. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect Immun. 2001;69:5329–5334. doi: 10.1128/IAI.69.9.5329-5334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maresso AW, Baldwin MR, Barbieri JT. Ezrin/Radixin/Moesin proteins are high affinity targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. J Biol Chem. 2004;279:38402–38408. doi: 10.1074/jbc.M405707200. [DOI] [PubMed] [Google Scholar]
- 56.Pederson KJ, Barbieri JT. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol Microbiol. 1998;30:751–759. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 57.Jia J, et al. c-Jun NH2-terminal kinase-mediated signaling is essential for Pseudomonas aeruginosa ExoS-induced apoptosis. Infect Immun. 2003;71:3361–3370. doi: 10.1128/IAI.71.6.3361-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alaoui-El-Azher M, et al. ExoS of Pseudomonas aeruginosa induces apoptosis through a Fas receptor/caspase 8-independent pathway in HeLa cells. Cell Microbiol. 2006;8:326–338. doi: 10.1111/j.1462-5822.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 59.Galle M, et al. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J Cell Mol Med. 2008;12:1767–1776. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garrity-Ryan L, et al. The ADP ribosyl-transferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. Infect Immun. 2004;72:546–558. doi: 10.1128/IAI.72.1.546-558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vance RE, Rietsch A, Mekalanos JJ. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ochoa CD, et al. Pseudomonas aeruginosa Exotoxin Y is a promiscuous cyclase that increases endothelial Tau phosphorylation and permeability. J Biol Chem. 2012;287:25407–25418. doi: 10.1074/jbc.M111.301440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yahr TL, et al. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vallis AJ, et al. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect Immun. 1999;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cowell BA, Evans DJ, Fleiszig SMJ. Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol Lett. 2005;250:71–76. doi: 10.1016/j.femsle.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 66.Sayner SL, et al. Paradoxical cAMP-induced lung endothelial hyperpermeability revealed by Pseudomonas aeruginosa ExoY. Circ Res. 2004;95:196–203. doi: 10.1161/01.RES.0000134922.25721.d9. [DOI] [PubMed] [Google Scholar]
- 67.Sayner SL, et al. Filamin A is a phosphorylation target of membrane but not cytosolic adenylyl cyclase activity. AJP Lung Cell Mol Physiol. 2011;301:L117–L124. doi: 10.1152/ajplung.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beste KY, et al. Nucleotidyl cyclase activity of soluble guanylyl cyclase α1 β1. Biochemistry. 2012;51:194–204. doi: 10.1021/bi201259y. [DOI] [PubMed] [Google Scholar]
- 69.Kulich SM, Frank DW, Barbieri JT. Purification and characterization of exoenzyme S from Pseudomonas aeruginosa 388. Infect Immun. 1993;61:307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masters SC, et al. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exo-enzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- 71.Xu Y, Barbieri JT. Pertussis toxin-mediated ADP-ribosylation of target proteins in Chinese hamster ovary cells involves a vesicle trafficking mechanism. Infect Immun. 1995;63:825–832. doi: 10.1128/iai.63.3.825-832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu PV. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
- 73.Schmalzer KM, Benson MA, Frank DW. Activation of ExoU phospholipase activity requires specific C-terminal regions. J Bacteriol. 2010;192:1801–1812. doi: 10.1128/JB.00904-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riese MJ, et al. Auto-ADP-ribosylation of Pseudomonas aeruginosa ExoS. J Biol Chem. 2002;277:12082–12088. doi: 10.1074/jbc.M109039200. [DOI] [PubMed] [Google Scholar]
- 75.Sato H, et al. Characterization of phos-pholipase activity of the Pseudomonas aerugi-nosa type III cytotoxin, ExoU. J Bacteriol. 2005;187:1192–1195. doi: 10.1128/JB.187.3.1192-1195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rabin SDP, Hauser AR. Functional regions of the Pseudomonas aeruginosa cyto-toxin ExoU. Infect Immun. 2005;73:573–582. doi: 10.1128/IAI.73.1.573-582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hemsath L, et al. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 78.Rocha CL, et al. Examination of the coordinate effects of Pseudomonas aeruginosa ExoS on Rac1. Infect Immun. 2005;73:5458–5467. doi: 10.1128/IAI.73.9.5458-5467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arnoldo A, et al. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet. 2008;4:e1000005. doi: 10.1371/journal.pgen.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castagnini M, et al. Arginine-specific mono ADP-ribosylation in vitro of antimicrobial peptides by ADP-ribosylating toxins. PLoS One. 2012;7:e41417. doi: 10.1371/journal.pone.0041417. [DOI] [PMC free article] [PubMed] [Google Scholar]