Abstract
Many mitochondrial outer membrane (MOM) proteins have a transmembrane domain near the C terminus and an N-terminal cytosolic moiety. It is not clear how these tail-anchored (TA) proteins posttranslationally select their target, but C-terminal charged residues play an important role. To investigate how discrimination between MOM and endoplasmic reticulum (ER) occurs, we used mammalian cytochrome b5, a TA protein existing in two, MOM or ER localized, versions. Substitution of the seven C-terminal residues of the ER isoform or of green fluorescent protein reporter constructs with one or two arginines resulted in MOM-targeted proteins, whereas a single C-terminal threonine caused promiscuous localization. To investigate whether targeting to MOM occurs from the cytosol or after transit through the ER, we tagged a MOM-directed construct with a C-terminal N-glycosylation sequence. Although in vitro this construct was efficiently glycosylated by microsomes, the protein expressed in vivo localized almost exclusively to MOM, and was nearly completely unglycosylated. The small fraction of glycosylated protein was in the ER and was not a precursor to the unglycosylated form. Thus, targeting occurs directly from the cytosol. Moreover, ER and MOM compete for the same polypeptide, explaining the dual localization of some TA proteins.
INTRODUCTION
Most mitochondrial proteins are synthesized in the cytosol and reach their final destination after release from the ribosome. This posttranslational targeting process has been studied in greatest detail for proteins directed to the matrix or to the inner membrane of mitochondria (Schatz and Dobberstein, 1996; Haucke and Schatz, 1997; Neupert, 1997). Matrix-directed precursors carry an N-terminal basic extension predicted to form an amphiphilic helix or β-sheet, whereas inner membrane proteins often lack a presequence. Protein complexes on the outer and inner mitochondrial membrane (translocases of the outer membrane [TOMs] and of the inner membrane, respectively), involved in the recognition and translocation of mitochondrial precursors, have been extensively characterized and the requirements for import have been defined.
Less information is available on the biogenesis of the mitochondrial outer membrane (MOM). The most studied MOM protein is porin, a deeply embedded membrane protein, whose insertion requires components of the TOM complex (Schleiff et al., 1999, Krimmer et al., 2001). The mechanisms of targeting and insertion of many other MOM proteins have not been as clearly defined yet.
Many proteins that reside on the MOM are anchored to the bilayer by a short hydrophobic domain close to the C terminus, with the N-terminal domain exposed to the cytosol. Proteins of this class, which have been called tail-anchored (TA) (Kutay et al., 1993), lack an N-terminal signal sequence and reach their destination within the cell by posttranslational mechanisms (Borgese et al., 1993; Kutay et al., 1993). TA proteins identified on the MOM include the proto-oncogene bcl-2 (Akao et al., 1994), the mitochondrial isoform of cytochrome b(5) [outer membrane b(5) or OM b(5); Lederer et al., 1983; D'Arrigo et al., 1993], TOM components TOM 5 and 6 (Neupert, 1997), a synaptojanin binding protein (Nemoto and De Camilli, 1999), an alternatively spliced isoform of vesicle associated membrane protein, VAMP-1B (Isenmann et al., 1998). An important problem is the mechanism through which these proteins discriminate between the MOM and the endoplasmic reticulum (ER) membrane, because the latter is the target for many polypeptides with similar topology.
In our laboratory, we have been using cytochrome (cyt) b(5) as a model to investigate how TA proteins choose their target membrane. Mammalian cyt b(5) exists in two homologous isoforms (Lederer et al., 1983), which are localized specifically to the MOM [OM b(5)] or to the ER [ER b(5)], with hardly any overlap between the two distributions (D'Arrigo et al., 1993). Work in our laboratory showed that the tail region of OM b(5) contains the information for localization to the MOM (De Silvestris et al., 1995). More recently, Kuroda et al. (1998) reported that, within the tail region, charged residues downstream to the transmembrane domain (TMD) play a crucial role, with basic residues favoring the targeting to MOM. A similar conclusion was reached for VAMP-1B (Isenmann et al., 1998).
In the present study, we have further investigated the mechanism by which TA proteins select their target membrane, first, by defining the minimal C-terminal sequence required to relocate ER b(5) to the MOM in vivo, and second, by analyzing the behavior of a construct containing an N-glycosylation consensus sequence appended to the C terminus of a MOM-targeted version of b(5). The results show that this construct, although fully competent to translocate its C terminus across ER membranes in vitro, prefers to insert into the MOM in vivo without first transiting through the ER. Thus, TA proteins appear to select their target membrane by independent, but competing pathways.
MATERIALS AND METHODS
Plasmid Constructions
DNA manipulations were carried out by standard techniques (Ausubel et al., 1987; Sambrook et al., 1989). The absence of errors in fragments generated by polymerase chain reaction (PCR) or in synthetic oligonucleotide cassettes was checked by sequencing.
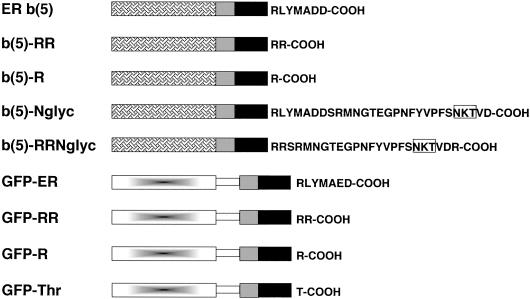
The constructs used in this study are schematized in Figure 1. The cyt b(5) forms were all derived from the plasmid pGb(5)AX, described in Pedrazzini et al. (2000). This plasmid contains the coding sequence of rabbit ER b(5), subcloned in the KpnI, XbaI sites of pGEM4, and having a unique AgeI site at the border between the regions specifying the TMD and the C-terminal polar residues, and a HindIII site immediately downstream to the XbaI site. To express this cDNA, as well as the other b(5) constructs, in mammalian cells, the b(5) insert was placed under the cytomegalovirus promoter by subcloning it into pCB6 (Brewer and Roth, 1991).
Figure 1.
Schematic representation of the constructs used in this study. Grilled box, heme-binding, cytosolic domain of ER b(5); gray box, polar flanking region of ER b(5) upstream to the TMD; black box, transmembrane domain of ER b(5); gradient-filled box, GFP; open rectangle, artificial linker (myc epitope followed by [Gly4-Ser]3. Residues downstream to the TMD are indicated with the single letter code. The N-glycosylation consensus site used in the b(5)-Nglyc and b(5)-RRNglyc constructs is boxed.
To construct the b(5)-RR mutant (Figure 1), pGb(5)AX was digested with Age1, and pCB6 with a modified polylinker (De Silvestris et al., 1995) was digested with ClaI. After filling the resulting sticky ends in the two plasmids, the b(5) sequence was excised from the linearized-blunted pGb(5)AX by digestion with KpnI in the polylinker upstream to the start codon. The b(5) fragment, which lacks 18 bp coding for the six C-terminal amino acids downstream to the Age1 site, was subcloned into the ClaI-digested and filled pCB6 cut with KpnI.
The b(5)-R mutant (Figure 1) was constructed by substituting the Age1/HindIII fragment of pGb(5)AX with a synthetic sequence obtained by pairing two complementary oligonucleotides (Table 1, oligonucleotides 1 and 2).
Table 1.
Oligonucleotides used in this study
| Oligo No. | Used to constructa | Strand | Sequence |
|---|---|---|---|
| 1. | Upper | 5′-CCGGTGATAATAGGATCCTCTAGA-3′ | |
| b(5)-R | |||
| 2. | Lower | 5′-AGCTTCTAGAGGATCCTATTATCA-3′ | |
| 3. | Upper | 5′-CCGGCGGAGCAGAATGAACGGAGGAACAGAAGGACCAAACTTCTACGTACCATTCAG- CAACAAAACAGTAGACAGCAGATGAA-3′ | |
| b(5)-RRNglyc | |||
| 4. | Lower | 5′-AGCTTTCATCTGCTGTCTACTGTTTTGTTGCTGAATGGTACGTAGAAGTTTGGTCCTTCT- GTTCCTCCGTTCATTCTGCTCCG-3′ | |
| 5. | Upper | 5′-TCGAAGGTGGAGGAGGTTCAGGAGGAGGTGGATCTGGAGGTGGAGGTGCGGATCCT-3′ | |
| Synthetic linker | |||
| 6. | Lower | 5′-TCGAAGGATCCGCCACCTCCAGATCCACCTCCTCCTGAACCTCCTCCACCT-3′ | |
| 7. | Upper | 5′-CTAGACTGCAGTCATTAACG-3′ | |
| GFP-RR | |||
| 8. | Lower | 5′-CCGGCGTTAATGACTGCAGT-3′ | |
| 9. | Upper | 5′-CTAGACTGCAGCTATCATTA-3′ | |
| GFP-R | |||
| 10. | Lower | 5′-CCGGTAATGATAGCTGCAGT-3′ | |
| 11. | Upper | 5′-CTAGACTGCAGTCATTACGTGTACATCAGCGCTACAACCAGAGCAGAGATCGCAGGG- ATAACCCAGTTAGTCCACCAA-3′ | |
| GFP-Thr | |||
| 12. | Lower | 5′-TCGAGTTGGTGGACTAACTGGGTTATCCCTGCGATCTCTGCTCTGGTTGTAGCGCTGAT- GTACACGTAATGACTGCA-3′ |
See Figure 1 for a schematic representation of the constructs.
The N-glyc tagged version of ER b(5) [Figure 1, b(5)-Nglyc] has been described in our previous work [called Nglyc-b(5) in Pedrazzini et al., 2000]. The b(5)-RRNglyc construct was obtained by substituting the Age1/HindIII fragment in pGb(5)AX with a synthetic sequence obtained by pairing two complementary oligonucleotides (Table 1, oligonucleotides 3 and 4).
The green fluorescent protein (GFP) fusion proteins were based on a cDNA coding for an enhanced version (F64L, S65T, V163A, I167T) of GFP obtained from A. Magee (Mill Hill, United Kingdom). This cDNA was engineered to contain at its 3′ end a myc epitope and subcloned into the mammalian expression vector pCDNA1 from which the BamHI site in the polylinker was deleted. A PCR fragment coding for the entire tail region of the ER isoform of rat cyt b(5) (Pro94-Asp134) was fused to the 3′ end of the GFP-myc construct. The tail region of ER b(5) was moved further from the 3′ extremity of the GFP cDNA by introduction of paired oligonucleotides coding for the sequence [Gly Gly Gly Gly Ser]3 (Table 1, oligonucleotides 5 and 6). This linker sequence contains a unique BamHI site to permit the addition of different tails to the GFP moiety. In addition, silent mutations, obtained by PCR, allowed the introduction of unique restriction sites at the 5′ and 3′ borders of the sequence coding for the TMD (XhoI and AgeI, respectively). The resulting plasmid codes for a fusion protein called GFP-ER (Figure 1).
To create the GFP-RR and GFP-R constructs, synthetic cassettes consisting of annealed oligonucleotides 7 and 8 (GFP-RR) or 9 and 10 (GFP-R) substituted the 3′ Age1/XbaI fragment. The plasmid coding for GFP-Thr (Figure 1) was obtained by substituting the XhoI/XbaI fragment of pCDNA-GFP-ER with paired oligonucleotides 11 and 12 (Table 1).
Antibodies
Affinity-purified polyclonal antibodies against bacterially expressed cyt b(5) are described in De Silvestris et al. (1995). In some experiments a new antiserum was used, raised against a Triton X-100-soluble purified rabbit b(5), cleaved with thrombin from a glutathione S-transferase fusion protein (GST gene fusion system; Amersham Pharmacia Biotech Italia, Cologno Monzese, Italy).
Other antibodies were obtained from the indicated sources: polyclonal antibodies against ribophorin I (Yu et al., 1989), Dr. Gert Kreibich, New York University School of Medicine, New York, NY; monoclonal antibody (mAb) against the NH2-terminal peptide of bovine opsin (Adamus et al., 1991), Dr. Paul Hargrave, University of Florida, Gainesville, FL; anti-calnexin mAb, Dr. Ari Helenius, Swiss Federal Institute of Technology, Zurich, Switzerland; polyclonal antibodies against bovine mitochondrial complex III (described in Borgese et al., 1996), Dr. R. Bisson, University of Padova, Italy. mAbs against mitochondrial 75-kDa glucose regulated protein (GRP-75) and polyclonal antibodies against GFP were from StressGen Biotechnologies (Victoria, BC, Canada) and Clontech (Palo Alto, CA), respectively.
Cell Culture and Transient Transfection
Most of the experiments in this study were performed with CV-1 cells, cultured and transiently transfected by the Ca2PO4 method, as previously described (De Silvestris et al., 1995). The efficiency of transfection was usually monitored by cotransfecting the cells with pEGFP (CLONTECH). For the subcellular fractionation experiment, we used HeLa cells, which were transfected with lipofectamine (Life Technologies, Gaithersburg, MD), with the use of 265 ng of total DNA/cm2 of culture dish surface [73.5 ng/cm2 of b(5)-Nglyc or b(5)-RRNglyc in pCB6 + 88.5 ng/cm2 of pEGFP + 103 ng/cm2 of carrier DNA pCB6], according to the instructions of the manufacturer.
Immunofluorescence
Paraformaldehyde-fixed cells were permeabilized with Triton X-100 and processed for immunofluorescence as previously described (De Silvestris et al., 1995). Mitochondria were stained either with appropriate antibodies or, before fixation, with Mitotracker CMX Rose (Molecular Probes, Eugene, OR).
In some experiments permeabilization was carried out with streptolysin O (SLO), obtained from Dr. S. Bhakdi (Johannes-Gutenberg Universität, Mainz, Germany; Bhakdi et al., 1993). CV-1 cells grown on coverslips were incubated with 40 U/ml SLO at 0°C in K+ containing buffer (ICB) as previously described (Pedrazzini et al., 2000). After removal of excess SLO, cells were incubated at 37°C for 30 min in ICB supplemented with antiopsin mAb and/or anti-b(5) antibodies. After fixation and permeabilization with Triton X-100 under standard conditions, the cells were incubated with secondary anti-mouse-tetramethylrhodamine B isothiocyanate and anti-rabbit-biotinylated antibodies, followed by incubation with streptavidin-fluorescein isothiocyanate (FITC) (Jackson Immunoresearch, West Grove, PA). To counterstain the mitochondrial marker GRP-75, SLO-permeabilized cells were first incubated with anti-b(5) antibodies, then fixed, permeabilized with Triton X-100, and incubated with anti-GRP-75 mAbs followed by fluorescent secondary antibodies.
Cells were observed either under a Zeiss Axioplan microscope equipped for epifluorescence or with a Bio-Rad MRC 1024 ES laser confocal microscope. Negatives obtained at the Zeiss microscope with a MC100 Zeiss camera were digitalized, and all images were processed with Adobe Photoshop software.
Subcellular Fractionation
Cell fractionation was performed on HeLa cells plated on two 15-cm Petri dishes and transfected with b(5)-Nglyc or b(5)-RRNglyc cDNAs. All operations were carried out at 4°C. Sixteen hours after transfection, cells were washed free of medium and detached with a rubber policeman. After collection by centrifugation, they were resuspended in 4.4 ml of hypotonic buffer (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-Cl pH 7.5, and protease inhibitors as previously described [Pedrazzini et al., 2000]), incubated on ice for 5 min, and then broken in a Dounce homogenizer with 20 strokes of pestle B. Then 3.2 ml of 2.5 times concentrated MS buffer (MS: 210 mM mannitol, 70 mM sucrose, 5 mM Tris-Cl pH 7.5, 1 mM EDTA) was added. The homogenate was brought to 10 ml with MS buffer and centrifuged at 2000 × g, 5 min. The postnuclear supernatant (PNS1) was further centrifuged at 2500 × g, 5 min, to obtain a second PNS (PNS2), which was centrifuged at 10,000 × g, 10 min, to obtain a mitochondria-enriched fraction (P10) and a supernatant (S10). S10 was centrifuged at 17,000 × g for 10 min, to obtain the ER-enriched fraction (P17). The P10 and P17 fractions were resuspended in 0.2 ml of MS.
In Vitro Transcription and Translation
b(5)-Nglyc and b(5)-RRNglyc in pGEM4 were transcribed from the SP6 promoter, and the resulting synthetic RNA was translated for 1 h at 32°C in 10 or 20 μl of reticulocyte lysate (Promega, Madison, WI) as previously described (Ceriotti et al., 1991). In some cases, microsomes were added posttranslationally. Translation was blocked with 2.4 mM cycloheximide and the translation products were incubated for a further hour at 32°C in the presence of 1 μl of dog pancreas microsomes (DPMs) (Promega). Translation products were immunoprecipitated with anti-opsin mAbs (Pedrazzini et al., 2000) and analyzed by SDS-PAGE fluorography or isoelectrofocusing (IEF) either as such or after treatment with glycanase (see below).
Metabolic Labeling Experiments
Metabolic labeling was carried out on CV-1 cells, plated on 10-cm dishes and transfected the day before with b(5)-Nglyc or b(5)-RRNglyc cDNAs. Labeling with 0.25 mCi/ml [35S]Met/Cys (Promix; Amersham Pharmacia Biotech) was carried out as previously described (Borgese et al., 1996). In one experiment 5 μg/ml tunicamycin (TM) was added during the 45 min of prelabeling and over the labeling time. b(5) constructs were immunoprecipitated from cleared lysates with anti-opsin mAbs as described in Pedrazzini et al. (2000), in some cases treated with glycanase, and analyzed by SDS-PAGE fluorography or isoelectric focusing (IEF).
Treatment with Glycanase
Peptide N-glycanase F (PNGase F) from New England Biolabs (Beverly, MA) was used according to the instructions of the manufacturer. Before digestion, immunocomplexes, containing metabolically labeled or in vitro synthesized b(5), were dissociated by incubation in 0.5% SDS at 100°C for 4 min.
Electrophoretic Techniques
Standard techniques are described in detail in previous publications (Borgese and Pietrini, 1986; Borgese et al., 1996). Enhanced chemiluminescence of Western blots was carried out with reagents Pico or Femto Supersignal (Pierce, Rockford, IL), according to the instructions of the manufacturer.
IEF was carried out with a Multiphor II apparatus (Amersham Pharmacia Biotech) on 1-mm-thick polyacrylamide gels. Immunoprecipitated, metabolically labeled, or in vitro translated b(5) constructs, digested or not with PNGase F, were supplemented with urea, NP-40, and pH 4–6.5 Ampholines (Amersham Pharmacia Biotech) to final concentrations of 8.5 M, 0.5 and 2%, respectively, in a volume of 35 μl. Horizontal IEF acrylamide gels (3.7% acrylamide, 0.21% bis-acrylamide, 8.5 M urea, 2% NP-40, 2% 4–6.5 pH Ampholines) were prerun for 45 min at 3.5 W with cooling at 12°C. Filter strips soaked in 1 M H3PO4 and 1 M NaOH were placed in contact with the positive and negative poles, respectively. Samples were loaded into wells close to the positive pole and focalization was carried out at 8 W (800–1000 V) for 3 h, followed by 1 h at 10 W (1000–1200 V) and 30 min at 11 W (∼1200 V). After focalization the gels were fixed in 7% acetic acid/20% methanol, dried, and exposed for autoradiography. The pH gradient profile was checked by dividing a strip of the gel (before fixation) into 1-cm squares, allowing these pieces to equilibrate with 7 ml of water, and then reading the pH of the resulting solution.
RESULTS
Positively Charged Residues at C Terminus Favor Targeting of cyt b(5) to Mitochondria
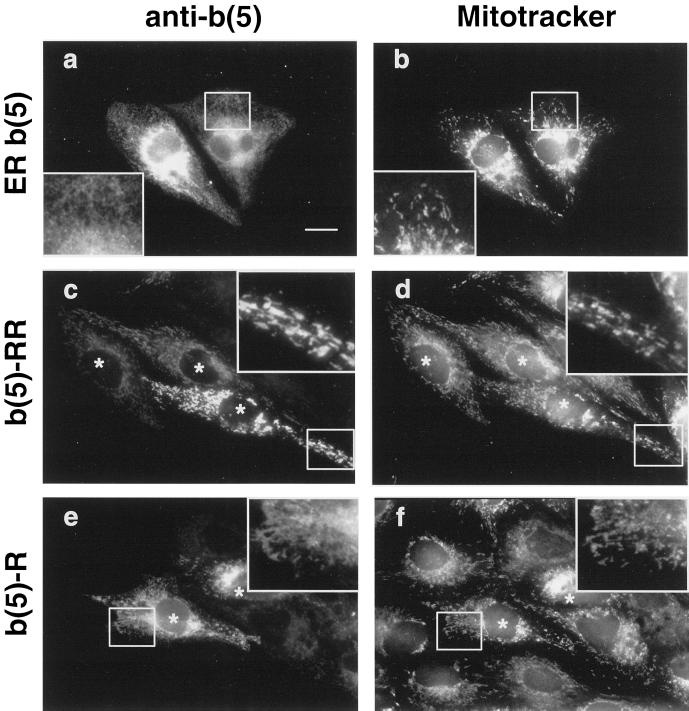
An important difference between OM- and ER b(5) resides is the opposite net charge of the C-terminal polar sequence, with a predominance of basic over acidic residues in the OM isoform, and vice versa, a predominance of negatively over positively charged amino acids in ER b(5) (see Table 2). Positive charges at the C terminus have been previously implicated in the targeting of TA proteins to the MOM (Isenmann et al., 1998; Kuroda et al., 1998). To investigate the minimal C-terminal sequence required for targeting of cyt b(5) to the MOM, we constructed two mutants, b(5)-RR and b(5)-R (Figure 1), in which the C-terminal polar peptide of ER b(5) was replaced with one or two arginines. The subcellular localization of ER b(5), b(5)-RR, and b(5)-R was analyzed by immunofluorescence in transiently transfected CV-1 cells, and compared with the distribution of mitochondria revealed by Mitotracker. As shown in Figure 2, ER b(5) showed the classical ER pattern and colocalization with mitochondria was not detectable (compare Figure 2, a and b). In contrast, the b(5)-RR mutant showed a clear mitochondrial localization (Figure 2, c and d and magnification in the inset). Also b(5)-R showed colocalization with mitochondria (Figure 2, e and f); however, a more diffuse reticular staining was also visible (Figure 2, e and f, insets), suggesting that this construct had a dual, ER plus MOM, localization. Thus, substitution of the C-terminal residues of ER b(5) with a single basic residue is sufficient to partially relocate the protein to mitochondria, whereas with two arginines the relocation appears complete.
Table 2.
Comparison of C-terminal sequences of cytochrome b(5) from different organisms
| Organism | Sequencea | Net chargeb |
|---|---|---|
| Mammals, ER isoform | ||
| Bovine | HLYTSEN | −1 |
| Pig | HFYTSEN | −1 |
| Horse | RIYTAED | −2 |
| Rabbit | RLYMADD | −2 |
| Rat | RLYMAED | −2 |
| Mouse | RLYMAED | −2 |
| Human | RLYMAED | −2 |
| Mammals, MOM isoform | ||
| Rat | RHFWADSKSS | +1 |
| Mouse | RHFWADSKSS | +1 |
| Human | RYYTSESKSS | 0 |
| Other animals | ||
| Chick | RSYYMSE | −1 |
| Fruit fly (Drosophila melanogaster) | KFFFGGAKQ | +1 |
| House fly | KFFFGTKSQ | +1 |
| Caenorhabditis elegans | KCMFN | 0 |
| Polyandrocarpa misakiensis | RYYISN | 0 |
| Ciona savignyi | RFYMSS | 0 |
| Plants | ||
| Common olive | RHYTKEK | +2 |
| Brassica oleracea | RQYTKKE | +1 |
| Southern Asian dodder (Cuscuta reflexa) | RFYKKQSSD | +1 |
| Borago officinalis | RFYTKSSA | +1 |
| Oryza sativa | RIYTKSESA | 0 |
| Common tobacco | RFYTKQSSA | +1 |
| Thale cress (Arabidopsis thaliana) | RKT | +1 |
| Fungi | ||
| Baker's yeast | E | −2 |
| Fission yeast | KLNK | +1 |
C-terminal residues, starting from the first charged residue downstream to the hydrophobic region, are shown.
For simplicity, H residues are considered to be 100% charged, with a consequent overestimation of their contribution to net positivity.
Figure 2.
Replacement of the C-terminal polar sequence of ER b(5) with a single or double arginine relocates the protein to mitochondria. CV-1 cells transfected with ER b(5) (a and b), b(5)-RR (c and d), or b(5)-R (e and f) were stained with Mitotracker CMX rose then fixed, permeabilized, and incubated with polyclonal anti-b(5) antibodies followed by FITC-conjugated secondary antibodies. The panels on the left and right side of the figure represent the same field of cells viewed by conventional epifluorescence under the fluorescein [cyt b(5)] or rhodamine (Mitotracker) filter, respectively. ER b(5) yields a classical ER pattern, and colocalization with mitochondria is not detectable (compare a and b). b(5)-RR and b(5)-R staining are mainly or partially superimposed with Mitotracker (compare c and d, or e and f, respectively). In cells transfected with b(5)-R, a residue of reticular staining is also evident (compare e and f, insets), that is not visible in b(5)-RR-transfected cells (compare c and e, insets). Bar, 20 μm. Insets, 2.3× magnification. Areas magnified in the insets are boxed. Asterisks in c–f are placed over the nuclei of transfected cells.
The Cytosolic Portion of cyt b(5) Is Not Involved in Targeting
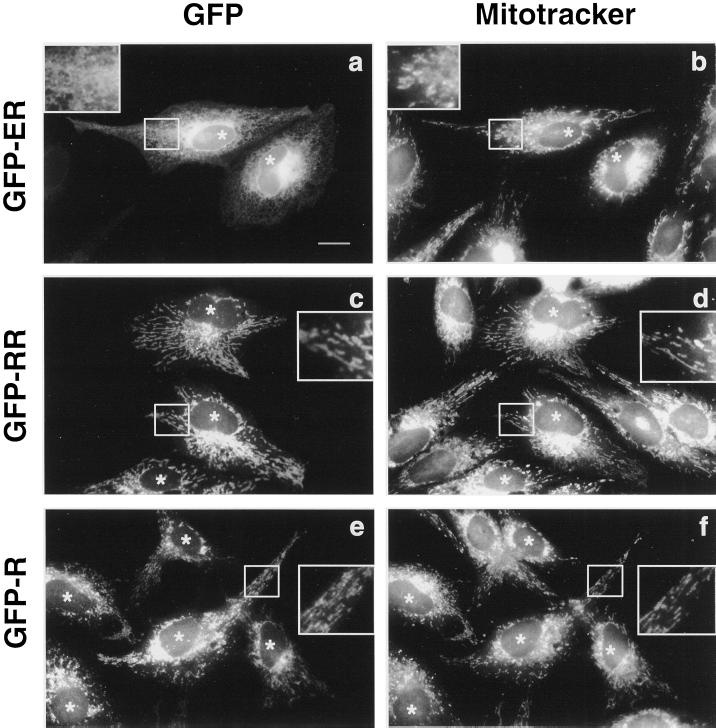
To investigate whether the C-terminal arginine residues are able to act as mitochondrial targeting determinants independently from the cytosolic portion of cyt b(5), we constructed fusion proteins, in which the catalytic domain of cyt b(5) was replaced with GFP (Figure 1). The fusion proteins consisted of GFP connected at its C terminus to a linker sequence followed by the entire tail region of ER b(5), b(5)-RR, or b(5)-R, including the upstream flanking region, the TMD, and the C-terminal polar residues (constructs GFP-ER, GFP-RR, and GFP-R, respectively; Figure 1). Figure 3 shows the intracellular localization of the three fusion proteins, analyzed by epifluorescence, compared with that of mitochondria visualized with Mitotracker. GFP-ER showed a classical reticular pattern; colocalization with mitochondria was not detectable (Figure 3, compare a and b). The localization of GFP-ER was not altered by deletion of the cyt b(5) polar sequence immediately upstream to the TMD (residues 92–105; Bulbarelli et al., manuscript in preparation). The localization of GFP-RR and GFP-R was strikingly different. Both constructs colocalized with mitochondria (Figure 3, c and d, e and f). No ER staining was visible for GFP-RR (Figure 3c), whereas some reticular staining sometimes appeared in the case of GFP-R. Thus, the C-terminal portion of ER b(5) is able to anchor a soluble reporter protein to the ER membrane and positive charges at the C terminus relocate this construct to the mitochondria, indicating that the cytosolic heme-binding domain of cytochrome b(5) is not involved in targeting.
Figure 3.
The heme-binding domain of cyt b(5) is not required for targeting. Mitotracker-loaded CV-1 cells expressing GFP-ER, GFP-RR, or GFP-R were fixed and observed by conventional epifluorescence. GFP fluorescence (a, c, and e) and Mitotracker labeling (b, d, and f) are compared in the same fields. GFP-ER retains the ER localization and colocalization with mitochondria is not detectable (compare a and b). GFP-RR and GFP-R are relocalized to mitochondria (compare c and d, or e and f, respectively). Bar, 20 μm; insets. Areas enlarged in the insets (2.3×) are boxed. Asterisks are placed over the nuclei of transfected cells.
A b(5) Tail without Charged Residues at the C Terminus Determines a Promiscuous Localization, on ER and Mitochondria
Although the results described until this point highlight the role of positively charged residues in determining the localization of a TA protein to mitochondria, they do not address the function of the C-terminal sequence (RLYMAE/DD) of ER b(5) in targeting. Is this sequence required for specific targeting to the ER and for exclusion of ER b(5) from mitochondria?
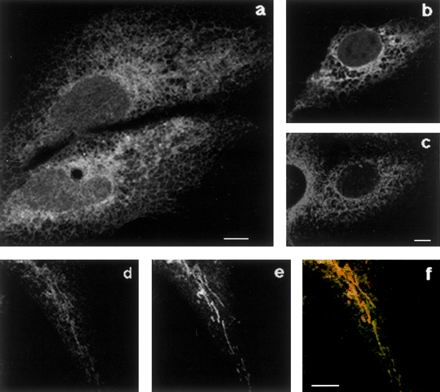
To investigate this question we substituted the C-terminal amino acids in GFP-ER with the polar, but uncharged, residue threonine, creating the GFP-Thr construct (Figure 1). On observation of CV-1 cells expressing this construct we first noticed a clear ER pattern (Figure 4a), and a good colocalization of the GFP fluorescence with the ER marker calnexin (Figure 4, b and c). However, a more accurate analysis revealed also a mitochondrial localization of GFP-Thr, as demonstrated by Mitotracker staining (Figure 4, d–f). It must be noted that, in some cells, GFP-Thr was also present in the cytosol and nucleus. By in vivo time course observation of GFP-Thr expressing cells, we concluded that this protein was unstable in the membrane and relocated to the cytosol with time (our unpublished observations). Optimal analysis of the compartmentalized distribution of this construct was obtained by observing a wave of newly synthesized protein, at short times after removal of a cycloheximide block, as was done for the experiment in Figure 4 (see legend for details). Under identical conditions, the wild-type ER b(5) showed an unmodified ER distribution with undetectable mitochondrial localization (our unpublished observations). We conclude that a charged C-terminal tail stabilizes cyt b(5) in the lipid bilayer and, in addition, that the negatively charged C-terminal sequence of ER b(5) increases the specificity of targeting to the ER.
Figure 4.
GFP-Thr localizes both to the ER and to mitochondria. CV-1 cells transfected with GFP-Thr were allowed to express the cDNA for 18 h, after which cells were further incubated in the presence of 20 μg/ml cycloheximide for 7 h. At this time the cells had nearly undetectable GFP fluorescence, as assessed by observation of the living cells. Cycloheximide was removed, and 45 min later cells were fixed (a–c) or loaded with Mitotracker and then fixed (d–f). The sample shown in b and c was stained with anti-calnexin antibodies followed by incubation with Texas Red-conjugated secondary antibodies. Single confocal sections are shown. (a) GFP fluorescence shows clear reticular staining. (b and c) GFP (b) and Texas Red (c, calnexin) fluorescence were acquired separately from the same field. Comparison of the two images shows a partial colocalization between GFP-Thr and the ER marker. (d–f) Comparison between GFP fluorescence (d) and Mitotracker (e) shows localization of GFP-Thr also to mitochondria (f, merge). Bars, 10 μm.
b(5)-RRNglyc, a b(5) Mutant with a Mitochondrial Targeting Determinant and an N-Glycosylation Consensus Close to C Terminus
Because b(5)-RR and other MOM-associated TA proteins do not have canonical signals recognized by the TOM complex, we considered the possibility that transport of these proteins to the MOM might occur via an alternate, indirect route through the ER. To investigate this possibility, we added to the C terminus of b(5)-RR a sequence corresponding to the 19 N-terminal amino acids of bovine opsin (followed by an additional arg) and containing a consensus site for N-glycosylation, previously used to tag ER b(5) (Pedrazzini et al., 2000); this construct was called b(5)-RRNglyc (Figure 1). We reasoned that if this construct reached the MOM via an indirect pathway, it might acquire an N-linked oligosaccharide while transiting through the ER.
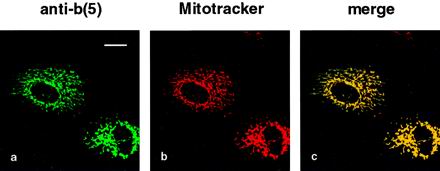
First, we controlled the localization of this new protein in transfected CV-1 cells. Double staining with anti-b(5) antibodies and Mitotracker showed an excellent colocalization (Figure 5, a and b, and merge in c), indicating that the targeting role of the basic residues downstream to the TMD was not impaired by the addition of the opsin sequence.
Figure 5.
b(5)-RRNglyc is targeted to mitochondria. Mitotracker-loaded CV-1 cells expressing b(5)-RRNglyc were fixed, permeabilized with Triton X-100, processed for immunofluorescence with anti-b(5) antibodies followed by FITC-conjugated secondary antibodies, and observed by confocal microscopy. Images of the same field were acquired separately with fluorescein [a, b(5)] and Texas Red (b, Mitotracker) filters. A single confocal section is shown. (c) Merged image of the two acquisitions. Bar, 20 μm.
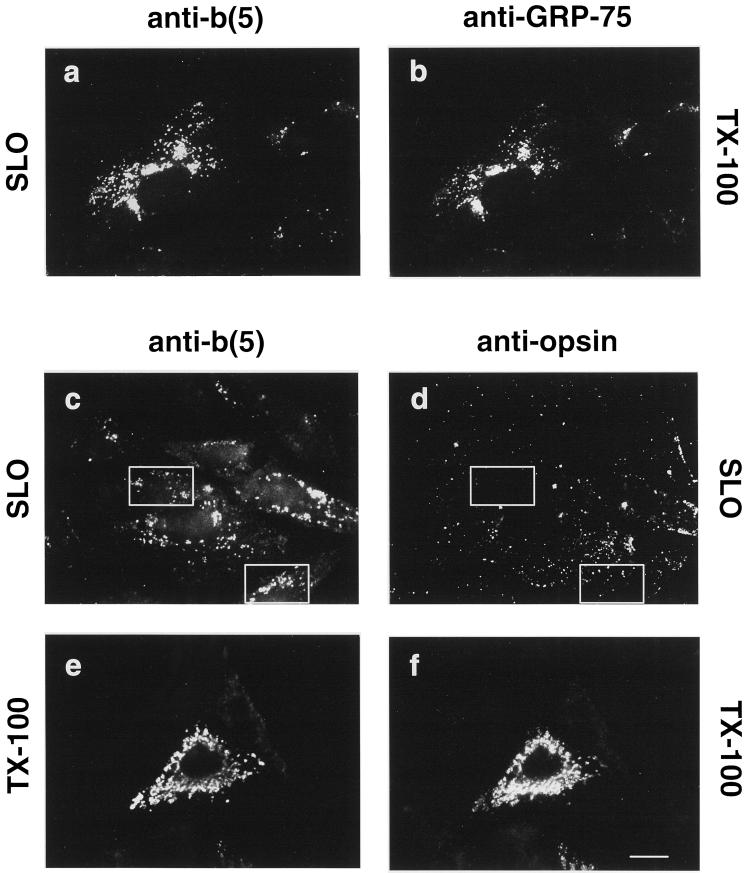
With the use of a selective permeabilization protocol, we next investigated whether the cytosolic domain of b(5)-RRNglyc was accessible to antibodies restricted to the cytosol, as expected for a MOM protein with TA topology. Moreover, because mAbs against the N-terminal bovine opsin sequence are available (Adamus et al., 1991), it became possible to investigate whether b(5)-RRNglyc has a transmembrane topology, with a translocated C terminus, as is the case for TA proteins on the ER membrane (Kutay et al., 1995; Kuroda et al., 1996; Masaki et al., 1996; Pedrazzini et al., 2000). As shown in Figure 6, anti-b(5) antibodies added to unfixed cells during selective SLO permeabilization of the plasma membrane recognized the heme-binding domain of b(5)-RRNglyc (Figure 6a). Because the structures stained under these conditions did not have the classical morphology of mitochondria, we identified them by counterstaining the mitochondrial matrix protein GRP-75 after fixation and permeabilization with Triton X-100. Comparison of Figure 6, a and b, shows that the structures labeled by anti-b(5) antibodies in unfixed SLO-permeabilized cells (Figure 6a) correspond to those recognized by the anti-GRP-75 mAbs (Figure 6b). These data indicate that in SLO-permeabilized cells anti-b(5) antibodies recognized the heme-binding domain of b(5)-RRNglyc on the surface of mitochondria, whose morphology was altered during the incubation required for permeabilization.
Figure 6.
b(5)-RRNglyc C-terminal polar residues are sequestered from antibodies restricted to the cytosol. CV-1 cells expressing b(5)-RRNglyc were permeabilized with 40 U/ml SLO (see MATERIALS AND METHODS) in the presence of anti-b(5) antibodies alone (a and b), or anti-b(5) antibodies and anti-opsin mAbs together (c and d), before fixation. Another sample was fixed and permeabilized with Triton X-100 (TX-100) without prior incubation with SLO, and doubly stained with anti-b(5) antibodies (e) and anti-opsin mAbs (f). Cells were visualized by conventional epifluorescence microscopy. (a and b) SLO-treated cells incubated with anti-b(5) antibodies were counterstained with anti-GRP-75 mAbs after fixation and permeabilization. Stained cells were examined under the fluorescein filter to visualize b(5) (a) or under the rhodamine filter to visualize anti-GRP-75-stained mitochondria (b). Anti-b(5) antibodies added to unfixed cells recognize b(5)-RRNglyc on the cytosolic side of the mitochondria as shown by colocalization with mitochondrial GRP-75. (c and d) In SLO-permeabilized cells, incubated with both anti-b(5) antibodies and anti-opsin mAbs together, the anti-opsin mAbs, revealed by rhodamine-conjugated secondary antibodies (d), shows only nonspecific staining that does not correspond to the anti-b(5) staining, shown in c. Boxes highlight areas in which the difference between staining with the two antibodies is clearly visible. After Triton X-100 permeabilization (e and f), anti-b(5) (e) and anti-opsin (f) antibodies stain the same structures. Bar, 20 μm. c–f were photographed with the same exposure. Each pair of images (c and d) and (e and f) was acquired from negatives and processed with Adobe Photoshop software under identical conditions.
A different result was obtained with anti-opsin mAbs. In SLO-permeabilized cells, these antibodies yielded only a nonspecific staining pattern visible on all cells (transfected and not transfected), which did not coincide with the anti-b(5) positive structures (compare Figure 6, c and d). When the incubation with primary antibodies were carried out after fixation and Trotpm X-100 permeabilization, both anti-b(5) and anti-opsin were able to recognize their epitopes and stained the same structures (Figure 6, e and f). Thus, the C-terminal residues of b(5)-RRNglyc become accessible to antibodies only after permeabilization of the MOM with Triton X-100, suggesting that they are sequestered in the intermembrane space.
The N-Linked Oligosaccharide of b(5)-RRNglyc as a Tool to Characterize Its Targeting Pathway to MOM
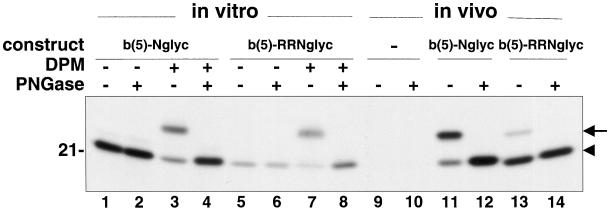
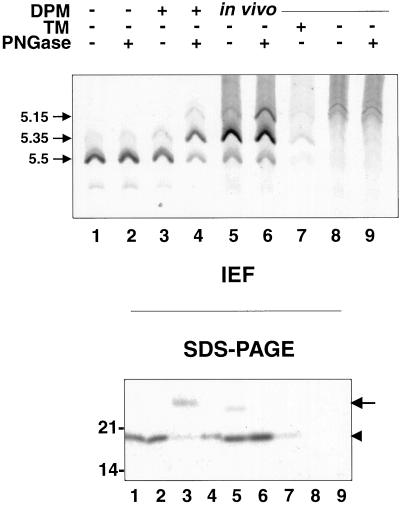
We first performed in vitro translation/translocation experiments, to investigate whether b(5)-RRNglyc is able to translocate its C terminus across the ER membrane and to become glycosylated. The behavior of b(5)-RRNglyc was compared with that of b(5)-Nglyc, a construct carrying the bovine opsin sequence appended to the unmodified C terminus of ER b(5) (Figure 1), which in vivo is localized on the ER (Pedrazzini et al., 2000). Synthetic mRNAs coding either for b(5)-Nglyc or b(5)-RRNglyc were translated in vitro in the reticulocyte lysate system (Figure 7, lanes 1–8), and the 35S-labeled translation products were further incubated either in the absence or presence of DPMs. After immunoprecipitation, samples were subjected to glycanase digestion (PNGase F) or left untreated, and then analyzed by SDS-PAGE autoradiography. From Figure 7 it can be seen that b(5)-RRNglyc was glycosylated by the posttranslationally added microsomes even more efficiently than b(5)-Nglyc, as shown by the induced shift of the majority of the translation product to a higher Mr species (arrow in Figure 7) sensitive to PNGase F treatment (Figure 7, lanes 4 and 8).
Figure 7.
Different degree of glycosylation of b(5)-RRNglyc obtained in vitro and in vivo. Lanes 1–8, b(5)-Nglyc (lanes 1–4) or b(5)-RRNglyc (lanes 5–8) mRNAs were translated in vitro in the reticulocyte lysate system in the presence of [35S]methionine. After blocking translation with cycloheximide, half of each sample was further incubated for another hour in the presence of DPMs, as indicated. The translation products were immunoselected with anti-opsin mAbs, treated (lanes 2, 4, 6, and 8) or mock treated (lanes 1, 3, 5, and 7) with PNGase F, and analyzed by SDS-PAGE/fluorography. The arrowhead indicates the position of the unglycosylated product. In the presence of DPMs, both constructs show an additional major band with lower electrophoretic mobility (lanes 3 and 7, arrow), which is converted to the more rapidly migrating polypeptide by PNGase F (lanes 4 and 8), demonstrating that it is glycosylated. Lanes 10–14, CV-1 cells were transfected with b(5)-Nglyc (lanes 11 and 12), b(5)-RRNglyc (lanes 13 and 14), or mock transfected (lanes 9 and 10) and labeled for 2 h with [35S]Met/Cys. Cleared cell lysates were subjected to anti-opsin mAb immunoselection. Immunocomplexes were digested with PNGase F (lanes 10, 12, and 14) or mock digested (lanes 9, 11, and 13). Note the lower proportion of glycosylated b(5)-RRNglyc in vivo (lane 13, arrow), compared with that obtained in vitro (lane 7). The numbers on the left of the panel indicates the migration of soybean trypsin inhibitor (21 kDa).
Having established that b(5)-RRNglyc is efficiently glycosylated in vitro, we investigated whether the same phenomenon occurs in vivo, in metabolically labeled transfected CV-1 cells (Figure 7, lanes 9–14). Although b(5)-Nglyc was more efficiently glycosylated in vivo than in vitro (Figure 7, lanes 11 and 12), only a minor fraction of b(5)-RRNglyc acquired oligosaccharide (Figure 7, lanes 13 and 14). Thus, in vitro the positive residues in the C-terminal region of b(5)-RRNglyc do not hinder its translocation across the ER membrane, however, the in vivo-expressed protein appears to be glycosylated very inefficiently.
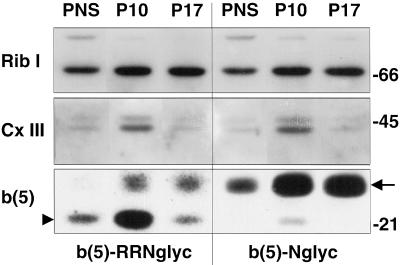
To investigate whether the small proportion of glycosylated b(5)-RRNglyc was localized on the ER or on the MOM, we performed a subcellular fractionation experiment on HeLa cells expressing either b(5)-Nglyc or b(5)-RRNglyc. By differential centrifugation, we isolated two fractions, containing respectively, particles sedimenting at 10,000 g (P10) and 17,000 g (P17). Equal amounts of protein of these fractions were analyzed by SDS-PAGE/Western blot and probed with antibodies against marker proteins (Figure 8). The ER marker ribophorin I (Mr of ∼66 kDa) was contained in the P10 and P17 fractions at approximately the same concentration (Figure 8, top), whereas an ∼45-kDa polypeptide from mitochondrial complex III was enriched in the P10 fraction (Figure 8, middle). Like the 45-kDa complex III protein, the band corresponding to the unglycosylated form of b(5)-RRNglyc (arrowhead in bottom panel) was highly enriched in P10 compared with P17, consistent with a mitochondrial localization. In contrast, the band corresponding to the glycosylated form of b(5)-RRNglyc and of b(5)-Nglyc (arrow in bottom panel) showed the same distribution as ribophorin I, suggesting that the small fraction of b(5)-RRNglyc that becomes glycosylated remains resident in the ER and is not transported to mitochondria.
Figure 8.
Subcellular distribution of glycosylated and unglycosylated b(5)-RRNglyc investigated by cell fractionation. HeLa cells, transfected with b(5)-RRNglyc or b(5)-Nglyc, were subjected to subcellular fractionation by differential centrifugation, to isolate P10 and P17 fractions as described in MATERIALS AND METHODS. The distribution of glycosylated (arrow) and unglycosylated (arrowhead) expression products was analyzed by Western blotting [b(5), bottom] and compared with that of a marker for the ER (ribophorin I, top) and for mitochondria (∼45-kDa complex III polypeptide, middle). The glycosylated forms of the constructs codistribute with ribophorin I, whereas the unglycosylated polypeptides have a distribution similar to the mitochondrial marker. Numbers on the right indicate the position and size (kDa) of markers (Bio-Rad, Richmond, CA).
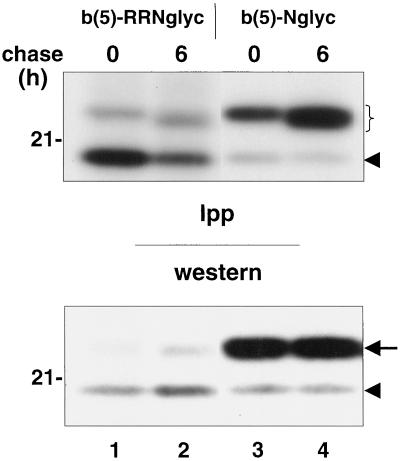
We considered the possibility that the unglycosylated b(5)-RRNglyc was the predominant form in vivo as a consequence of the instability of the glycosylated form or of its enzymatic deglycosylation. To investigate the metabolic stability of our constructs, and possible precursor–product relationships, we performed a pulse/chase analysis on CV-1 cells transfected either with b(5)-RRNglyc or b(5)-Nglyc cDNAs (Figure 9). Cleared lysates were prepared from [35S]Met/Cys-labeled cells. A part was used for immunoprecipitation of b(5) constructs followed by SDS-PAGE fluorography (Figure 9, top), whereas another part was analyzed directly by Western blotting (Figure 9, bottom). During a 6-h chase period, the glycosylated form of both b(5)-RRNglyc and b(5)-Nglyc was stable (Figure 9, top, bracket), with no indication of conversion of the glycosylated form of b(5)-RRNglyc into the more rapidly migrating unglycosylated form (Figure 9, top, arrowhead). In addition, after a 6-h chase, a small increase in the electrophoretic mobility of both the b(5)-RRNglyc and b(5)-Nglyc glycosylated forms was observed. This was probably due to mannose trimming, which occurs in the ER lumen. This result strengthens the conclusion, based on cell fractionation, that the glycosylated form of b(5)-RRNglyc remains a resident of the ER membrane.
Figure 9.
Glycosylated b(5)-RRNglyc is stable on the ER membrane. CV-1 cells transfected with b(5)-RRNglyc or b(5)-Nglyc cDNAs were labeled for 2 h with [35S]Met/Cys. Expression products were immunoselected with anti-opsin mAbs from cleared cell lysates prepared at the end of pulse or after 6 h of chase and subjected to SDS-PAGE/fluorography (top, Ipp). An aliquot of each lysate was also analyzed directly by Western blot with the use of enhanced chemiluminescence to reveal bound antibodies (bottom, Western). The bracket in the top panel and the arrow in the bottom panel indicate the position of the glycosylated form of both constructs. The arrowhead in both panels indicates the unglycosylated form. For both constructs, the glycosylated form is stable, although a slight downward mobility shift is observed at 6 h of chase, probably due to mannose trimming. The position of the 21-kDa size marker is indicated on the left.
Although the results described above suggest that the major in vivo-produced species of b(5)-RRNglyc is not a product of deglycosylation of the oligosaccharide-bearing form, it remained possible that a deglycosylation event was occurring immediately after synthesis and glycosylation of the protein, on a time scale not investigated in the pulse-chase experiment of Figure 9. To test this hypothesis, we analyzed in vitro- and in vivo-synthesized b(5)-RRNglyc by IEF (Figure 10). Enzymatic removal of N-linked oligosaccharide by cellular glycanases results in a shift of the pI of the deglycosylated protein to a more acidic value compared both with the glycosylated form and with the polypeptide, which has never undergone glycosylation, due to the conversion of the GlcNAc-linked asparagine to aspartate (Tarentino and Plummer, 1994; Wiertz et al., 1996). b(5)-RRNglyc was immunoprecipitated either from in vitro translation mixtures (Figure 10, lanes 1–4) or from lysates obtained from metabolically labeled transfected (lanes 5–7) or mock-transfected (lanes 8 and 9) CV-1 cells. Immunoprecipitates were either treated with PNGase F or left untreated and then analyzed both by IEF (Figure 10, top) and by SDS-PAGE (Figure 10, bottom). The majority of b(5)-RRNglyc translated in vitro without subsequent addition of microsomes, focused at pH 5.55, as predicted by its amino acid composition (Figure 10, top, lanes 1 and 2). In addition, a small amount of b(5)-RRNglyc with a more acidic isoelectric point (5.35) was also present, which might be due to a posttranslational modification, such as removal of the C-terminal arginine by a carboxypeptidase. When b(5)-RRNglyc was posttranslationally incubated with microsomes, it was efficiently glycosylated (Figure 10, bottom, lane 3) and retained the same IEF pattern as the unglycosylated protein (Figure 10, top, lane 3), as expected. On the other hand, when the in vitro translated and glycosylated b(5)-RRNglyc was deglycosylated by treatment with PNGase F (Figure 10, bottom, lane 4), the IEF pattern changed (Figure 10, top, lane 4). The major band now focused at pH 5.35 as expected for conversion of an asparagine in the pI 5.5 form to an aspartate residue. In addition, a weak band with a pI of 5.15 appeared, which was presumably due to the deglycosylation of the pI 5.35 form present in the sample not treated with PNGase F. Finally, a small amount of in vitro-translated product retained a pI of 5.5 and can be correlated to the fraction of b(5)-RRNglyc, which did not undergo glycosylation (Figure 10, bottom, lane 3, arrowhead).
Figure 10.
b(5)-RRNglyc does not undergo intracellular deglycosylation. Lanes 1–4, b(5)-RRNglyc was translated in vitro in the presence of [35S]methionine. After blocking translation with cycloheximide, half of each sample was further incubated for another hour in the presence of DPMs, as indicated. After immunoprecipitation with anti-opsin mAbs, the immunoselected products were digested with PNGase F (lanes 2 and 4) or mock digested (lanes 1 and 3) then analyzed by isoelectrofocusing (top) or SDS-PAGE followed by autoradiography (bottom). Numbers on the left of the top panel indicate the pIs of the observed species. The pI 5.5 and pI 5.35 forms produced in the presence of DPMs (lane 3) are shifted to pI 5.35 and 5.15 by PNGase F treatment (lane 4), whereas those produced in the absence of DPMs are unaffected by PNGase digestion (lanes 1 and 2). Lanes 5–9, CV-1 cells were transfected with b(5)-RRNglyc cDNA (lanes 5–7) or mock transfected (lanes 8 and 9) and labeled for 2 h with [35S]Met/Cys in the presence (lane 7) or absence (lanes 5, 6, 8, and 9) of TM. Immunoprecipitates obtained with anti-opsin mAbs were treated with PNGase F (lanes 6 and 9) or mock digested (lanes 5, 7, and 8). Samples were analyzed by IEF or SDS-PAGE. Both TM-treated and untreated cells yield a major band focused at pH 5.35, indicating that b(5)-RRNglyc has not undergone deglycosylation in vivo. Arrow and arrowhead in the bottom panel indicate the positions of the glycosylated and unglycosylated polypeptide, respectively. Molecular weight standards are indicated as in Figures 7–9. See text for further explanation.
The pattern obtained for the untreated in vivo-expressed b(5)-RRNglyc is shown in Figure 10, lane 5. As usual, SDS-PAGE analysis showed a preponderance of the unglycosylated form (arrowhead). The IEF pattern resembled that of the PNGase F-treated, in vitro translated and translocated product (Figure 10, top, lane 4), with species focusing at pH 5.15, 5.35, and 5.5. Although this result might have suggested that the in vivo-expressed protein had been deglycosylated by an intracellular glycanase, this interpretation could be ruled out for two reasons: 1) the pI 5.15 band was also present in nontransfected cells (Figure 10, lanes 8 and 9) and thus was presumably mainly nonspecific; and 2) importantly, b(5)-RRNglyc produced in TM-treated cells focused at pH 5.35 (Figure 10, lane 7), like the major band in untreated cells (Figure 10, lane 5). Because tunicamycin inhibits N-glycosylation, the b(5)-RRNglyc product from treated cells must contain asparagine in the glycosylation consensus sequence. Thus, the increased proportion of pI 5.35 form produced in vivo compared with in vitro (Figure 10, lane 7 vs. lanes 1–3) can most likely be attributed to a higher efficiency of the same posttranslational modification responsible for the presence of the pI 5.35 band in the in vitro-translated samples (Figure 10, lanes 1–3). Finally, when the immunoprecipitate from in vivo-labeled transfected cells was treated with PNGase F, an increase in the pI 5.15 band was observed (Figure 10, lane 6), presumably as a consequence of deglycosylation of the small fraction of the pI 5.35 oligosaccharide-bearing protein. Although these results do not exclude the presence of a minor population of deglycosylated b(5)-RRNglyc molecules, they do show that the majority of the protein present on MOM has not undergone a cycle of glycosylation/deglycosylation.
DISCUSSION
Protein targeting to the ER and to mitochondria generally occurs by distinct pathways, in which specific signals for one or the other destination are recognized by different import machinery. The targeting of TA proteins appears to constitute an exception to this generalization, because at least some proteins with this topology are localized to both ER and MOM (bcl-2; Akao et al., 1994). On the other hand, some other TA proteins, e.g., the two cyt b(5) isoforms, have a very specific localization on only one kind of membrane (D'Arrigo et al., 1993). What is the molecular basis for these differences in specificity of TA protein targeting? During the past few years, some progress has been made toward answering this question.
First, it was firmly established that TA proteins constitute a class of true transmembrane proteins, in that the C terminus of ER-directed members is translocated across the phospholipid bilayer (Kutay et al., 1995; Honsho et al., 1998; Pedrazzini et al., 2000). The translocation occurs posttranslationally, probably in a translocon-independent manner (Kutay et al., 1995; Pedrazzini et al., 2000). Second, it was found that the tail region is responsible for discrimination between ER and MOM (De Silvestris et al., 1995), and, more recently, that the extreme C-terminal polar sequence, predicted to be exposed at the exoplasmic side of the membrane, plays a crucial role in this process, with basic residues favoring targeting to the MOM (Isenmann et al., 1998; Kuroda et al., 1998). In the case of cyt b(5), substitution of only one basic residue in the C-terminal sequence of OM b(5) (see Table 2) with an alanine resulted in targeting of the mutated protein to the ER, whereas substitution of an acidic residue in the tail of ER-b(5) with a lysine resulted in partial relocation of the protein to mitochondria (Kuroda et al., 1998).
In the present study, we have further investigated, both in vitro and in vivo the role of the C-terminal residues in targeting of TA proteins, with the use of mammalian cyt b(5) as model. We first defined the minimal sequence at the C terminus required for targeting to the MOM, and found that a truncated form of ER-b(5) with a single arginine downstream to the TMD was partially relocated to mitochondria, whereas a similar protein with two arg residues at the C terminus was observed, by immunofluorescence, exclusively on mitochondria. The same behavior was seen for GFP fusion proteins, in which GFP replaced b(5)'s heme-binding domain, indicating that this part of the cytochrome is not involved in targeting.
It has been reported that the C-terminal, negatively charged sequence of ER b(5) is required for its targeting to the ER (Mitoma and Ito, 1992). Therefore, we analyzed another GFP-based TA construct, in which the extreme C terminus of ER-b(5) was replaced with a single Thr residue. Under appropriate conditions, this construct could be observed both on mitochondria and on the ER. This result suggests that the more strongly negatively charged C-terminal sequence of ER-b(5) is not strictly required for insertion of the protein into the ER membrane, but that it does confer increased specificity to its targeting. The apparent contradiction between our result and that of Mitoma and Ito (1992) is probably due to the different constructs used. The latter studies analyzed a truncated ER-b(5) lacking the last 10 C-terminal amino acids, a deletion that invades the downstream extremity of the TMD, whereas our construct, GFP-Thr, has the full-length hydrophobic domain.
The results discussed so far suggest that the degree of localization of TA proteins to the MOM is a function of the net positive charge in the C-terminal region. It should be noted, however, that the lack of a negatively charged C-terminal region is not sufficient to determine MOM targeting of TA proteins, because features of the TMD also play a role. In their study on an alternatively spliced, mitochondrially localized, isoform of VAMP-1B, Isenmann et al. (1998) found that a short TMD (18 residues), in conjunction with C-terminal positively charged residues, was required for targeting to the MOM. Similarly, we found that a b(5) construct with an extended TMD (22 instead of 17 residues) and a double arginine motif at the C terminus was targeted mainly to the ER and not to the MOM (Pedrazzini, unpublished results).
A short TMD together with a lack of net negative charge at the C terminus is present in a number of MOM-localized TA proteins. Of the proteins listed in INTRODUCTION, all have this combination of features, with the exception of TOM 6. The MOM localization of this protein may depend on its assembly into the TOM complex. The requirement for these two features together can explain why ER-targeted TA proteins do not necessarily have a negatively charged C-terminal polar peptide. It is interesting that similar properties (a mildly hydrophobic region followed by positively charged residues) are also present in N-terminal signal-anchor sequences involved in MOM targeting and are required for their function (Millar and Shore, 1996; Kanaji et al., 2000), suggesting that common machinery may be involved in inserting both N- and C terminally-anchored proteins into the MOM.
To further investigate the relationship between MOM and ER targeting, we turned to another construct, in which the positively charged amino acids downstream to the TMD were followed by a 22-residue sequence (corresponding to the N terminus of bovine opsin) containing a consensus for N-glycosylation [b(5)-RRNglyc construct]. Our initial idea was to test the possibility that MOM-localized TA proteins reach their final destination after transiting through the ER. Points of close contact between the MOM and the ER have been observed and are thought to be of physiological importance in calcium signaling (Rizzuto et al., 1998). ER-like membranes that are associated with mitochondria have been implicated as intermediates in the import of lipids from the ER to mitochondria (Shiao et al., 1998), and it has been suggested that these mitochondria-associated ER membranes may also be involved in the transport of proteins (Vance et al., 1997). In addition, an N-glycosylated protein has been isolated from a mitochondrial inner membrane fraction, and pulse-chase experiments suggested that it reached its final destination after transiting through the ER (Chandra et al., 1998). If transport of proteins from the ER to mitochondria occurs, TA polypeptides would seem to be good candidates for this pathway, because of the extremely small size of their exoplasmic region.
We first investigated the localization of b(5)-RRNglyc in transfected cells, and obtained evidence that it is localized on the MOM, with the C-terminal opsin sequence sequestered within the intermembrane space. Next, we analyzed whether the N-glycosylation consensus was used, and found that in vitro the construct could be efficiently posttranslationally glycosylated by DPMs, whereas in vivo, the degree of glycosylation was very low. This contrasts with the results for the ER-localized b(5)-Nglyc construct, which is glycosylated in vivo and in vitro with comparable efficiency (Pedrazzini et al., 2000; this study). Cell fractionation showed that the small proportion of glycosylated b(5)-RRNglyc was contained in the ER. Pulse-chase metabolic labeling experiments and analysis by IEF indicated that the unglycosylated b(5)-RRNglyc localized on mitochondria is not a product of deglycosylation of an oligosaccharide-bearing precursor. We conclude that our construct (and probably all TA proteins) reaches the MOM by an independent targeting pathway.
Although b(5)-RRNglyc is targeted to mitochondria by a pathway that does not involve the ER, as occurs for all well-characterized matrix and inner membrane-directed proteins (Neupert, 1997), our results also highlight important differences between the classical targeting of mitochondrial precursors and TA proteins. In vitro studies on the targeting of TA proteins have often been hampered by the tendency of these proteins to associate nonspecifically to any lipid bilayer (Enoch et al., 1979). The use of a C-terminal N-glycosylation consensus allowed us to monitor the bona fide insertion of the TMD into the ER membrane, with translocation of the C terminus into the lumen. The striking difference in the degree of glycosylation attained by b(5)-RRNglyc in vitro and in vivo that we observed indicates that, notwithstanding its full competence to insert into ER membranes, when faced with a choice in vivo, this protein prefers the MOM. Thus, it appears that ER and MOM are in competition for TA proteins, a phenomenon that can explain the dual localization of some members of this class of polypeptides.
In principle, positively charged residues could act as negative signals for the ER, i.e., decrease the binding of the protein to the ER with a consequent default localization to the MOM, or vice versa, as positive signals that promote binding to the MOM, thus sequestering the targeted protein from the ER. The data presented in this article favor the latter view, because b(5)-RRNglyc was even more efficient in inserting into microsomes in vitro than b(5)-Nglyc, which in vivo localizes to the ER. Reasoning along the same lines, we suggest that the C-terminal residues of ER-b(5) do not increase targeting to the ER because of a positive effect, but simply because they are shunned by the mitochondrial targeting system. In other words, the ER is probably permissive for the insertion of all TA proteins, but, in vivo, positive signals for other organelles can result in targeting to alternative destinations.
If targeting of TA proteins to the MOM occurs via a signal-based mechanism, it should be possible to identify specific chaperones and/or surface receptors on the mitochondrial surface, involved in recognition and insertion of the C-terminal tails. The studies carried out in cell free systems until now (Nguyen et al., 1993; Janiak et al., 1994; Millar and Shore, 1996; Lan et al., 2000) have yielded conflicting results with respect to temperature, energy, and chaperone requirements, as well as to the involvement of components of the TOM system or of other surface receptors. At least some of the inconsistencies may be due to the difficulty in distinguishing nonspecific binding of TA proteins from physiological targeting.
As a consequence of the results of this and previous (Isenmann et al., 1998; Kuroda et al., 1998) studies, it seems feasible to make predictions on the intracellular localization of TA proteins on the basis of their C-terminal sequence. We have examined cyt b(5) sequences from different organisms, found in the data banks with a Blast search (Table 2). Interestingly, a net negative charge in the extreme C-terminal portion appears to be characteristic only of the mammalian ER isoform. In other organisms, with the exception of the chick and of Saccharomcyes cerevisiae, the C-terminal net charge is neutral or positive, as is the case for the mammalian MOM isoform. In addition, all b(5)s (with the possible exceptions of Caenorhabditis elegans, and Sshizosaccharomyces pombe) have TMDs of <20 residues, compatible with targeting to the MOM (not shown in the Table). One might therefore speculate that in many organisms, cyt b(5) is a MOM, and not an ER, protein, as has always been assumed. Alternatively, the charge-based discrimination between MOM and ER may not operate, or may operate at lower efficiency, in most species, and a single b(5) isoform would carry out its functions both on the MOM and on the ER. The division of labor between two differently targeted isoforms seems to be in any case a recent development in evolution.
ACKNOWLEDGMENTS
In addition to the people who kindly donated antibodies, we thank Roberto Sitia and Alessandro Vitale for helpful suggestions and Francesco Clementi for continued support. We are particularly grateful to Angelo Viotti and Christian Cosentino (Istituto C.N.R. di Biosintesi Vegetali) for hospitality and help with the isoelectric focusing experiment. This work was supported by a grant from Associazione Italiana per la Ricerca sul Cancro (AIRC) and Telethon (Grant E734) to N.B. E.P. was supported by an AIRC fellowship.
Abbreviations used:
- cyt
cytochrome
- DPM
dog pancreas microsome
- ER
endoplasmic reticulum
- ER b(5)
endoplasmic reticulum isoform of cytochrome b(5)
- FITC
fluorescein isothiocyanate
- IEF
isoelectric focusing
- mAb
monoclonal antibody
- MOM
mitochondrial outer membrane
- OM b(5)
outer membrane isoform of cytochrome b(5)
- PNGase F
peptide N-glycanase F
- PNS
postnuclear supernatant
- SLO
streptolysin O
- TA
tail-anchored
- TMD
transmembrane domain
- TOM
translocase of the outer membrane
- TM
tunicamycin
REFERENCES
- Adamus G, Arendt A, Hargrave PA. Genetic control of antibody response to bovine rhodopsin in mice: epitope mapping of rhodopsin structure. J Neuroimmunol. 1991;34:89–97. doi: 10.1016/0165-5728(91)90118-q. [DOI] [PubMed] [Google Scholar]
- Akao Y, Otsuki YS, Kataoka S, Ito Y, Tsujimoto Y. Multiple subcellular localization of bcl-2: detection in nuclear outer membrane, endoplasmic reticulum, and mitochondrial membrane. Cancer Res. 1994;54:2468–2471. [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987. [Google Scholar]
- Bhakdi S, Weller U, Walev I, Martin E, Jonas D, Palmer M. A guide to the use of pore-forming toxins for controlled permeabilization of cell membranes. Med Microbiol Immunol. 1993;182:167–175. doi: 10.1007/BF00219946. [DOI] [PubMed] [Google Scholar]
- Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not endoplasmic reticulum membranes. J Cell Biol. 1996;135:1501–1513. doi: 10.1083/jcb.135.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, D'Arrigo A, De Silvestris M, Pietrini G. NADH-cytochrome b5 reductase and cytochrome b5 - The problem of postranslational targeting to the endoplasmic reticulum. In: Borgese N, Harris JR, editors. Subcellular Biochemistry. Vol. 21. New York: Plenum Press; 1993. pp. 313–341. [DOI] [PubMed] [Google Scholar]
- Borgese N, Pietrini G. Distribution of the integral membrane protein NADH-cytochrome b5 reductase in rat liver cells, studied with a quantitative radioimmunoblotting assay. Biochem J. 1986;239:393–403. doi: 10.1042/bj2390393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CB, Roth MG. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriotti A, Pedrazzini E, Fabbrini MS, Zoppè M, Bollini R, Vitale A. Expression of the wild-type and mutated vacuolar storage protein phaseolin in Xenopus oocytes reveals relationships between assembly and intracellular transport. Eur J Biochem. 1991;202:959–968. doi: 10.1111/j.1432-1033.1991.tb16456.x. [DOI] [PubMed] [Google Scholar]
- Chandra NC, Spiro MJ, Spiro RG. Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J Biol Chem. 1998;273:19715–19721. doi: 10.1074/jbc.273.31.19715. [DOI] [PubMed] [Google Scholar]
- D'Arrigo A, Manera E, Longhi R, Borgese N. The specific subcellular localization of two isoforms of cytochrome b5 suggests novel targeting pathways. J Biol Chem. 1993;268:2802–2808. [PubMed] [Google Scholar]
- De Silvestris M, D'Arrigo A, Borgese N. The targeting information of the mitochondrial outer membrane isoform of cytochrome b5 is contained within the carboxyl-terminal region. FEBS Lett. 1995;370:69–74. doi: 10.1016/0014-5793(95)00797-d. [DOI] [PubMed] [Google Scholar]
- Enoch HG, Fleming PJ, Strittmatter P. The binding of cytochrome b5 to phospholipid vesicles and biological membranes. Effect of orientation on intermembrane transfer and digestion by carboxypeptidase Y. J Biol Chem. 1979;254:6483–6488. [PubMed] [Google Scholar]
- Haucke V, Schatz G. Import of protein into mitochondria and chloroplasts. Trends Cell Biol. 1997;7:103–106. doi: 10.1016/S0962-8924(96)10052-0. [DOI] [PubMed] [Google Scholar]
- Honsho M, Mitoma J-Y, Ito A. Retention of cytochrome b(5) in the Endoplasmic Reticulum is transmembrane and luminal domain-dependent. J Biol Chem. 1998;273:20860–20866. doi: 10.1074/jbc.273.33.20860. [DOI] [PubMed] [Google Scholar]
- Isenmann S, Khew-Goodall Y, Gamble J, Vadas M, Wattenberg B. A splce-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting. Mol Biol Cell. 1998;9:1649–1660. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak F, Leber B, Andrews DW. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J Biol Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer T, Rapaport D, Ryan MT, Meisinger C, Kassenbrock CK, Blachly-Dyson E, Forte M, Douglas MJ, Neupert W, Nargang FE, Pfanner N. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J Cell Biol. 2001;152:289–300. doi: 10.1083/jcb.152.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda R, Ikenoue T, Honsho M, Tsujimoto S, Mitoma JY, Ito A. Charged aminoacids at the carboxyl-terminal portions determine the intracellular location of two isoforms of cytochrome b(5) J Biol Chem. 1998;273:31097–31102. doi: 10.1074/jbc.273.47.31097. [DOI] [PubMed] [Google Scholar]
- Kuroda R, Kinoshita J-Y, Honsho M, Mitoma J-Y, Ito A. J. Biochem. 120, 828–833. 1996. In situ topology of cytochrome b(5) in the endoplasmic reticulum membrane. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilgen G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Lan L, Isenmann S, Wattenberg BW. Targeting and insertion of C-terminally anchored proteins to the mitochondrial outer membrane is specific and saturable but does not strictly require ATP or molecular chaperones. Biochem J. 2000;349:611–621. doi: 10.1042/0264-6021:3490611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer F, Ghrir R, Guiard B, Cortial S, Ito A. Two homologous cytochromes b5 in a single cell. Eur J Biochem. 1983;132:95–102. doi: 10.1111/j.1432-1033.1983.tb07330.x. [DOI] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Membrane topology and retention of microsomal aldehyde dehydrogenase in the endoplasmic reticulum. J Biol Chem. 1996;271:16939–16944. doi: 10.1074/jbc.271.28.16939. [DOI] [PubMed] [Google Scholar]
- Millar DG, Shore GC. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J Biol Chem. 1996;271:25823–25829. doi: 10.1074/jbc.271.42.25823. [DOI] [PubMed] [Google Scholar]
- Mitoma J-Y, Ito A. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 1992;11:4197–4204. doi: 10.1002/j.1460-2075.1992.tb05513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Millar DG, Yong VW, Korsmeyer SJ, Shore GC. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J Biol Chem. 1993;268:25265–25268. [PubMed] [Google Scholar]
- Pedrazzini E, Villa A, Longhi R, Bulbarelli A, Borgese N. Mechanism of residence of cytochrome b(5), a tail-anchored protein, in the endoplasmic reticulum. J Cell Biol. 2000;148:899–913. doi: 10.1083/jcb.148.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Silvius JR, Shore G. Direct membrane insertion of voltage-dependent anion-selective channel protein catalyzed by mitochondrial Tom20. J Cell Biol. 1999;145:973–978. doi: 10.1083/jcb.145.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao Y-J, Balcerzak B, Vance JE. A mitochondrial membrane protein is required for translocation of phosphatidylserine from mitochondria-associated membranes to mitochondria. Biochem J. 1998;331:217–223. doi: 10.1042/bj3310217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH. Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificities of oligosaccharide-cleaving enzymes from Flavobacterium menigosepticum. Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- Vance JE, Stone SJ, Faust JR. Abnormalities in mitochondria-associated membranes and phospholipid biosynthetic enzymes in the mnd/mnd mouse model of neuronal ceroid lipofuscinosis. Biochim Biophys Acta. 1997;1344:286–299. doi: 10.1016/s0005-2760(96)00153-1. [DOI] [PubMed] [Google Scholar]
- Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhang Y, Sabatini DD, Kreibich G. Reconstitution of translocation-competent membrane vesicles from detergent-solubilized dog pancreas rough microsomes. Proc Natl Acad Sci USA. 1989;86:9931–9935. doi: 10.1073/pnas.86.24.9931. [DOI] [PMC free article] [PubMed] [Google Scholar]