Abstract
γ-Tubulin is a conserved essential protein required for assembly and function of the mitotic spindle in humans and yeast. For example, human γ-tubulin can replace the γ-tubulin gene in Schizosaccharomyces pombe. To understand the structural/functional domains of γ-tubulin, we performed a systematic alanine-scanning mutagenesis of human γ-tubulin (TUBG1) and studied phenotypes of each mutant allele in S. pombe. Our screen, both in the presence and absence of the endogenous S. pombe γ-tubulin, resulted in 11 lethal mutations and 12 cold-sensitive mutations. Based on structural mapping onto a homology model of human γ-tubulin generated by free energy minimization, all deleterious mutations are found in residues predicted to be located on the surface, some in positions to interact with α- and/or β-tubulins in the microtubule lattice. As expected, one class of tubg1 mutations has either an abnormal assembly or loss of the mitotic spindle. Surprisingly, a subset of mutants with abnormal spindles does not arrest in M phase but proceeds through anaphase followed by abnormal cytokinesis. These studies reveal that in addition to its previously appreciated role in spindle microtubule nucleation, γ-tubulin is involved in the coordination of postmetaphase events, anaphase, and cytokinesis.
INTRODUCTION
The microtubule lattice is composed of a head-to-tail assembly of heterodimeric protein subunits, α-tubulin and β-tubulin (Amos and Klug, 1974). The two ends of a microtubule are different in composition, and this difference is reflected in the assembly and disassembly kinetics at the two ends. For example, the microtubule plus end is much faster at addition and dissociation of tubulin subunits during its growth and shrinkage compared with the minus end. In the mitotic spindle, minus ends are anchored to the spindle poles, whereas the plus ends are captured by the kinetochores of chromosomes (Euteneuer and McIntosh, 1981). The protein that binds and anchors microtubules to the spindle pole is another protein of the tubulin family, γ-tubulin, which was originally discovered in a suppressor screen of a β-tubulin mutant in the filamentous fungus Aspergillus nidulans (Oakley and Oakley, 1989). γ-Tubulin is an essential protein that is highly conserved (reviewed in Oakley, 2000). Furthermore, γ-tubulin binds with high affinity to microtubule minus ends in vitro (Li and Joshi, 1995; Leguy et al., 2000) and is required for the assembly of the mitotic spindle microtubules both in vivo and in vitro (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Joshi et al., 1992; Stearns and Kirschner, 1994; Felix et al., 1994; Sunkel et al., 1995, Sobel and Snyder, 1995; Spang et al., 1996, Marschall et al., 1996, reviewed in Oakley, 2000; Wiese and Zheng, 2000).
The mitotic spindle, a bipolar assembly of microtubules, is the macromolecular machine that is responsible for chromosome segregation and their faithful transmission into daughter cells during mitosis. To accomplish this task, paired duplicated sister chromatids must obtain a bipolar orientation at the mid plate of the metaphase spindle, via the attachment of their kinetochores to the spindle microtubules from two opposite spindle poles. On successful attachment of kinetochore microtubules, and perhaps upon the application of the resulting poleward tension between the sister kinetochores generated by pulling forces, a biochemical signaling cascade is activated. The pathway culminates in the proteolytic cleavage of the sister chromatid cohesin subunit, thus triggering a decisive and irreversible metaphase-anaphase transition (Uhlmann et al. 1999; reviewed in Amon, 1999; Burke, 2000; Nasmyth et al., 2000). Subsequent to the completion of anaphase, another signal transduction pathway involving the components of the septation initiation network is activated by a spindle pole-associated GTP binding protein Spg1p (reviewed in Gould and Simanis, 1997). This pathway culminates in the events that lead to the physical constriction of the actomyosin cytokinetic ring that pinches the plasma membrane at the center followed by the deposition of septal components and fission. The precise coordination of this sequence of events is achieved by feedback loops that ensure the completion of the previous steps before the initiation of the next steps (Balasubramanian et al., 2000).
After successful bipolar attachment of kinetochores to microtubules, one feedback loop monitors the connection between the kinetochore and the plus ends of the microtubules before triggering anaphase (reviewed in Shah and Cleveland, 2000). At the end of anaphase another feedback loop triggers subsequent cytokinesis (reviewed in Amon, 1999; Burke, 2000; Hoyt 2000). The role of minus end attachment to the spindle pole in coordinated triggering of these mitotic steps is not known. To investigate whether the microtubule minus end binding protein of the spindle pole, γ-tubulin, plays any role in mitotic events, we carried out a systematic alanine-scanning mutagenesis of the human γ-tubulin TUBG1 and generated 27 mutations, 12 of which confer cold-sensitive (cs−) growth defects in Schizosaccharomyces pombe, whereas 11 were lethal. Based on structural mapping onto a homology model of human γ-tubulin, all deleterious mutations are found in residues predicted to be located on the surface, often in a position to interact with α-tubulins and/or β-tubulins in the microtubule lattice. Although some of the tubg1 cs− phenotypes at the restrictive temperature showed an expected loss of mitotic spindle, many tubg1 mutants could assemble a mitotic spindle, although it often appeared abnormal. Despite abnormal spindle assembly, four of these mutants accomplished aberrant completion of mitosis and septation despite spindle defects. These results suggest a role for γ-tubulin in monitoring proper spindle assembly before anaphase and cytokinesis (see DISCUSSION).
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Manipulations
The following yeast strains were used in the characterization and genetic manipulation of the γ-tubulin mutants. NC377 (h+/h−, leu1-32/leu1-32, his3D1/+, ura4-D18/ura4-D18, ade6-M210/ade6-M216, +/gtb1::ura4+) (Horio and Oakley, 1994)-derived haploids were used to identify mutations that are conditional in the absence of endogenous γ-tubulin, whereas JLP201 (h+, leu1-32, ura4-D18, ade6-210) (Paluh and Clayton, 1996) was used to identify mutations that are conditional in the presence of endogenous γ-tubulin. PY321 (h+, leu1-32, ura4-D18, lys1+::lacO, GFP-lacI-NLS) (Watanabe and Nurse, 1999) was used to monitor sister chromatid separation in γ-tubulin mutants. PY321 is a yeast strain that encodes a green fluorescent protein (GFP)-LacI fusion protein at the his1+ locus. This fusion protein binds to a tandem repeat of LacO sequences, which is inserted near the centromere of chromosome I at the lys1+ locus (Nabeshima et al., 1998; Watanabe and Nurse, 1999). Binding of the GFP-LacI fusion protein to LacO at the his1+ locus provides an efficient tool for marking the centromeric region (Nabeshima et al., 1998). Cells were grown in either rich medium (YPD), or minimal medium (MM) with the indicated supplements (Alfa et al., 1993). All yeast transformations were performed by the lithium acetate method (Alfa et al., 1993). We screened for cold-sensitive mutants at 18°C and temperature-sensitive mutants at 36°C. Strains were maintained at the permissive temperatures of 30 and 26°C, respectively.
Alanine-scanning Mutagenesis of TUBG1
TUBG1 cDNA was subcloned into the NdeI site of the pALTER-EX1 vector (Promega, Madison, WI) downstream from the SP6 promoter to create pTWH101. The amino acid sequence of TUBG1p was scanned for clusters of charged residues. Clusters were defined as a group of charged amino acids occurring within two to five residues of each other. Twenty-seven charged clusters were identified. The charged amino acids (Arg [R], Asp [D], Glu [E], Lys [K]) in these clusters were changed to alanine one cluster at a time. The mutagenesis was carried out with the use of oligonucleotide-directed mutagenesis, with the use of pTWH101 as the template. pALTER-EX1 contains a tetracycline resistance gene and an inactivated ampicillin resistance gene. The ampicillin-repair oligonucleotide restores the activity of the inactivated AmpR gene, whereas the tetracycline-knockout oligonucleotide inactivates the TetR gene. The inactivation of the TetR gene and the activation of the AmpR gene provided an effective method for selecting potential alanine-scanning mutants. Oligonucleotides synthesized by Integrated DNA Technologies (Coralville, IA) were used to change each cluster independently. Because contiguous alanine codons can be used to create a PstI recognition site, we were able to use PstI digestion to further screen for potential alanine-scanning mutants. Each of the 27 mutations was sequenced to verify that the intended mutation was present and that it was the only difference between the mutant (tubg1) and the wild type (TUBG1). Additionally, we could produce stable protein of the predicted size from all of the mutations generated in vitro.
Generation of γ-Tubulin Homology Model
The human γ-tubulin homology model was generated with the use of the Swiss-Model homology algorithm (www.expasy.ch/swissmod/SWISS-MODEL.html). This modeling algorithm requires a target sequence that shares at least 25% identity to a template with a known three-dimensional structure (Peitsch et al., 1995; Peitsch, 1996; Guex and Peitsch, 1997). The TUBG1p sequence was first processed by Swiss-Model, which BLASTs (Altschul et al., 1990) it against other sequences derived from the Brookhaven Protein Data Bank. In this process, porcine α-tubulin and β-tubulin (structures solved by Nogales et al., 1998, 1999) were selected as templates for the homology model construction. The TUBG1p sequence was then aligned to the template sequences with the use of a structurally corrected multiple sequence alignment. This was done with the use of the best-scoring diagonals as determined by SIM (Huang et al., 1991). A three-dimensional match was then performed by superimposing Cα atom pairs from the highest scoring local sequence alignment to construct a model. The structure was then optimized by maximizing the number of Cα atom pairs in the common core and minimizing their relative mean square deviation. The secondary structures and loops were further modified with the use of energy considerations such as van der Waals radii, hydrogen bonds, hydrophobic, polar, and ionic interactions to generate a minimal free energy conformation.
Flow Cytometry Analysis of Alanine-scanning Mutants
Wild-type cells, gtb1Δ cells with wild-type TUBG1, wild-type cells with wild-type TUBG1, wild-type cells with dominant tubg1 alleles, and gtb1Δ cells with recessive tubg1 alleles were grown at 30°C until mid-log phase and then shifted to 18°C for 10 h. Cells were then fixed with 70% cold ethanol. Cells were then processed with 0.1 mg/ml RNase A in 50 mM M Na citrate for 2 h at 37°C. For DNA staining, cells were suspended in 1 μM SYTOX Green (Molecular Probes, Eugene, OR) in 50 mM Na citrate. Cells were then immediately analyzed with the use of fluorescence-activated cell sorter (FACS).
Analysis of Alanine-scanning Mutants of TUBG1 in Fission Yeast
The plasmids used to transform yeast strains were constructed by subcloning each of the tubg1 alleles into pREP1 at the NdeI site downstream of the nmt1+ promoter (Maundrell, 1990, 1993). Wild-type JLP201 (Paluh and Clayton, 1996) cells were transformed with either one of the tubg1-pREP1 plasmids or TUBG1-pREP1 and grown on minimal media supplemented with adenine, histidine, and uracil. Transformants were then screened at 18 and 36°C to identify conditional mutants in the presence of endogenous γ-tubulin. The strains were maintained at 30 and 26°C, respectively. A diploid strain, NC377 (Horio et al., 1991; Horio and Oakley, 1994), bearing one endogenous wild-type copy of S. pombe γ-tubulin, gtb1+ and one disrupted copy, gtb1::ura4+, was also transformed with the mutant plasmids. The resulting transformants were then randomly sporulated, selected for ura+, leu+, and the spores were tested for conditional growth.
Immunofluorescence Microscopy
To obtain cultures enriched for mitotic cells, cells were grown to mid-log phase and then synchronized in S phase for 3 h by the addition of 20 mM hydroxyurea (HU) (Sigma Chemical Co., St. Louis, MO). The HU was then removed and the cells were shifted to 18°C for 8–12 h. Cells were then fixed and processed for immunofluorescence microscopy.
To prepare for microtubule staining, cells were grown to a density of 2–4 × 106 cells/ml at the permissive temperature and then shifted to the restrictive temperature for 8–12 h. Cells were fixed with formaldehyde and gluteraldehyde according to Alfa et al. (1993). After fixation and preincubation with PEMBAL [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 1 mM EGTA, 1 mM MgSO4, pH 6.9, 1% bovine serum albumin, 0.1% NaN3, 100 mM l-lysine hydrochloride] for 30 min at room temperature, the cells were incubated overnight at 26°C with the anti-α-tubulin monoclonal antibody TAT1 (1:25) in PEMBAL (Woods et al., 1989). The cells were then washed and incubated overnight at 26°C with the secondary antibody Cy3-conjugated goat anti-mouse (1:200) in PEMBAL (Jackson Laboratories, Bar Harbor, ME). The cells were then washed with PEMBAL and phosphate buffered saline (PBS). To visualize the DNA, cells were incubated in 1 μg/ml 4′, 6′-diamidino-2-phenylindole (DAPI) in PBS at 26°C for 30 min or incubated with 10 μg/ml Hoechst in PBS for 1 h. Cells were viewed under a Ziess Axiovert 135 microscope.
To determine γ-tubulin localization, cells were fixed with methanol. Briefly, cells were filtered onto a membrane disk and then fixed in cold methanol at −80°C for 10 min or longer. After fixation the cells were washed with PEM [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 1 mM EGTA, 1 mM MgSO4, pH 6.9] then permeabilized with 100T Zymolase 1.3 mg/ml (Seikagu, Japan) in PEMS (PEM + 1.2 M sorbitol). The cells were then blocked with PEMBALG (PEMBAL + glysine) for 30 min and then incubated overnight with the anti-γ-tubulin antibody, GTU-88 (1:200) (Sigma Chemical Co.) in PEMBALG. The after day, cells were washed and incubated overnight with the secondary antibody goat anti-mouse-Cy3 (1:1000) (Jackson Laboratories) in PEMBALG. DNA was visualized by either DAPI or Hoechst staining as previously described. Cells were viewed under a Ziess Axiovert 135 microscope.
To monitor separation of sister chromatids, PY321 cells were transformed with tubg1 mutations. The transformants were grown to mid-log phase and then incubated with 10 μg/ml Hoechst for 1 h. The cells were visualized with the use of an Olympus BX60 Epifluorescence microscope equipped with a Photometrics Quantix digital camera and IP-Lab Spectrum software.
RESULTS
Alanine-scanning Mutants Map to Surface of γ-Tubulin Homology Model
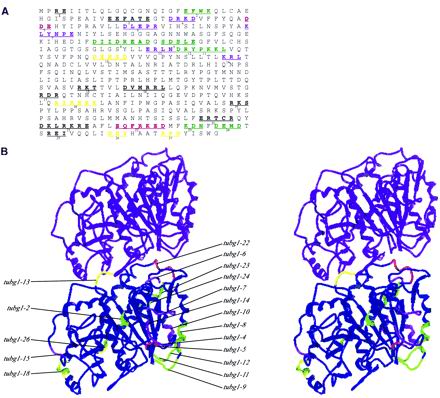
The logic for with the use of alanine-scanning mutagenesis is that by changing charged clusters of amino acids, which are hydrophilic and thus usually on the surface of the protein, subtle changes can be created in the protein structure that could prove to be useful in studying structure–function relationships. The amino acid sequence of γ-tubulin is as similar to α-tubulin and β-tubulin as the sequence of α-tubulin is to that of β-tubulin (Oakley and Oakley, 1989). Although the atomic structures of α-tubulin and β-tubulin are very similar to one another, there must exist some subtle differences in the structure of γ-tubulin. This is because γ-tubulin is not embedded in the microtubule lattice, but instead binds the component(s) of the microtubule-organizing center, in addition to the tubulin subunit(s) at microtubule minus ends. Thus, a simple substitution of γ-tubulin residues onto the α-tubulin or β-tubulin structural model was not sufficient for our mapping studies. Therefore, we generated a free energy, minimized homology model of γ-tubulin taking into consideration the allowable van der Waals radii of atoms, hydrogen bonds, and hydrophobic, polar, and ionic interactions. Figure 1 shows the locations of the charged amino acids that were changed to alanine on the γ-tubulin amino acid sequence (Figure 1A), and mutations that confer deleterious growth phenotypes are shown on the stereo image of the homology model of human γ-tubulin (Figure 1B). For orientation, we placed the equivalent structural model of the most likely tubulin subunit exposed at the minus end of a microtubule, α-tubulin (Fan et al., 1996). As expected, most of the alanine-scanning mutations are on the external surface of the protein where it is exposed to the aqueous environment of the cytoplasm. Many of the mutations that are deleterious to S. pombe growth are interestingly located at sites that might be involved in lateral (tubg1-8, tubg1-9, tubg1-11, tubg1-12, tubg1-15, and tubg1-18) or longitudinal (tubg1-4, tubg1-9, tubg1-13, and tubg1-22) interactions with the α-tubulin and/or β-tubulin subunits of the microtubule lattice (Inclán and Nogales, 2001). These observations suggest that at least some of the lateral and longitudinal contact sites of γ-tubulin must be equivalent to those of α-tubulin and β-tubulin within the microtubule lattice. The mutations, such as tubg1-2, tubg1-7, tubg1-24, and tubg1-26, that are not located at either the putative lateral or longitudinal interaction sites are localized to regions that may be involved in interactions with other nontubulin proteins. This homology model of human γ-tubulin can be used to further investigate the structure–function relationship of γ-tubulin. The alanine-scanning mutants will be useful to further study the interactions of γ-tubulin with other tubulins or other components of the microtubule-organizing center.
Figure 1.
Locations of the charged clusters of human γ-tubulin. (A) Amino acid sequence of human γ-tubulin is shown with the charged clusters underlined and the residues that were changed to alanine in bold letters. A cluster was defined as two or more charged residues within five residues of each other. Twenty-seven such charged clusters were altered by site-directed mutagenesis, replacing the residues with the neutral amino acid with a small side chain, alanine. The color of each cluster represents the phenotypic outcome after its expression in either wild-type S. pombe (hence a dominant phenotype), or in haploid S. pombe cells in which the endogenous γ-tubulin gene is disrupted (hence the recessive phenotype). Green, recessive lethal; purple, recessive cold sensitive; red, dominant cold sensitive; yellow, recessive lethal and dominant cold sensitive. (B) Stereo pair of the minimal free energy γ-tubulin homology model shown in longitudinal contact with the α-tubulin subunit of the microtubule minus end. The model is oriented to reveal a view from inside the lumen of a microtubule. The left and right sides thus represent putative lateral contact sites of γ-tubulin with other tubulin subunits. The colors of these clusters correspond to the color scheme showed in A.
Growth of Alanine-scanning Mutants in Fission Yeast
It has been previously shown that human γ-tubulin (TUBG1) can functionally replace the S. pombe γ-tubulin gene gtb1+ (Horio and Oakley, 1994). Because we were interested in identifying evolutionarily conserved features of γ-tubulin, we have examined the functional consequences of human TUBG1 alanine-scanning mutants in fission yeast. All experiments were performed with the use of either haploid cells carrying a disrupted γ-tubulin allele or wild-type haploid cells each transformed with one of the alanine-scanning mutant human γ-tubulin alleles. In both cases, the wild-type TUBG1 transformants served as controls.
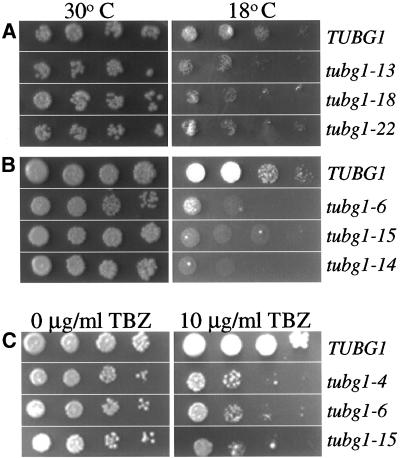
The amino acid changes and growth phenotypes of the alanine-scanning mutants of TUBG1 are summarized in Table 1. All of the conditional mutants identified were cold sensitive (cs−) to varying degrees, six in the presence of endogenous gtb1+ and six in the absence of gtb1+ indicated as recessive cs− (Table 1). There were also 11 mutants that were unable to support growth in the absence of endogenous gtb1+ (referred to as recessive lethal in Table 1). Representative growth phenotypes at the permissive and restrictive temperatures are shown in Figure 2. Because the alanine-scanning mutations are predicted to affect microtubule assembly, we hypothesized that some of these mutations could be sensitive to microtubule-depolymerizing drugs, such as thiabenzadole (TBZ). To test this hypothesis, serial dilutions of all cs− mutants were plated on media containing 10 μg/ml TBZ. All six recessive cold-sensitive mutants showed increased sensitivity to TBZ. A few representative examples of these data of the cold-sensitive growth phenotypes and TBZ sensitivity are shown in Figure 2.
Table 1.
Growth phenotypes of alanine-scanning mutants of TUBG1 in S. pombe
| Mutant | Amino Acid Change | with gtb1+ | without gtb1+ |
|---|---|---|---|
| tubg1-1 | R3A, E4A | wt | wt |
| tubg1-2 | E20A, K23A | wt | Recessive lethal |
| tubg1-3 | E38A, E39A, E43A | wt | wt |
| tubg1-4 | D46A, R47A, K48A, D49A | wt | cs− |
| tubg1-5 | D56A, D57A, E58A | cs− | wt |
| tubg1-6 | D68A, E70A, P71A, R72A | wt | cs− |
| tubg1-7 | K84A, L85A, Y86A, N87A, P88A, E89A | wt | cs− |
| tubg1-8 | D120A, D123A, R124A, E125A, D127A | wt | Recessive lethal |
| tubg1-9 | S129A, D130A, S131A, E133A | wt | Recessive lethal |
| tubg1-10 | E155A, R156A, L157A, D159A, R160A | wt | cs− |
| tubg1-11 | K163A, K164A, L165V | wt | Recessive lethal |
| tubg1-12 | D159A, R160A, K163A, K164A | wt | Recessive lethal |
| tubg1-13 | D176A, E177A, S179A, D180A | cs− | Recessive lethal |
| tubg1-14 | K193A, R194A, L195A | wt | cs− |
| tubg1-15 | R286A, K287A, T288A | wt | cs− |
| tubg1-16 | D292A, R295A, R296A, L297A | wt | wt |
| tubg1-17 | R309A, D310A, R311A | wt | wt |
| tubg1-18 | R339A, R341A, E342A, R343A, K344A | cs− | Recessive lethal |
| tubg1-19 | R362A, K363A, S364A | wt | wt |
| tubg1-20 | E389A, R390A, T391A, R393A | wt | wt |
| tubg1-21 | D396A, K397A, R399A, K400A, R401A, E402A | wt | wt |
| tubg1-22 | E406A, R409A, K410A, E411A, D412A | cs− | wt |
| tubg1-23 | K415A, D416A, N417A | wt | Recessive lethal |
| tubg1-24 | D419A, E420A, D422A | wt | Recessive lethal |
| tubg1-25 | R425A, E426A, I427A | wt | wt |
| tubg1-26 | D433A, E434A, Y435A | cs− | Recessive lethal |
| tubg1-27 | R440, P441A, D442A | cs− | Recessive lethal |
wt, wild type.
Figure 2.
tubg1 alanine-scanning mutants that show conditional growth. A 10-fold dilution series containing 105–102 cells of (A) dominant cold-sensitive (cs−) or TUBG1, (B) recessive cs− mutants or TUBG1 in the background of a γ-tubulin null mutant (C) recessive cs− mutants that were sensitive to the microtubule-depolymerizing drug TBZ or TUBG1 in the background of a γ-tubulin null mutant were plated on appropriate selective media with or without 10 μg/ml TBZ. The plates were then incubated at a permissive temperature of 30°C or a restrictive temperature of 18°C for 7 d. Panels shown in C were all incubated at the permissive temperature of 30°C.
Mutant γ-Tubulin Localization Is Comparable to Wild-Type γ-Tubulin Localization
By creating a mutation on the surface of γ-tubulin that may disrupt protein-protein interactions, one might also disrupt the interactions that are responsible for the intracellular localization of γ-tubulin. To determine whether any of the cs− alanine-scanning mutants differ in the γ-tubulin localization from that of the wild-type protein, we performed immunolocalization of γ-tubulin in strains transformed with either the mutant alleles or the wild-type γ-tubulin. We did not detect any differences in the intracellular localization of γ-tubulin in the cs− mutants compared with the wild-type cells (Figure 3). It is therefore likely that mutations that do disrupt γ-tubulin localization confer lethality.
Figure 3.
Normal γ-tubulin localization is not perturbed in the conditional tubg1 mutants. Exponentially growing cells at 30°C were either shifted at 18°C for 8 h, fixed in methanol, and then processed for immunolocalization with the use of the mouse monoclonal GTU-88 followed by Cy3-conjugated goat anti-mouse antibodies. DNA was stained with 10 μg/ml Hoechst for 1 h. Arrows indicate dividing cells.
Mutant γ-Tubulins Display Aberrant Cell Cycle Profiles
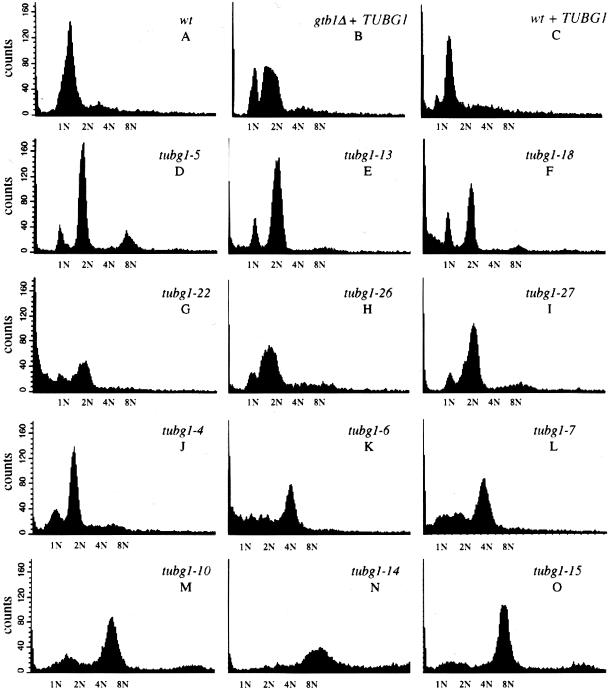
To gain a general insight into the progression of the cell cycle in the cs− alleles, we performed a flow cytometric analysis (FACS) of DNA content (Figure 4). Wild-type cells (Figure 4A), gtb1Δ cells expressing TUBG1 (Figure 4B), and wild-type cells expressing TUBG1, and the dominant (Figure 4, D–I) and the recessive (Figure 4, J–O) cs− cells were grown to mid-log phase at 30°C and then shifted to 18°C for 10 h. Cells were prepared for FACS analysis by staining with the fluorescent DNA dye SYTOX Green. For each strain, 10,000 events were analyzed. The profiles reveal a striking difference between wild-type controls and tubg1 mutants. As shown in Figure 4A, wild-type cells seem to accumulate in the G1 phase of the cell cycle with an unduplicated, 1N DNA content. gtb1Δ cells expressing wild-type TUBG1 seem to retard the cell cycle with 2N DNA content (Figure 4B). Wild-type cells expressing wild-type TUBG1 also display this delay as judged by an increased number of cells with 2N DNA content (Figure 4C). Although human γ-tubulin can functionally complement gtb1Δ cells (Horio and Oakley, 1994), there are some subtle abnormalities in the cell cycle timing of these cells (Figure 4, B and C). Also, it should be noted that a very small fraction of the cell population in all the wild-type controls show cells with 4N and 8N DNA content. FACS profiles of some mutants display striking deviations from the FACS profiles of their appropriate control counterparts. For example tubg1-5, tubg1-14, and tubg1-15 (8N) and tubg1-7 and tubg1-10 (4N) have a pronounced increased in the population of cells with more than 2N DNA content. Additionally, tubg1-6, tubg1-7, tubg1-18, and tubg1-22 have cells with less than 1N DNA content. The remaining mutants, like their respective wild-type controls, have a predominant population of cells with 2N DNA content with the exception of tubg1-14 and tubg1-15.
Figure 4.
Flow cytometric analysis of DNA content of the cold sensitive tubg1 mutants. Wild-type cells (A), gtb1Δ cells expressing TUBG1 (B), and wild-type cells expressing TUBG1, and the dominant (D–I) and the recessive (J–O) cold-sensitive cells were grown to mid-log phase at 30°C and then shifted to 18°C for 10 h. Cells were fixed with ethanol and stained with the fluorescent DNA dye SYTOX Green. For each strain, 10,000 events were analyzed. The relative fluorescence corresponding to 1N, 2N, 4N, and 8N is indicated on the x-axis. The profiles reveal a striking difference between wild-type controls and tubg1 mutants.
Penetrance and Expressivitiy of Mutant Phenotypes
The strikingly aberrant cell cycle profiles of most tubg1 mutants persuaded us to examine the cytological phenotypes of each of the strains. To do this, both dominant and recessive cs− mutants were grown at the permissive temperature of 30°C and then shifted to the restrictive temperature of 18°C for 8–12 h. Cells were then fixed and stained to visualize chromosomes and microtubules. All of the mutants shared defects in chromosome segregation to various degrees (Table 2; Figure 5). These defects include asymmetrically localized nuclei (asym nuc), lagging DNA during anaphase (DNA seg), cells with more than one nucleus (>1 nucleus), and anucleate cells (0 nuc). Other defects include altered cellular morphologies: pear, banana, and oval shapes. We also observed defects in septation and cell separation. Septation defects include asymmetric placement of septa (asym sept), multiple septa (mult sept), and/or obliquely placed septa (obliq sept). Defects in cell separation are consistent with the FACS profiles with an accumulation of more than 2N DNA content. The percentages for each of the phenotypes observed for each of the 12 mutants is tabulated in Table 2 and the most striking DNA segregation defects are shown in Figure 5 along with cellular microtubules.
Table 2.
Summary of the expressivity of various phenotypes of the tubg1 cold-sensitive mutants
| Mutant | Pear (%) | Banana (%) | Oval (%) | Asym nuc (%) | DNA seg (%) | >1 Nucleus (%) | 0 Nucleus (%) | Asym sept (%) | Mult sept (%) | Obliq sept (%) | Total (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TUBG1 | 0.49 | 1.48 | 2.96 | 0.49 | 5.42 | ||||||
| tubg1-4 | 24.77 | 1.80 | 4.50 | 0.90 | 1.80 | 33.78 | |||||
| tubg1-5 | 1.01 | 3.54 | 3.03 | 20.71 | 2.02 | 15.15 | 3.54 | 11.62 | 3.03 | 63.64 | |
| tubg1-6 | 0.48 | 31.03 | 0.48 | 8.57 | 3.81 | 0.43 | 45.24 | ||||
| tubg1-7 | 2.00 | 0.50 | 6.00 | 6.50 | 3.50 | 4.00 | 1.00 | 1.00 | 24.50 | ||
| tubg1-10 | 0.83 | 7.50 | 10.00 | 4.17 | 3.33 | 4.17 | 30.00 | ||||
| tubg1-13 | 0.49 | 4.37 | 4.00 | 15.05 | 1.00 | 1.94 | 2.43 | 17.48 | 8.74 | 54.58 | |
| tubg1-14 | 2.04 | 4.08 | 7.65 | 1.02 | 27.04 | 0.51 | 4.08 | 2.04 | 48.47 | ||
| tubg1-15 | 1.65 | 4.13 | 4.55 | 0.41 | 10.74 | ||||||
| tubg1-18 | 0.51 | 2.02 | 1.52 | 21.21 | 4.55 | 3.54 | 1.01 | 34.34 | |||
| tubg1-22 | 2.00 | 2.00 | 18.41 | 4.48 | 0.50 | 4.48 | 31.84 | ||||
| tubg1-26 | 9.47 | 12.63 | 6.32 | 10.00 | 2.63 | 41.05 | |||||
| tubg1-27 | 3.57 | 17.34 | 1.53 | 9.69 | 5.10 | 7.14 | 2.55 | 46.94 |
pear, pear shaped; banana, curved shaped; oval, oval shaped; asym nuc, asymmetric localization of the nucleus; DNA seg, abnormal DNA segregation; >1 nuc, reduced nuclear material; 0 nucleus, absence of nucleus; asym sept, misplaced septum; mult sept, multiple septa; and obliq sept, formation of septum that is not perpendicular to the long axis of the cell.
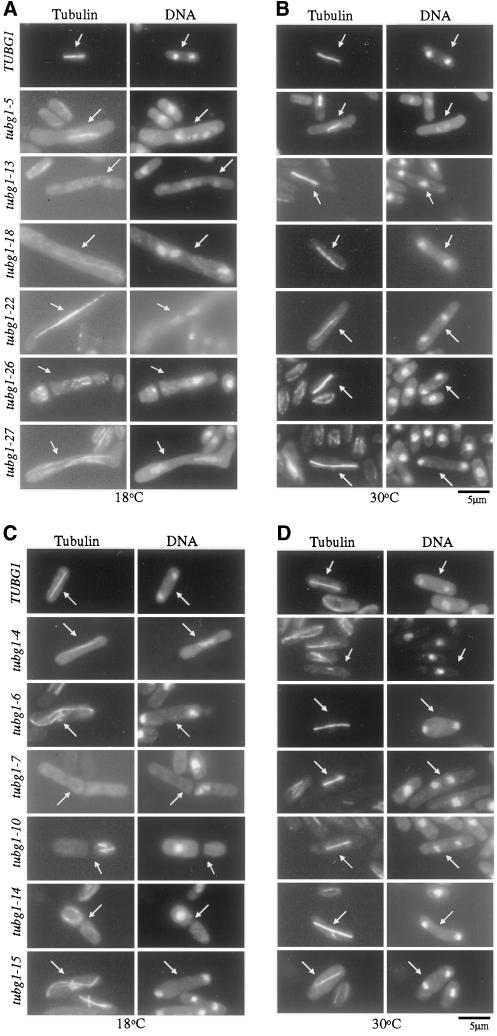
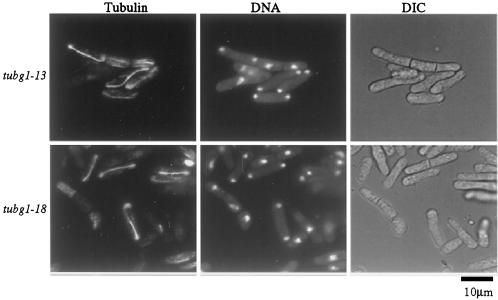
Figure 5.
Cold-sensitive tubg1 mutants undergo abnormal mitosis. Dominant cs− cells (A and B), recessive cs− cells (C and D), and wild-type cells expressing TUBG1 were grown to mid-log phase at 30°C and then shifted to 18°C (A and C) or left at 30°C for 10 h (B and D). Cells were then fixed and microtubules were stained with the anti-α-tubulin, monoclonal antibody TAT-1 (1:50) followed by Cy3-conjugated goat anti-mouse secondary antibody. DNA was stained with 1 μg/ml DAPI. Arrows indicate mitotic cells.
To our surprise, none of our mutants showed a complete loss of microtubules. However, tubg1-7, tubg1-10, tubg1-13, and tubg1-18 showed an overall reduction in total tubulin polymer. The most pronounced defects in microtubules included curved and S-shaped spindles (tubg1-6 and tubg1-15, Figure 5). The number of cells showing microtubule defects varied from experiment to experiment but in general, 5–10% of the cells expressed this defect.
To determine how many of the observed phenotypes represented terminal events, we wanted to enrich for cells that synchronously enter mitosis. To do this, we enriched for synchronous cells with the use of hydroxyurea, to induce an S phase block, and then followed cells 4–6 h after their release from arrest. This strategy yielded cells that entered mitosis in a partially synchronous manner as shown in Figure 6. These studies further confirmed that the defects observed appear as early as 4–6 h after the temperature shift and allowed further detailed observations of the phenotypes that were not apparent from studying nonsynchronous cell populations. Multiple septa were visible in cells that had failed to undergo fission but the presence of DNA spots suggested that the cells progressed through another round of DNA replication and segregation (Figure 6). Additionally, the daughter cells that remained attached to each other often lost synchrony as revealed by the presence of the elongated anaphase spindle in one cell and the interphase microtubules in the other (Figure 6, tubg1-13).
Figure 6.
Early phenotypes in partially synchronized cells (20 mM HU, 3 h) reveal disrupted coordination of mitosis and cytokinesis. After partial synchronization, HU was removed and cells were shifted to 18°C for 4–6 h. Cells were then fixed and stained for microtubules and DNA as previously described.
Metaphase-Anaphase Transition Defects in tubg1 Mutants
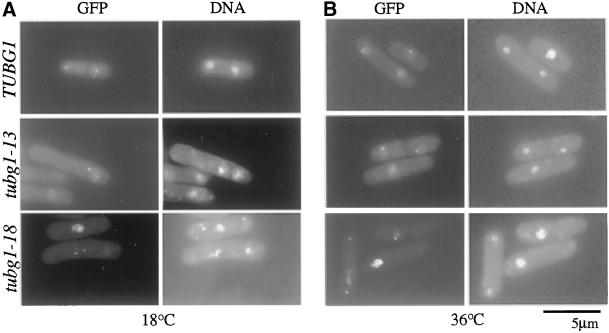
A striking observation in chromosome segregation was dispersed chromosomes that were left behind at the metaphase plate despite elongated anaphase B spindles (Figure 5A, tubg1-5 and tubg1-22). Furthermore as shown in Table 2, many cells showed the formation of multiple septa, indicative of exit from mitosis. These observations combined with perturbed FACS profiles compelled us to investigate further the nature of the chromosome segregation defects. The appearance of elongated spindles and improper chromosome segregation might arise due to simple mechanical defects in the microtubule attachment at the spindle pole and thus lead to an accidental elongation of the spindle. Alternatively, it might represent a premature anaphase spindle elongation before the proper biorientation of chromosomes. To distinguish between these possibilities, we monitored sister chromatid separation by transforming PY321 with the alanine-scanning mutants of tubg1. PY321 is an S. pombe strain that contains a GFP-LacI marker that binds to a region proximal to the centromere of chromosome I, which contains a tandem, repeat of LacI-binding LacO sequences (Nabeshima et al., 1998). By monitoring the movement of the GFP signal, one can monitor sister chromatid segregation. If the first possibility is correct, the missegregated chromosomes despite elongated spindles might simply represent unseparated metaphase chromatid pairs. If, however, a premature anaphase is triggered, the centromeres must separate even though the chromosomes fail to segregate to the poles. For tubg1-13 and tubg1-18, we found that the sister chromatids clearly separated while the chromatids failed to segregate to the poles (Figure 7). These data combined with the occurrence of abnormal septation suggests a bypass of the spindle assembly checkpoint and a possible role for γ-tubulin in the coordination of sister chromatid segregation and cytokinesis.
Figure 7.
Sister chromatid separation in tubg1 mutants. GFP-marked centromere containing cells (PY321) were transformed with tubg1 alleles and the cultures were grown at 30°C and then either left at 30°C (B) or shifted to 18°C for 10 h (A). DNA was stained with DAPI.
DISCUSSION
We have taken advantage of the genetically amenable model system, S. pombe, in an attempt to answer the question “Does γ-tubulin have other functions besides microtubule nucleation?” This analysis yielded 12 cold-sensitive mutants of TUBG1, thus providing powerful means to analyze γ-tubulin function during the fission yeast cell cycle. Six of these mutants were cold sensitive in the presence of the endogenous S. pombe gtb1+, and thus were dominant. The other six mutants conferred cold sensitivity in the absence of wild-type gtb1+, and thus were recessive. Of the dominant cs− mutants, tubg1-13, tubg1-18, tubg1-26, tubg1-27 were unable to support growth of S. pombe cells in the absence of endogenous γ-tubulin, and thus conferred recessive lethality. Of the 27 alanine-scanning mutants of γ-tubulin, eight were able and 19 were unable to sustain growth of S. pombe cells.
To understand the nature of γ-tubulin interactions that are perturbed in these deleterious mutations, we mapped each mutation on a minimal free energy homology model of human γ-tubulin constructed by with the use of the known core structures of α-tubulin and β-tubulin and then minimizing the free energy. Of the mutants that were unable to support the growth of S. pombe cells, tubg1-9, tubg1-11, tubg1-13, tubg1-22, are located at the putative longitudinal contact sites of γ-tubulin with other tubulin subunits. Two mutations, tubg1-8 and tubg1-18, that conferred recessive lethality are located at the putative lateral contact sites of γ-tubulin with other tubulins of the microtubule lattice. The recessive cs− mutant tubg1-10 is also located at the putative lateral tubulin contact site. Collectively, the results from these analyses provide strong support to the hypothesis that at least some of the γ-tubulin interactions with the microtubule lattice are equivalent to those of α-tubulin and β-tubulin.
There are also other deleterious mutations such as tubg1-2, tubg1-4, tubg1-5, tubg1-6, tubg1-7, tubg1-14, tubg1-15, tubg1-24, tubg1-26, and tubg1-27 that map to sites that do not lie at either the lateral or the longitudinal contact sites of γ-tubulin with other tubulin subunits. These mutant proteins can be produced as soluble and stable proteins in an in vitro rabbit reticulocyte transcription/translation system. Thus, mutations at these sites might not denature or destabilize the protein. It is possible that these mutations either affect γ-tubulin interaction with other tubulin subunits by subtle long-range perturbations in its structure, or these sites on γ-tubulin might be in contact with other nontubulin proteins. Hence, these mutations offer a possible means to further explore the interactions of γ-tubulin with other heterologous subunits of the γ-tubulin complexes or other spindle pole body components.
Aside from the deleterious mutations, there are eight mutations that do not appear to affect γ-tubulin function. Surprisingly, some of these mutations such as tubg1-1, tubg1-3, tubg1-16, tubg1-19, and tubg1-21 lie in regions that are highly evolutionarily conserved in γ-tubulin between fission yeast and humans. How can cells tolerate these changes in the regions of γ-tubulin that were not allowed to change in 1 billion years due to the naturally ticking evolutionary clock? It is possible that these conserved regions might provide some selective advantage to the growth of S. pombe cells in the wild, but not in the amenities of a laboratory Petri dish. It is also possible that the conservation of these sites is due to the coevolution of a γ-tubulin binding protein that imposed constraints on the mutation of these conserved sites.
Because γ-tubulin is a component of the microtubule-organizing center that is required for the proper orientation and organization of microtubules, we expected that these alanine-scanning γ-tubulin mutants would have microtubule organization defects (Stearns et al., 1991; Zheng et al., 1991; Joshi et al., 1992). Similarly, because it had been determined that microtubule organization affects cell morphology and position of the interphase nucleus, we also expected those defects in the γ-tubulin mutants (Walker, 1982; Verde et al., 1995; Hagan and Yanagida, 1997; Mata and Nurse, 1997; Sawin and Nurse, 1998). Finally, we expected to find mutations that affected the spindle would also block the subsequent M phase events such as sister chromatid separation and cytokinesis because a damaged or abnormal spindle activates spindle assembly checkpoint mechanisms responsible for delaying these subsequent postmetaphase events. Surprisingly, in some mutants, despite the presence of abnormal spindles, anaphase and cytokinesis progressed, leading to the generation of aneuploid or aploid cells. These data suggest that in addition to its previously identified role in the nucleation of spindle microtubules, γ-tubulin may also play a crucial role in the coordination of sister chromatid separation in anaphase followed by cytokinesis. Real-time analysis will be necessary to determine the precise extent of the loss of the coordination between different mitotic stages. The defects in the coordination of mitosis are reflected in the gross perturbations of the cell cycle profiles in the γ-tubulin mutants compared with their wild-type control counterparts (Figure 4).
Our hypothesis that γ-tubulin is involved in chromosome separation, septation, and cytokinesis is consistent with some curious unexplained observations in the literature. For example, Oakley et al. (1990) disrupted γ-tubulin in A. nidulans to generate a recessive lethal mutant that blocked nuclear division and the spindle formation. However, there was an increase in DNA staining while the number of cells in mitosis did not increase. This indicated that although the mutation resulted in a lack of a mitotic spindle, the cells did not arrest in mitosis. This observation suggested that depletion of γ-tubulin not only inhibited spindle microtubule assembly but also allowed exit from mitosis in the absence of a mitotic spindle. This scenario is different from the mitotic arrest that results from microtubule disassembly induced either by treatment with microtubule-depolymerizing drugs (Hoyt et al., 1991; Li and Murray, 1991) or by mutations in β-tubulin (Hiraoka et al., 1984). In addition, Paluh et al. (2000) have reported that in the presence of a mutant γ-tubulin allele in S. pombe, proper chromosome segregation is dependent on the presence of a checkpoint gene mad2+. Furthermore, a mutant allele of the S. cerevisiae homolog of γ-tubulin, TUB4, requires a mitotic checkpoint control gene, BUB2, for cell cycle arrest and survival (Spang et al., 1996). In addition, a C-terminal deletion in Tub4p causes mitotic defects, apparently unrelated to microtubule nucleation (Vogel and Snyder, 2000). Together, these data suggest a possible role for γ-tubulin in regulating not only spindle microtubule assembly but also the coordination of anaphase and exit from mitosis.
The cell cycle is closely monitored to verify that DNA is properly replicated before the cell enters mitosis as well as to ensure that once the cell has entered mitosis, sister chromatids are attached to microtubules from both poles before anaphase and subsequent cytokinesis are allowed to progress. These surveillance mechanisms, which monitor the timely and proper progression of the cell cycle are called checkpoints (Hartwell and Weinert, 1989). Two prominent checkpoints that monitor mitosis are the spindle assembly checkpoint (reviewed in Amon, 1999) and the exit from mitosis (reviewed in Cerutti and Simanis, 1999; Balasubramanian et al., 2000; Hoyt, 2000). This system of mitotic checkpoints has been well elucidated in the budding yeast and homologs have been identified in other systems, including fission yeast, Xenopus, and mammalian systems (reviewed in Amon, 1999). The spindle checkpoint monitors attachment of the sister chromatids to properly assembled spindle microtubules, thereby preventing premature anaphase from occurring. One of the major players involved in this system of checkpoints is a spindle pole protein, Cdc16p (the fission yeast homolog of Bub2p). Cdc16p is a negative regulator of the septation initiation pathway involved in ensuring that sister chromatids are properly segregated to opposite poles before the cells undergo septation, signaling exit from mitosis (Fankhauser et al., 1993). Cdc16p is also required to prevent septation from occurring during interphase (Cerutti and Simanis, 1999). Mutations in γ-tubulin, particularly tubg1-13, are reminiscent of mutations in the genes, such as cdc16+, involved in the septation initiation pathway (reviewed in Gould and Simanis, 1997). The tubg1-13 phenotype of septum formation without cytokinesis is also similar to that of ppb1 (Yoshida et al., 1994) and sep1 (Sipiczki et al., 1993), both of which are also defective in cytokinesis. Thus, these data suggest that γ-tubulin may be involved in the spindle assembly checkpoint that prevents sister chromatid separation and cytokinesis in response to spindle damage.
γ-Tubulin is required for the nucleation of microtubule assembly in vivo in mammalian cells (Joshi et al., 1992) and for spindle assembly and function in S. pombe, S. cerevisiae, A. nidulans, and Drosophila (Oakley et al., 1990; Horio et al., 1991; Sobel and Snyder, 1995; Sunkel et al., 1995; Spang et al., 1996; Marschall et al., 1996). Therefore, we predicted that the cs− γ-tubulin mutant alleles might display defects in microtubule assembly and organization at the restrictive temperature. Like other mutations that affect microtubule assembly, tubg1 mutant alleles might block the progression of anaphase and cytokinesis that would normally arrest cells in M phase. To our surprise, none of our mutants showed a complete loss of microtubules although some mutants had fewer microtubules. The most pronounced defects in microtubules were curved spindles (Figure 6). One surprising result that emerged from our observations was that four mutants, tubg1-7, tubg1-13, tubg1-18, and tubg1-26, did not seem to arrest in mitosis but seemed to segregate chromosomes and accomplish cytokinesis, albeit abnormally, producing aneuploid or aploid cells with misplaced septa. Although three of these, tubg1-13, tubg1-18, and tubg1-26, arrest after the completion of mitosis, presumably in the G1 phase of the next cell cycle, tubg1-7 seemed to accumulate 4N DNA content (Figure 4). Collectively, our data suggest that γ-tubulin might be involved not only in microtubule nucleation and organization but also, surprisingly, in the coordination of postmetaphase events such as chromosome segregation and septation during cytokinesis.
ACKNOWLEDGMENTS
We thank Drs. Ken Downing, Eva Nogales, James Snyder, Dennis Liotta, James Nettles, and Minmin Wang for help with homology modeling; Dr. Berl Oakley for communicating unpublished results; Drs. Janet Paluh, Yoshinori Watanabe, Paul Nurse, Zachus Cande, Berl Oakley, and Keith Gull for yeast strains and reagents; and Drs. Sara Leung and Satoru Uzawa, Janet Paluh, John Shanks, Kate Crawford, and Bob Karrafa for technical help and advice at the early stages of this project. We thank Dr. Richard J. McIntosh and two anonymous reviewers for valuable comments. We also thank Drs. Winfield Sale and Victor Faundez for carefully reading the manuscript. This work was supported by grants to H.C.J. from the National Institutes of Health, the American Cancer Society, and the Emory University Research Committee, and T.W.H. was supported by a Predoctoral fellowship from the National Institutes of Health (GM-18037).
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Amos LA, Klug A. Arrangement of subunits in flagellar microtubules. J Cell Sci. 1974;14:523–549. doi: 10.1242/jcs.14.3.523. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- Burke DJ. Complexity in the spindle checkpoint. Curr Opin Genet Dev. 2000;10:26–31. doi: 10.1016/s0959-437x(99)00040-4. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Simanis V. Asymmetry of the spindle pole bodies and Spg1p GAP segregation during mitosis in fission yeast. J Cell Sci. 1999;112:2313–2321. doi: 10.1242/jcs.112.14.2313. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Polarity of some motility-related microtubules. Proc Natl Acad Sci USA. 1981;78:372–376. doi: 10.1073/pnas.78.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Griffiths AD, Lockhart A, Cross RA, Amos LA. Microtubule minus ends can be labeled with a phage display antibody specific to α-tubulin. J Mol Biol. 1996;259:325–330. doi: 10.1006/jmbi.1996.0322. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16+ gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1997;110:1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The nda3+ gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Horio T, Oakley BR. Human gamma-tubulin functions in fission yeast. J Cell Biol. 1994;126:1465–1473. doi: 10.1083/jcb.126.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Huang X, Miller M. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–367. [Google Scholar]
- Inclán YF, Nogales E. Structural models for the self-assembly and microtubule interactions of γ-, δ- and ε-tubulin. J Cell Sci. 2001;114:413–422. doi: 10.1242/jcs.114.2.413. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Leguy R, Melki R, Pantaloni D, Carlier MF. Monomeric gamma-tubulin nucleates microtubules. J Biol Chem. 2000;275:21975–21980. doi: 10.1074/jbc.M000688200. [DOI] [PubMed] [Google Scholar]
- Li Q, Joshi HC. gamma-tubulin is a minus end-specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Nurse P. tea1+ and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1+ of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison JM. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1979;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: dis1+ is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Oakley BR. Gamma-tubulin. Curr Top Dev Biol. 2000;49:27–54. doi: 10.1016/s0070-2153(99)49003-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Clayton DA. Mutational analysis of the gene for Schizosaccharomyces pombe RNase MRP RNA, mrp1+, using plasmid shuffle by counterselection on canavanine. Yeast. 1996;12:1393–1405. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1393::AID-YEA29%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol Biol Cell. 2000;11:1225–1239. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modeling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- Peitsch MC, Wells TN, Stampf DR, Sussman JL. The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem Sci. 1995;20:82–84. doi: 10.1016/s0968-0004(00)88963-x. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Sipiczki M, Grallert B, Miklos I. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J Cell Sci. 1993;104:485–493. doi: 10.1242/jcs.104.2.485. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. Gamma-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Gomes R, Sampaio P, Perdigao J, Gonzalez C. Gamma-tubulin is required for the structure and function of the microtubule organizing center in Drosophila neuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Snyder M. The carboxy terminus of Tub4p is required for γ-tubulin function in budding yeast. J Cell Sci. 2000;113:3871–3882. doi: 10.1242/jcs.113.21.3871. [DOI] [PubMed] [Google Scholar]
- Walker GM. Cell cycle specificity of certain antimicrotubular drugs in Schizosaccharomyces pombe. J Gen Microbiol. 1982;128:61–71. doi: 10.1099/00221287-128-1-61. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Toda T, Yanagida M. A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J Cell Sci. 1994;107:1725–1735. doi: 10.1242/jcs.107.7.1725. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]