Abstract
There is currently a crisis in science related to highly publicized failures to reproduce large numbers of published studies. The current work proposes, by way of case studies, a methodology for moving the study of reproducibility in computational work to a full stage beyond that of earlier work. Specifically, it presents a case study in attempting to reproduce the reports of two R libraries for doing text mining of the PubMed/MEDLINE repository of scientific publications. The main findings are that a rational paradigm for reproduction of natural language processing papers can be established; the advertised functionality was difficult, but not impossible, to reproduce; and reproducibility studies can produce additional insights into the functioning of the published system. Additionally, the work on reproducibility lead to the production of novel user-centered documentation that has been accessed 260 times since its publication—an average of once a day per library.
Keywords: reproducibility, natural language processing, PubMed/MEDLINE
1. Introduction
The general crisis of (non-)reproducibility in science extends into natural language processing research (Pedersen, 2008; Branco, 2012; Fokkens et al., 2013). The authors of this paper are well aware that we ourselves have made software publicly available that no longer runs, or no longer functions completely, or is no longer available, despite having published URLs for it. The goal of this paper is to help to establish a methodology for exploring issues of reproducibility in the field.
It is not hyperbole to say that there is a crisis in science related to highly publicized failures to reproduce large numbers of published studies. The phenomenon has been observed in fields as diverse as computer science (Collberg et al., 2014b; Collberg et al., 2014a; Proebsting and Warren, 2015), psychology (Collaboration and others, 2012), signal processing (Kovacevic, 2007; Vandewalle et al., 2009), cancer biology (Barrows et al., 2010; Prinz et al., 2011), medicine (Begley and Ellis, 2012; Mobley et al., 2013), and biomedical research in general (Collins and Tabak, 2014), with implications even for fields that bridge the social and humanistic sciences, such as classic linguistic field research (Bisang, 2011; Berez, 2015).
Does this crisis really extend to natural language processing? There is some reason to think that it does not. A number of libraries, executables, and architectures for natural language processing have been published on and subsequently used extensively by large numbers of other researchers. These artifacts have been subjected to extensive “testing” in the form of their uses “in the wild,” and some of them have held up to intensive use. However, these encouraging facts might not be representative of the state of the natural language processing software ecosystem as a whole. Impressionistically, in addition to this set of highly successful natural language processing distributions, there are myriad applications reported in the literature that turn out to be uncompilable, unusable, unobtainable, or otherwise not reproducible (Pedersen, 2008; Poprat et al., 2008). This paper attempts to move the field beyond those impressionistic observations to a rational approach to assessing reproducibility in natural language processing. We report on our experiences with the pubmed.mineR (Rani et al., 2015) and rentrez libraries for the R programming language. These libraries provide a number of affordances for doing text mining of the PubMed/MEDLINE repository of biomedical publications. PubMed/MEDLINE is a prominent part of the biomedical research milieu, with 23 million entries and new ones being added at a rate of 2,700 per day. Text mining, especially of the PubMed/MEDLINE repository, is a booming field in the bioscience and bioinformatics communities. Pubmed.mineR and rentrez attempt to facilitate that work with the R language. R provides an extensive range of affordances for statistics and graphing, and is one of the fastest-growing languages in the world (Ihaka and Gentleman, 1996).
To see the motivation for the approach that we describe here, consider Figure 1. It shows the increase in processing time as a popular publicly available language processing API is given increasingly large inputs. This is one of the systems mentioned above, and it has been used extensively. Note that processing times increase logarithmically (correlation coefficient of fit to log model = 0.80) up to about 18,000 words, followed soon by a program crash at 19,000 tokens. Under what conditions can we say that the many publications on this system’s performance are reproducible? At the most, we can assume them to be reproducible only up to about 18,000 words of input (assuming memory and other configuration similar to the machine that we used at the time). Input size under which the reported performance numbers hold is not something that is reported or (as far as the authors can tell) considered in those publications, but the data reported in Figure 1 suggests that the reported performance is not, in fact, reproducible for all possible inputs, and the logarithmic increase in processing times suggests that as the memory on the machine reaches its limits, the application is not robust in the face of phenomena like swapping memory to disk.
Figure 1.
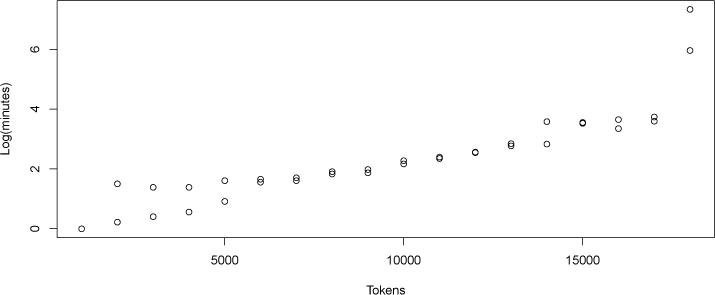
Processing time with increasing size of input for a popular publicly available natural language processing API. Processing time increases logartithmically with input size until 18,000 tokens, and then the program crashes. The experiment was run twice, resulting in two different run times at most input sizes.
1.1. Problems of reproducibility in natural language processing
A number of factors conspire to make reproducibility in any traditional sense difficult to impossible in the domain of natural language processing.
The data is often not available (Branco, 2012). In some cases, the shared task model has made great progress towards addressing this issue. In other cases, such as natural language processing in the medical, intelligence, and law enforcement domains, the problem of unavailability of data will probably never be addressed in such a way as to facilitate reproducibility.
Natural language processing research is primarily published in conference proceedings, not journals. Because conference papers routinely have page limits, there is typically not enough space to give all information on the methodology that would be necessary to replicate the work (see, for example, (Fokkens et al., 2013)).
There is little or no tradition in the community of publishing reproduction attempts—the bias is strongly in favor of novel methods (Fokkens et al., 2013).
1.1.1. The reproducibility hierarchy of needs
There are a number of conditions that must be met in order to reproduce a study in natural language processing. These form a hierarchy—if the most basic conditions cannot be met, then the higher ones cannot, either. We consider here a typical natural language processing paper reporting a new tool and/or method.
Availability: the system must be available, or there must be sufficient detail available to reconstruct the system, exactly.
Builds: the code must build.
Runs: The built code must run.
Evaluation: it must be possible to run on the same data and measure the output using the same implementation of the same scoring metric.
Most of the sociology of natural language processing militates against all steps in the hierarchy being met. Limits on conference paper lengths assure that there will rarely be enough information available in the methodology section to reconstruct the system. GitHub and similar distribution mechanisms have, of course, made it easier to distribute versioned code, but many people still report not being able to find code, not being able to remember how to build it, etc. Maven has made progress in ensuring that build processes are repeatable, but most projects in NLP are not distributed as Maven projects, which in any case are not appropriate for every language and architecture used in NLP research. Even given a built program, it may not run, e.g. due to undocumented platform dependencies, configuration files, input requirements, memory requirements, processor requirements, graphics card requirements, etc.
An experiment by (Collberg et al., 2014a) looked at levels 1 and 2 of this hierarchy in a study of systems reported at computer science conferences. The results are discussed elsewhere in this paper. The work resulted in a principled approach to evaluating the extent of buildability in CS research reproduction. That was clearly difficult to do in a reasonably methodologically sound way. The work reported here attempts to go to the next level–evaluating not buildability, but executability–and it is not immediately clear what that methodology should be. This paper is a step towards developing such a methodology, or a framework for developing such a methodology. The novelty of the work reported here is that it takes the effort to level 3—that is, executability. More specifically, it explores the possibilities for working at level 3, and shows some results for work of that nature.
1.1.2. An expanded conception of reproducibility
Collberg et al. (Collberg et al., 2014a) suggest that in the context of computer science research, the notion of reproducibility—defined by them as the independent confirmation of a scientific hypothesis through reproduction by an independent researcher/lab—can usefully be replaced by the concept of repeatability. In particular, they define three types of what they call weak repeatability. The highest level is the ability of a system to be acquired, and then built in 30 minutes or fewer. The next level is the ability of a system to be acquired, and then built, regardless of the time required to do so. The lowest level is the ability of a system to be acquired, and then either built, regardless of the time required to do so, or the original author’s insistence that the code would build, if only enough of an effort were made.
This notion of weak reproducibility is demonstrably useful; Table 1.1.2. shows how effective it was in quantifiying reproducibility issues in computer science. However, as the authors point out, it leaves out crucial elements of reproducibility. For one thing, it does not take versioning into consideration: assuming that code is available and builds, can we necessarily assume that it is the same version as the code that was used in the paper? For another, the definition does not take into consideration what (Collberg et al., 2014a) call executability: will the code not just build, but run? For example, even examples from papers don’t always work as advertised. We suggest here other work that can be useful in evaluating the claims of a paper. The work reported here tries to tackle the executaility issue specifically. We suggest here some more things to think about:
Processing time: it can be revealing to measure processing times as increasingly large data sets are treated. For example, we found one widely used system that showed a linear increase in processing time with input text size until at some (repeatable) input size the processing time began increasing rapidly, and then (with more increases in input size) the system crashed.
Validating through debugging output: we found one library that produced debugging output that could clearly be demonstrated to be wrong with simple UNIX commands.
Metamorphic testing: natural language processing applications are obvious candidates for metamorphic testing (defined below, in the Methods section).
Table 1.
Summary of results on 402 papers whose results were backed by code, from (Collberg et al., 2014a).
| Code available and builds within 30 minutes | 32.3% |
| Code available and builds | 48.3% |
| Either code builds, or original authors insist that it would | 54.0% |
1.2. Related literature
(Collberg et al., 2014a) reviews two previous studies of code-sharing in computer science research. (Kovacevic, 2007) began with 15 IEEE papers and evaluated the presence of proofs, availability of code, and availability of data. They found that all papers presented proofs, none of the papers made code available, and 33% of papers were based on data that was available (probably due to the wide use of publicly available data sets in that field). In our hierarchy of needs, this would be level 1. (Vandewalle et al., 2009) looked at 134 IEEE papers in terms of availability of code and of data, and found that code was available in 9% of cases and data was available in 33% of cases (same field). Again, this corresponds to level 1 of the hierarchy of needs. (Collberg et al., 2014a), discussed elsewhere in this paper, took the work to level 2; this paper advances to level 3, or executability.
2. Materials and methods
Two R libraries for text mining from PubMed/MEDLINE, the primary repository for biomedical publications, were selected for the case study. They are interesting case studies in that they allow the examination of what we are calling level 3 of the hierarchy. Since we know in advance, due to their availability on CRAN, that they are available and they build, they allow us to take the next step of studying their run-time behaviors. They were also selected due to their intended domain of application. While many systems that are reported in the general natural language processing literature are avowedly research systems, and it could be argued that their run-time characteristics were not a focus of the research, the situation is different in biomedical natural language processing. In this specialized domain, the stated purpose of the work is often not research per se, but the goal of providing a working tool to biologists or physicians (Hirschman et al., 2007). Thus, investigations of reproducibility at the level of run-time behavior are clearly relevant in biomedical natural language processing.
2.1. Pubmed.mineR
Pubmed.mineR is a library for doing text mining from the PubMed/MEDLINE collection of documents. PubMed/MEDLINE contains references to about 23 million articles in the domain of biomedical science, broadly construed. Pubmed.mineR provides a clean interface to named entity recognition and normalization web services provided by the National Center for Biotechnology Information via the PubTator application (Wei et al., 2013). Pubmed.mineR was released with documentation for the various and sundry methods that it provides, but no manual.
Two tests are designed to evaluate the performance of pubmed.mineR package. In Test 1, we built four document collections of different sizes and tested the speed of named entity recognition and normalization by using the package. In Test 2, we examined the stability of performance by running 10 iterations of the largest document set.
All of the experiments were carried out on a Mac desktop with installation of Mac OS X (version 10.7.5). The processor was a 2.93 Ghz Intel Core 2 DUO. The memory was 4 GB 1067 MHz DDR3. All results regarding performance should be interpreted as valid only for this (typical) machine configuration.
3. Results
3.1. Level of the reproducibility hierarchy reached
The system was available, as advertised. It installed without problems. The code did not run as advertised in the documentation, but the authors responded quickly to requests for help, and it was possible to get it working relatively quickly. Thus, level 3—executability—was reached.
3.2. Performance of pubmed.mineR package
The tester was unable to load any documents by following the documentation provided with the library. The authors responded quickly to requests for help, and the tester was successful in using one of the methods for loading data. Thus, level 3—executability—was reached for this library, as well.
3.2.1. Test 1. Performance of pubmed.mineR package on diverse document collections
In order to get a broad picture of pubmed.mineR performance, we evaluated it on four sets of data from PubMed. We varied the size of the data sets from quite small (about 2200 abstracts) to reasonably large (about 640,000 abstracts). It is not possible to keep the contents constant while varying size, so we tried instead to maximize variability of content by using four different queries to retrieve the abstracts. We then evaluated the mean processing time per document for each of the data sets.
Table 2 shows the queries for the four data sets.
Table 2.
Queries and sizes for the four data sets.
| Number of abstracts | Query |
|---|---|
|
| |
| 2K (2,283 abstracts) | synthetic lethal |
| 60K (59,854 abstracts) | human drug disease “blood cell” |
| 172K (171,955 abstracts) | human drug disease cell |
| 638K (637,836 abstracts) | human drug disease |
The results are not problematic at all, but they are certainly unexpected. Processing time per document decreases quite a bit as document set size goes up. To evaluate the likely explanation that this was due to the length of the connection time to PubTator being amortized over a successively larger number of documents, we revisited the R code to ensure that we were only measuring the document processing time per se. Indeed, we found that the probable explanation was not the case at all, and that the unusual result does, in fact, represent the actual document processing times. We have no explanation for the behavior.
3.2.2. Test 2. Performance of pubmed.mineR package concerning its own stability
The second test evaluated the pattern of increase in processing time for the entire document collection, as well as variability in that processing time. The largest data set was used as input to pubmed.mineR, and the processing time was measured for every 10,000 abstracts. To evaluate variability, the process was repeated 10 times.
Figure 2 shows the cumulative processing time for the document collection and the mean processing time per document. The cumulative processing time for the document collection increases linearly. Figure 3 shows the variability in cumulative processing time over the course of the 10 repetitions. There are a few outliers, but the variation is generally small.
Figure 2.
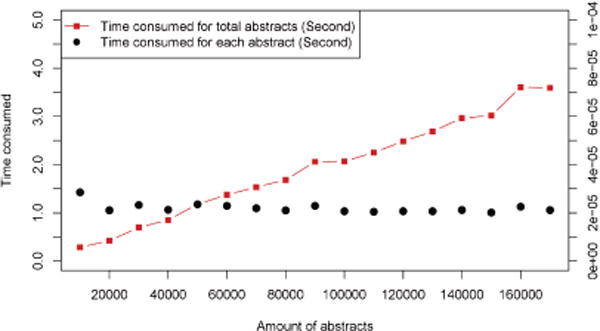
Cumulative processing time for the entire document collection and per-document processing time.
Figure 3.
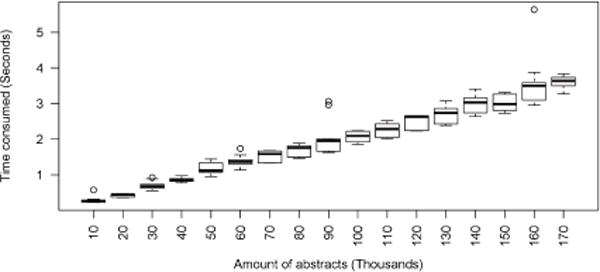
Variability in cumulative processing time.
3.3. Metamorphic testing and exploring the parameter space with rentrez
The functionality for pubmed.mineR and for rentrez are quite different. Rentrez’s functionality is oriented less towards processing documents than towards retrieving them–more specifically, towards retrieving document identifiers. For an information retrieval library, different kinds of validation are applicable. In the case of rentrez, we tried metamorphic testing, and exploration of the parameter space. Metamorphic testing (Murphy et al., 2008; Chen et al., 2009; Xie et al., 2009; Xie et al., 2011) is applied in situations where we have no “oracle”—situations where we cannot know in advance what the exact output of a function or of a program should be. The general approach of metamorphic testing is to change some aspect of the input for which we can predict in a general way whether or not there should be a change in the output, and what the overall trend in the change should be. For example, if we calculate the mean and the standard deviation for some data set, and then add 100 to every value in the data set, the mean should change, and it should increase. In contrast, the standard deviation should not change at all.
The metamorphic test that we applied to rentrez was to give it two different queries, where we had a priori reason to think that the two queries should give us result sets of different sizes, and that the size of the second result set should be considerably smaller. For the first (and presumably larger) result set, we used the query apoptosis. (Apoptosis is a cellular process that is very important in embryological development and in the maintenance of proper cell states in the adult. Failure of apoptosis is a common cause of cancer, and due to the importance of apoptosis in development and in disease, it has been studied extensively (Hunter, 2012).) For the second (and presumably smaller) result set, we used the query judo. (Judo is a sport of Japanese origin that is not widely practiced in the United States, the source of most PubMed/MEDLINE publications, and it has not been widely studied in the English-language scientific literature.)
3.4. Rentrez methods: Exploring the parameter space
A search of PubMed/MEDLINE can return millions of article identifiers, but by default, the rentrez interface only returns 20 of them. This number can be changed by setting the appropriate variable when the search function is called. We varied the value of the variable systematically through a segment of the parameter space for that variable, which has no explicitly stated range, from 100000 to 1. We used the apoptosis query described in the methods for metamorphic testing.
3.5. Rentrez results: exploring the parameter space
We tried a realistic value for the variable that controls the maximum number of identifiers returned in a result set, as described above in the Methods section. We immediately found a bug. When the variable is set to 100,000 declaratively, the search function returns no identifiers. On the other hand, if it is set programmatically (e.g. in a for-loop from 100,000 down to 1), then the search function works well. After communication with the library author, a bug report has been filed.
3.6. New documentation
As one product of the study, new documentation was written for the two R libraries. It is available at https://zipfslaw.org/2015/10/19/pubmed-miner/ and at https://zipfslaw.org/2015/12/24/rentrez/, and has been accessed an average of once or more per day for the past several months. It is hoped that the documentation itself will add to the reproducibility of the work, by providing clear guidelines for running the code that were absent in the original publications—that is, by making it easier for future users to reach level 3 of the reproducibility hierarchy of needs.
4. Discussion and conclusions
4.1. Summary of the results of the two case studies
4.1.1. Pubmed.mineR
The library is available and installs without problems. Problems with running the code were quickly resolved by communication with the authors, and new documentation addresses those issues. Level 3, or executability, was reached.
Processing time for the document collection increases linearly with the size of the input data set.
Processing time per individual document decreases with the size of the input data set.
Processing time is relatively stable across multiple repetitions of the same input.
New documentation responding to the problems with running the code may increase the chances for reproducibility in the future.
4.1.2. Rentrez
The library is available and installs without problems. There were no problems with getting the code to run. Again, Level 3, or executability, was reached.
The author was very responsive to requests for help with the library.
Metamorphic testing did not reveal any issues.
Exploring the parameter space quickly revealed an issue.
4.2. Conclusions
We have laid out some of the issues that pose problems for the notion of reproducibility in natural language processing. We showed how previous work on reproducibility in computer science can be used to establish a hierarchy of desiderata regarding reproducibility even in the face of these restrictions. We then showed how how those desiderata could be extended with conceptually straightforward, easily implementable tests of program performance.
The work reported here examined two R libraries for natural language processing in the biomedical domain. Both of those libraries presented the same problems for reproducibility testing: unavailability of data, and inadequate space for documentation of the experimental methodology in the original publications. We explored a number of possibilities for an expanded notion of reproducibility that we suggest might be appropriate for natural language processing research. In so doing, we found that all of those exploratory methods made a contribution to understanding the extent to which the research was or was not reproducible, whether by finding issues with the library (varying input sizes until a library crashed; exploring the parameter space) or by providing reassuring sanity checks (metamorphic testing, stability). There is a possible counterargument to the entire methodology of this paper. This counter-argument would be that papers in natural language processing describe research, not engineering efforts, and that therefore it is not relevant to study systems with respect to performance characteristics like processing times, the ability of the code to build, and the like. This counterargument does not hold, because unlike the case in general natural language processing, the stated motivation in paper after paper in the biomedical natural language processing field is to provide a tool that meets some need of either physicians or biologists. The selection of biomedical natural language processing libraries for the work reported here was quite deliberate, as issues of run-time repeatability are quite relevant in this domain.
In principle, reproducibility in computational systems can be achieved easily without really addressing the right underlying issue. It should be possible to package arbitrary environments in a self-contained virtual machine that will execute for a long time to come. However, it may still not be possible to change anything about it or to use it for any actual task, and the fact that it produces the same output every time one pushes “run” is of little reassurance with respect to correctness, or even with respect to doing what is described in the paper. What one wants of a scientific result is to be able to (1) rely on it, and (2) put it to new uses (Fokkens et al., 2013). So, although the methodology described here improves on the lesser alternatives of not running, not building, or not even being available, evaluating the extent to which a program meets the goals of reliability and applicability to new uses remains for future work.
It is not our intention to point fingers or to lay blame at anyone’s feet. As pointed out in the introduction to this paper, we are well aware that we ourselves have made software publicly available that no longer runs, or no longer functions completely, or is no longer available, despite our having published URLs for it. Rather, the hope of this paper is to help to establish a methodology for exploring issues of reproducibility in the field of natural language processing. It’s clear that such a methodology is needed, and this paper does not claim to be the last word on the subject: two hours after we submitted this paper for review, one of the libraries stopped working completely—it returned only empty vectors. Perhaps the back end to which it connects had changed its interface—we really don’t know, and the authors of the library have been silent on the specifics. After some communication with the authors, the former functionality was restored—the vectors that are returned are no longer empty. However, the behavior has not been the same since—that is, the contents of the vectors are different—and so far, we have been unable to reproduce our previous results. The research that relied on the library is at a standstill.
Table 3.
Per-document processing time varies with input size.
| Data set | 2K | 60K | 172K | 638K |
|---|---|---|---|---|
| Size of file | 4,148Kb | 111,595Kb | 153.5Mb | 322.7Mb |
| Mean processing time per abstract (seconds) | 0.0025 | 0.00025 | 0.000022 | NA |
Acknowledgments
KBC’s work was supported by grants NIH 2R01 LM008111-09A1 NIH 2R01 to Lawrence E. Hunter, LM009254-09 NIH to Lawrence E. Hunter, 1R01MH096906-01A1 to Tal Yarkoni, and NSF IIS-1207592 to Lawrence E. Hunter and Barbara Grimpe.
Bibliographical References
- Barrows NJ, Le Sommer C, Garcia-Blanco MA, Pearson JL. Factors affecting reproducibility between genome-scale sirna-based screens. Journal of biomolecular screening. 2010;15(7):735–747. doi: 10.1177/1087057110374994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Berez AL. On valuing reproducibility in science and linguistics. Research, Records and Responsibility: Ten years of PARADISEC. 2015:39. [Google Scholar]
- Bisang W. Variation and reproducibility in linguistics. Linguistic Universals and Language Variation. 2011;231:237. [Google Scholar]
- Branco A. Reliability and meta-reliability of language resources: Ready to initiate the integrity debate? Proceedings of the twelfth workshop on treebanks and linguistic theories. 2012:27–36. [Google Scholar]
- Chen TY, Ho JW, Liu H, Xie X. An innovative approach for testing bioinformatics programs using metamorphic testing. BMC Bioinformatics. 2009;10(1):24. doi: 10.1186/1471-2105-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaboration O.S et al. An open, large-scale, collaborative effort to estimate the reproducibility of psychological science. Perspectives on Psychological Science. 2012;7(6):657–660. doi: 10.1177/1745691612462588. [DOI] [PubMed] [Google Scholar]
- Collberg C, Proebsting T, Warren A. Repeatability and benefaction in computer systems research: A study and a modest proposal. Department of Computer Science, University of Arizona, Tech Rep TR. 2014a:14–04. [Google Scholar]
- Collberg C, Proebsting T, Moraila G, Shankaran A, Shi Z, Warren AM. Measuring reproducibility in computer systems research. Department of Computer Science, University of Arizona, Tech Rep 2014b [Google Scholar]
- Collins FS, Tabak LA. Nih plans to enhance reproducibility. Nature. 2014;505(7485):612. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens A, Van Erp M, Postma M, Pedersen T, Vossen P, Freire N. Offspring from reproduction problems: What replication failure teaches us. Association for Computational Linguistics. 2013:1691–1701. [Google Scholar]
- Hirschman L, Bourne P, Cohen KB, Yu H. Translating Biology: text mining tools that work. 2007 [PMC free article] [PubMed] [Google Scholar]
- Hunter LE. The Processes of Life: An Introduction to Molecular Biology. The MIT Press; 2012. [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of computational and graphical statistics. 1996;5(3):299–314. [Google Scholar]
- Kovacevic J. Acoustics, Speech and Signal Processing, 2007 ICASSP 2007 IEEE International Conference on. Vol. 4. IEEE; 2007. How to encourage and publish reproducible research; pp. IV–1273. [Google Scholar]
- Mobley A, Linder SK, Braeuer R, Ellis LM, Zwelling L. A survey on data reproducibility in cancer research provides insights into our limited ability to translate findings from the laboratory to the clinic. PLoS One. 2013;8(5):e63221. doi: 10.1371/journal.pone.0063221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C, Kaiser GE, Hu L. Properties of machine learning applications for use in metamorphic testing 2008 [Google Scholar]
- Pedersen T. Empiricism is not a matter of faith. Computational Linguistics. 2008;34(3):465–470. [Google Scholar]
- Poprat M, Beisswanger E, Hahn U. Software Engineering, Testing, and Quality Assurance for Natural Language Processing. Columbus, Ohio: Association for Computational Linguistics; 2008. Jun, Building a BioWordNet using WordNet data structures and WordNet’s software infrastructure–a failure story; pp. 31–39. [Google Scholar]
- Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nature reviews Drug discovery. 2011;10(9):712–712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- Proebsting T, Warren AM. Repeatability and benefaction in computer systems research 2015 [Google Scholar]
- Rani J, Shah AR, Ramachandran S. pubmed.mineR: An R package with text-mining algorithms to analyse PubMed abstracts. Journal of bio-sciences. 2015;40(4):671–682. doi: 10.1007/s12038-015-9552-2. [DOI] [PubMed] [Google Scholar]
- Vandewalle P, Kovačević J, Vetterli M. Signal Processing Magazine. 3. Vol. 26. IEEE; 2009. Reproducible research in signal processing; pp. 37–47. [Google Scholar]
- Wei CH, Kao HY, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt441. gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ho J, Murphy C, Kaiser G, Xu B, Chen TY. Quality Software, 2009 QSIC’09 9th International Conference on. IEEE; 2009. Application of metamorphic testing to supervised classifiers; pp. 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ho JW, Murphy C, Kaiser G, Xu B, Chen TY. Testing and validating machine learning classifiers by metamorphic testing. Journal of Systems and Software. 2011;84(4):544–558. doi: 10.1016/j.jss.2010.11.920. [DOI] [PMC free article] [PubMed] [Google Scholar]


