Summary
PGC1α is a pleiotropic co-factor that affects angiogenesis, mitochondrial biogenesis and oxidative muscle remodeling via its association with multiple transcription factors including the master oxidative nuclear receptor ERRγ. To decipher their epistatic relationship, we explored ERRγ gain-of-function in muscle-specific PGC1α/β double knockout mice (PKO). ERRγ-driven transcriptional reprogramming largely rescues muscle damage and improves muscle function in PKO mice, inducing mitochondrial biogenesis, antioxidant defense, angiogenesis, and a glycolytic-to-oxidative fiber-type transformation independent of PGC1α/β. Furthermore, in combination with voluntary exercise, ERRγ gain-of-function largely restores mitochondrial energetic deficits in PKO muscle, resulting in a 5-fold increase in running performance. Thus, while PGCs can interact with multiple transcription factors, these findings implicate ERRs as the major molecular target through which PGC1α/β regulates both innate and adaptive energy metabolism.
Introduction
Originally identified as a co-activator for PPARγ in brown fat (Puigserver et al., 1998), PPAR Gamma Co-activator 1α (PGC1α) is a master regulator of mitochondrial energy metabolism (Mouchiroud et al., 2014). In skeletal muscle, PGC1α and its closely related homologue PGC1β are required for maintaining basal mitochondrial energy metabolism and muscle functions (Rowe et al., 2013; Zechner et al., 2010). In addition, numerous studies have implicated muscle PGC1α in exercise-induced oxidative muscle remodeling, including glycolytic-to-oxidative fiber type transformations, increased vasculature development (angiogenesis), elevated mitochondrial biogenesis and OXPHOS activity, and a shift from glucose to fatty acid as the energy source (Egan and Zierath, 2013; Geng et al., 2010; Handschin et al., 2007; Holloszy and Booth, 1976; Lin et al., 2005; Lin et al., 2002). It is now clear that exercise training activates and induces muscle PGC1α, likely through both AMPK and SIRT1 signaling pathways, which subsequently induces downstream genes involved in oxidative muscle remodeling through its interaction with a number of transcription factors (TFs) (Canto and Auwerx, 2009).
As PGC1α interacts with over 20 TFs, the role and importance of specific downstream effectors of exercise-induced oxidative muscle remodeling is not clear (Villena, 2015). With regard to mitochondrial oxidative metabolism, the estrogen-related receptors ERRα, β, and γ have been suggested as key partners (Fan et al., 2013) (Mouchiroud et al., 2014). In particular, expression of ERRγ is highly specific to oxidative muscle fibers, and is enhanced during exercise-induced oxidative remodeling (Narkar et al., 2011) (Rangwala et al., 2010). Furthermore, due to its intrinsic transcriptional activity ectopic expression of ERRγ in glycolytic muscle is sufficient to drive oxidative muscle remodeling and increase endurance performance in the absence of PGC1α induction (Narkar et al., 2011) (Rangwala et al., 2010) (Gan et al., 2013). However, unlike exercise, ERRγ overexpression does not induce the expression or activation of PGC1α/β, suggesting PGC1-independent transcriptional regulation may naturally occur during ERRγ-driven muscle remodeling. However, while PGC1α/β are not induced, it is possible that basal levels of PGC1α/β are sufficient to drive ERRγ activity. Moreover, it remains to be demonstrated what features of PGC1α induction are dependent on ERR and what aspects of PGC1α/β deficient muscle can be rescued by ERRγ overexpression.
In addition to PGC1α, transcriptional co-repressors such as NCoR and RIP140 also participate in oxidative muscle remodeling induced by exercise, whereby reductions in their expression and the resulting de-repression of downstream TFs activates oxidative gene expression (Seth et al., 2007) (Yamamoto et al., 2011) (Fan et al., 2013). Furthermore, exercise is known to directly induce the expression of TFs such as ERRγ and PPARδ, which can activate their target genes without changes in expression or activity of co-factors (Wang et al., 2004) (Narkar et al., 2011) (Rangwala et al., 2010). Therefore, exercise training could beneficially affect muscle remodeling independent of PGC1α/β. Indeed, it has been shown that adult muscle PGC1α and β are dispensable for endurance exercise-induced oxidative muscle remodeling. However, the short-term (5 day) induction of a mature muscle cell cre driver in this model would allow for the incorporation of wild-type satellite cells during exercise-induced muscle regeneration, confounding the conclusion (Ballmann et al., 2016).
Here we show that ERRγ overexpression largely rescues autonomous muscle damage in a germ-line muscle-specific PGC1α/β double knockout mouse model (PKO). Despite the complete loss of muscle PGC1α/β, ERRγ overexpression induces multiple aspects of oxidative muscle remodeling, including increased mitochondrial biogenesis and fatty acid oxidation, a glycolytic-to-oxidative fiber-type transformation, and angiogenesis. Consistent with this, genomic analyses identify gene networks in these pathways that are directly bound and activated by ERRγ independent of PGC1α/β. Furthermore, when combined with exercise training, ERRγ overexpression almost completely restores the mitochondrial energetic deficiency of PKO muscle. These findings identify ERRγ as a pro-oxidative transcription factor that directly regulates oxidative remodeling in a PGC1-independent fashion, and implicate the ERR subfamily as the dominant mediators of PGC1α in mitochondrial biogenesis and adaptive energy metabolism.
Results
Defects in PGC1-deficient muscle are improved by ERRγ
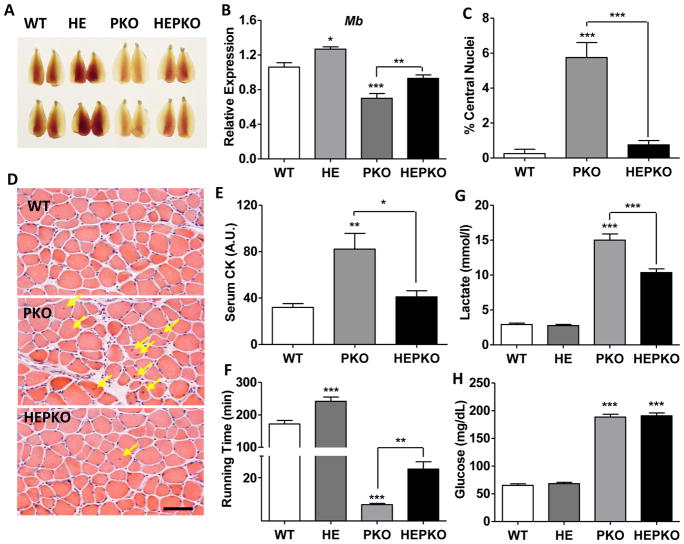
To determine the regulatory hierarchy in oxidative remodeling and mitochondrial function in muscle, we generated muscle-specific PGC1α/β double knockout (PKO) mice with comprehensive depletion of both PGC1α and β in skeletal muscle (>90% depletion, Fig. S1A–B and RNA-seq data in SRP110311). Loss of PGC1α/β caused a pronounced change in the color of the skeletal muscle, suggestive of an oxidative-to-glycolytic transformation (Fig. 1A). Consistent with this color change, the expression of myoglobin (Mb), a major determinant of muscle color, was reduced ~35% in PKO muscle (Fig. 1B). Notably, PKO mice showed evidence of severe muscle damage under sedentary conditions, as seen by the increased number of centrally localized nuclei (Fig. 1C–D), the marked increases in expression of developmental myosin genes Myh3 and Myh8 (Fig. S1C–D), and the elevated serum levels of muscle protein creatine kinase (CK, Fig. 1E). Furthermore, the endurance capacity of PKO mice was severely compromised. When subjected to a treadmill endurance test, the running time of PKO mice was reduced to ~5% of WT mice (Fig. 1F). The dramatic reduction in running performance was likely caused by a sharp elevation in lactate production (Fig. 1G) (Robergs et al., 2004), as circulating glucose – an independent determinant of endurance capacity – was not exhausted in PKO mice (Fan et al., 2017) (Fig. 1H).
Figure 1. ERRγ improves running defect and muscle damage in PGC1-null mice.
(A) Images of Tibialis anterior (TA) muscle from wild-type (WT), ERRγ transgenic (HE), muscle-specific PGC1α/β knockout (PKO), and HEPKO mice; (B) Relative expression of Myoglobin (Mb) in plantaris muscle; (C–D) Percentage centralized-nuclei in muscle and representative H&E staining (E) serum creatine kinase (CK) levels in sedentary mice; (F) running time during low-speed endurance test; (G) blood lactate levels after 6 minutes of endurance running (upon failure of the first mouse); and (H) blood glucose level at failure in endurance test. n=5. Data represent mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar, 50μm. See also Figure S1.
To determine the capacity of ERRγ to rescue muscle defects, we crossed PKO mice with muscle-specific ERRγ transgenic (HE) mice and examined muscle function and running performance between WT, HE, PKO, and HEPKO mice. While overexpression of ERRγ intensified the reddish color of transgenic muscle as previously described (HE compared to WT muscle) (Narkar et al., 2011), it also largely restored WT-like color to PKO muscle (HEPKO vs PKO in Fig. 1A). Consistent changes in the expression of Mb were seen, with the overexpression of ERRγ increasing its level in HEPKO muscle to those similar to WT mice (Fig. 1B). Of note, the muscle damage seen in sedentary PKO mice was almost completely rescued in HEPKO mice, with a marked reduction in centrally localized nuclei and normalized expression of Myh3 and Myh8, as well as circulating CK levels (Fig. 1C–E and Fig. S1C–D). Furthermore, the overexpression of ERRγ nearly tripled the running time of PKO mice (HEPKO, 24 minutes compared to PKO, 8.8 minutes, Fig. 1F), although the persistently elevated lactate levels (not fully rescued by ERRγ overexpression) eventually resulted in pre-exhaustion running failure (Fig. 1F–H).
ERRγ improves mitochondrial energetic defects in PGC1-deficient muscle
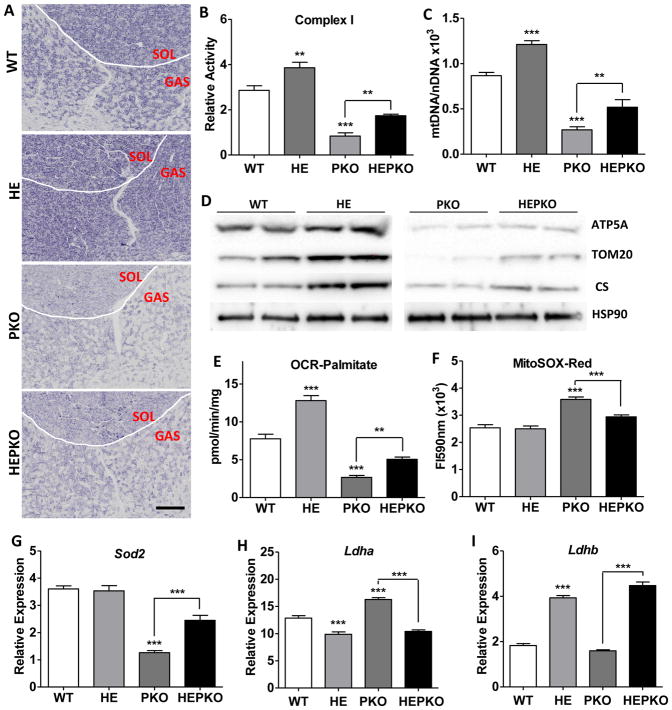
Severe defects in mitochondrial energetics were apparent in muscle from PKO mice, consistent with previous findings (Zechner et al., 2010). In particular, the activities of the electron transport chain (ETC) complexes were dramatically reduced (Fig. 2A–B and S2A–B), as was the TCA cycle enzyme citrate synthase (CS, Fig. S2C). Consistent with this, the expression of core genes involved in OXPHOS and the TCA cycle including Cox6a2, mt-Nd4, Idh2, Atp5a, and Cs, as well as the mtDNA copy number, were all reduced in PKO muscle, with equivalent reductions seen in mitochondrial protein levels (Fig. 2C–D and S2D–F). As a consequence, mitochondrial oxygen consumption rates determined using succinate and palmitate as substrates were reduced 45% and 65%, respectively (Fig. 2E and S2G). Notably, each of these mitochondrial energetic defects was significantly restored by the overexpression of ERRγ. Interestingly, the magnitude of the changes induced by ERRγ in the PKO mice was comparable, and in some cases greater than those induced in WT mice (HEPKO vs PKO compared to HE vs WT, Fig. 2A–E and S2A–G). This suggests that ERRγ-driven improvements in mitochondrial energy metabolism are not contingent on the function of PGC1α/β.
Figure 2. ERRγ improves mitochondrial energetic defects in PGC1-null muscle.
(A) Immunohistochemical staining for mitochondrial complex I activity in cryosections from soleus (SOL) and gastrocnemius (GAS); (B) mitochondrial complex I activity measured in isolated mitochondrial from quadriceps muscle; (C) relative mtDNA copy numbers in plantaris; (D) western blots showing the levels of mitochondrial proteins ATP5A, TOM20, and CS, as well as a cytoplasmic control HSP90 in plantaris; (E) oxygen consumption rates measured in freshly isolated quadriceps mitochondria using palmitoylcarnitine as substrate; (F) ROS measured as MitoSOX fluorescence intensity in freshly isolated quadriceps mitochondria; and (G–I) Relative expression levels of Sod2 (G), Ldha (H), and Ldhb (I) in plantaris. n=5. Data represent mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar, 500μm. See also Figure S2.
Compromised mitochondrial function is commonly associated with elevated production of reactive oxygen species (ROS) (Fan et al., 2012), which is known to cause oxidative muscle damage (Powers et al., 2011). Indeed, mitochondria isolated from PKO muscle showed >40% increase in ROS production compared to those from WT muscle (Fig. 2F), while their membrane potential remained unaffected (Fig. S2H). In addition to the mitochondrial energetic defects described above, the increase in ROS is also attributed to a marked reduction in the expression of the key mitochondrial anti-oxidant gene Sod2 (Fig. 2G). In contrast, the levels of ROS in mitochondria isolated from HEPKO muscle approached those seen in WT muscle (Fig. 2F), consistent with the doubling of Sod2 expression (Fig. 2G) and increases in additional anti-oxidant genes such as Gpx3 upon ERRγ overexpression (Fig. S2I).
The dramatic increase in circulating lactate levels in PKO mice upon exercise (Fig. 1G) suggests that lactate metabolism may be dysregulated in PKO mice. The inter-conversion of lactate and pyruvate is controlled by the relative expression of the two subunits of lactate dehydrogenase (LDH), with LDHA driving lactate formation and LDHB promoting pyruvate production (Summermatter et al., 2013). Indeed, Ldha was significantly increased in PKO muscle (Fig. 2H). Notably, ERRγ overexpression coordinately suppressed Ldha and activated Ldhb expression in both WT and PKO muscle (Fig. 2H–I), consistent with the reduction in serum lactate levels in HEPKO mice (Fig. 1G).
ERRγ directly controls PGC1 target genes in mitochondrial energy metabolism
To understand the privileged interplay between PGC1α/β and ERRγ in regulating the expression of genes involved in mitochondrial energy metabolism, we compared the transcriptomic changes induced in muscle in the above described loss- and gain-of-function mouse models. Consistent with a central role of PGC1α/β in mitochondrial function, the down-regulated gene set in the oxidative soleus (SOL) muscle from PKO mice was enriched in gene ontology (GO) categories related to mitochondrial energetic functions, including OXPHOS, TCA cycle, and fatty acid oxidation (FAO) (Fig. S3A). These same GO categories were enriched in the 1321 genes upregulated by ERRγ overexpression in the glycolytic white quadriceps (WQ) (Fig. S3A), providing mechanistic insight into the functional rescue of the mitochondrial energetic defects in PKO muscle (Fig. 2 and S2), and confirming the overlapping roles of both PGC1α/β and ERRγ in regulating mitochondrial energetic functions. Of note, two GO categories (muscle protein and vasculature development) were uniquely enriched in the ERRγ-upregulated gene set (Fig. S3A), suggesting PGC1α/β play a minimal role in these pathways.
To further delineate the roles of PGC1α/β and ERRγ in muscle, we identified the genome-wide binding sites (cistrome) of ERRγ by performing chromatin immunoprecipitation coupled with high-throughput deep sequencing (ChIP-Seq) assays. While 9387 ERRγ binding sites were identified in HE skeletal muscle, only three peaks were found in WT glycolytic muscle where ERRγ is minimally expressed. Motif analysis identified a highly enriched consensus ERR-response element (ERRE) present in over 40% of the total peaks (P=10−1594) (Fig. S3B and Table S1). The second most enriched motif is the myocyte enhancer factor 2c (MEF2C, P=10−422) (Fig. S3B and Table S1), suggesting a potential interaction between this critical muscle transcription factor and ERRγ (Estrella et al., 2015). The majority of the ERRγ binding sites are located in introns and intergenic regions with about 15% in promoters (Fig. S3B), suggesting much of its transcriptional regulation is mediated through distal enhancers. Interestingly, ~15% of ERRγ binding sites coincide with the H3K27Ac active enhancer mark, however, no correlation with ERRγ-induced gene expression changes is evident, suggesting H3K27Ac-independent transcriptional mechanisms (Fig. S3C). Overlaying the ERRγ cistromic and transcriptomic data revealed that 48% (629 out of 1321) of the genes up-regulated by ERRγ overexpression contain ERRγ binding sites, based on the proximity to the closest transcriptional start site. Notably, this directly regulated ERRγ target gene set is enriched for the same GO categories as found in the total ERRγ-induced gene set, including OXPHOS, TCA cycle, FAO, vasculature, and muscle proteins (Fig. S3D). In addition, the lactate metabolism genes Ldha and Ldhb that are repressed and activated by ERRγ overexpression, respectively, both contain strong ERRγ binding sites and are also direct ERRγ target genes (Fig. S3E). Together the gene expression and the ERRγ DNA binding data sets demonstrate that ERRγ directly binds to and regulates not only PGC1-targeted genes in mitochondrial energetic metabolism but also PGC1-independent genes in vasculature development and fiber-type determination.
ERRγ-specified oxidative remodeling is independent of PGC1α/β
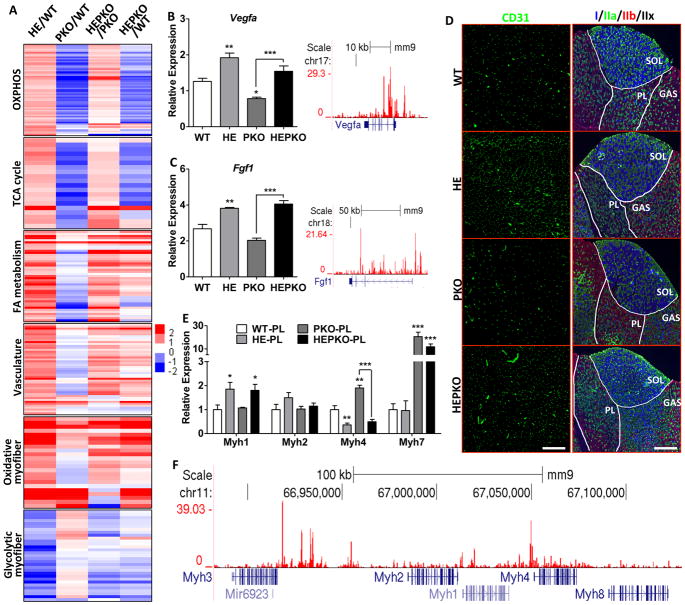
Interestingly, while PGC1α/β are required for the basal expression of mitochondrial energetic genes in OXPHOS, TCA cycle, and FAO (Fig. 3A and S3A), ERRγ overexpression was able to significantly induce the expression of these genes in PGC1α/β-deficient muscle (HEPKO/PKO, Fig. 3A). Furthermore, ERRγ directly activated genes involved in vasculature development including Vegfa and Fgf1, independent of PGC1α/β expression (Fig. 3A–C and S3D). Loss of PGC1α/β minimally affected angiogenic gene expression (Fig. 3A–C and S3A), and the vasculature density (CD31 staining) in PKO muscle was not significantly lower than that in WT (Fig. 3D and S3F), consistent with the lack of vasculature gene enrichment in the downregulated gene set in PKO muscle (Fig. S3A). However, overexpression of ERRγ in PKO muscle induced the expression of angiogenic genes to levels comparable to those in HE muscle (Fig. 3A–C), and the extent of vascularization in HEPKO muscle was similar to that seen in HE muscle (Fig. 3D and S3F), suggesting PGC1α/β are dispensable for both innate and adaptive angiogenesis in skeletal muscle.
Figure 3. ERRγ induces oxidative muscle remodeling independent of PGC1.
(A) Heat maps showing relative changes in expression of selected genes involved in OXPHOS, TCA cycle, FAO, vasculature, oxidative-specific myofiber, and glycolytic-specific myofiber in plantaris; (B–C) Relative expression of Vegfa and Fgf1 in plantaris (left) and genome browser tracks showing ERRγ binding (right); (D) Immunostaining of CD31 for vasculature (left) and Myh I/IIa/IIb for fiber-typing (right); (E) Relative expression of Myh1/2/4/7 in plantaris; and (F) genome browser track showing ERRγ binding at the Myh cluster locus. n=5. Data represent mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars, 300μm (CD31) and 750μm (Myh). See also Figure S3.
ERRγ-induced oxidative muscle remodeling also involves muscle fiber-type changes through its direct regulation of muscle genes, including the different myosin heavy chain (MHC) genes such as Myh1 (intermediate type IIx), Myh2 (oxidative IIa), and Myh4 (glycolytic IIb) (Fig. 3A, 3E, and S3D). Overall ERRγ drives a glycolytic-to-oxidative fiber type switch in different ways dependent on specific muscle types: in the oxidative SOL, ERRγ mainly switches intermediate type IIx fibers to oxidative type IIa (reduced Myh1 and increased Myh2 expression) (Fig. 3D and S3G); while in the more glycolytic WQ and PL (plantaris) it induces a switch from glycolytic IIb to intermediate IIx and oxidative IIa fibers (reduced Myh4 and increased Myh1/2 expression) (Fig. 3D–E and S3H–I). Surprisingly, the slow-twitch oxidative type I fiber (Myh7) is not affected by ERRγ overexpression in all muscles examined (SOL, WA, and PL) (Fig. 3D–E and S3G–I). These effects are consistent with direct and extensive ERRγ binding throughout the genomic locus containing the Myh gene cluster comprised of Myh1, 2, 3, 4, and 8 (Fig. 3F) but not to the loci containing the slow-twitch Myh7 (Fig. S3J).
It has been previously reported that the knockout of muscle PGC1α/β increases type I fibers (Myh7) (Zechner et al., 2010), although the use of a whole-body PGC1α knockout model in that study raised concerns as to whether loss of PGC1α in other tissues contributed to the phenotype (Handschin and Spiegelman, 2011) (Zechner et al., 2011). In our muscle-specific PGC1α/β knockout mice, both SOL and PL consistently show increased type I fibers and Myh7 expression (Fig. 3D–E and S3F–G), clearly demonstrating that loss of PGC1α/β in muscle drives an increase in type I fibers. Although the exact mechanism remains elusive, ERRγ is unlikely involved in this effect since its overexpression has minimal effect on Myh7 expression in both WT and PKO mice (Fig. 3D–E and S3F–H).
Combined effects of exercise and ERRγ improve PKO muscle function
Endurance exercise induces oxidative muscle remodeling and improves performance (Holloszy, 1967). Given our finding that overexpression of ERRγ improved PKO muscle function by inducing genes involved in oxidative muscle remodeling, we explored whether exercise could also rescue muscle dysfunction in PKO mice and whether exercise could combine with the benefits of ERRγ overexpression in the absence of PGC1α/β.
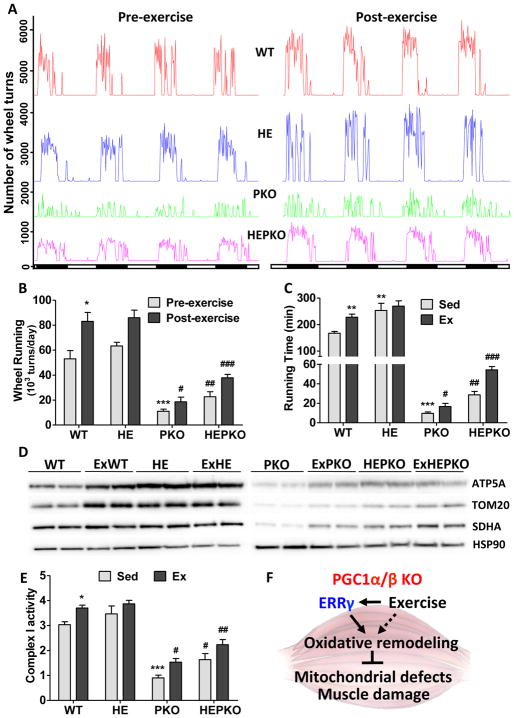
Voluntary wheel running was used as an exercise training regimen for WT, HE, PKO, and HEPKO mice. Consistent with our treadmill studies, the initial wheel-running activities of PKO mice were dramatically reduced compared to WT mice (green and red lines, respectively, Fig. 4A–B). However, after 8 weeks of exercise, significant improvements in voluntary running activity and treadmill sustained running performance were seen in PKO mice (Fig. 4A–C). Mechanistically, while both PGC1α and β as well as ERRα are not affected by exercise in PKO muscle, we found that ERRγ itself is induced and its co-repressor NCOR significantly reduced by exercise (Fig. S4A), potentially contributing to the improvements. Another exercise-related transcriptional co-factor, RIP140, however is not changed (Fig. S4A), which could be due to the relatively low exercise activity of PKO mice. By comparison, these exercise-induced improvements were further potentiated by the overexpression of ERRγ (Fig. 4A–B), such that exercised HEPKO mice ran ~5 times longer than PKO mice on the treadmill running test (55 vs 9.5 minutes, respectively) (Fig. 4C). This improvement is in agreement with the lower serum lactate levels measured after forced running, where the reduction attributed to ERRγ overexpression (sedentary HEPKO compared to PKO mice) was further attenuated by exercise training (Fig. S4B). We found that exercise training and ERRγ overexpression coordinately affected the expression of Ldha and Ldhb in PKO muscle (down- and up-regulated, respectively, Fig. S4C–D), to suppress glycolysis and promote oxidative metabolism to improve muscle function. Furthermore, in PKO muscle, additive effects of exercise and ERRγ overexpression were seen on the levels of mitochondrial proteins (Fig. 4D), mitochondrial complex I activity (Fig. 4E), and the expression of OXPHOS and FAO genes such as Ndufa4 and Acadl (Fig. S4E–F), suggesting the presence of additional transcriptional regulators that are independent of ERRγ and PGC1α/β. It is noteworthy that exercise training fails to further induce oxidative remodeling in the already highly oxidative HE muscle (Fig. 4C–E and S4B–F).
Figure 4. Exercise and ERRγ synergistically improve oxidative functions in PKO muscle.
(A) Running wheel activity of WT (red), HE (blue), PKO (green), and HEPKO (purple) mice before (left) and after (right) 6 weeks of voluntary wheel running; (B) Cumulative daily wheel running from (A); (C) Total running time of sedentary and exercised (8 weeks of voluntary wheel running) mice in endurance test; (D) Western blots showing the levels of mitochondrial proteins ATP5A, TOM20, and SDHA, as well as a cytoplasmic control HSP90 in plantaris; (E) Mitochondrial complex I activity in isolated mitochondrial from quadriceps; and (F) Diagram showing ERRγ- and exercise-induced oxidative muscle remodeling in PKO mice that improves muscle damage and mitochondrial defects. Data represent mean ± SEM. * and # show statistical difference from WT and PKO, respectively. n=5. Data represent mean ± SEM, * and # p < 0.05, ** and ## p < 0.01, *** and ### p < 0.001. See also Figure S4.
Discussion
The intrinsic pleiotropic nature of co-regulatory factors such as PGC1α/β creates a mechanistic challenge in deconstructing the role of individual TF targets. As PGC1 broadly impacts muscle oxidative metabolism and performance it becomes key to decipher principal targets that mediate these effects. The muscle-specific PGC1α/β KO mouse provides a means to identify defining factors by virtue of their ability to rescue defects resulting from PGC1α/β deficiency. Thus, via gain of function studies, we genetically establish a pivotal role for ERRγ in mitochondrial energy metabolism as well as its epistatic relationship with PGC1α/β in driving a broad oxidative platform. For example, while PKO mice show reduced gene expression in major mitochondrial energetic pathways (including OXPHOS, TCA cycle, and FAO metabolism), ERRγ overexpression significantly boosts expression of these genes in PKO muscle, as well as restoring a multitude of the above-mentioned mitochondrial energetic dysfunctions (Fig. S4F). ERRγ overexpression also significantly, though not completely, improves exercise performance in PKO mice (by about 3-fold). Unexpectedly, PGC1 deficiency in muscle shows little change in vasculature or oxidative myofibers such that ERRγ overexpression enhances both into the realm of highly trained animals. This indicates that baseline vasculature and oxidative fiber determination are PGC1-independent pathways (Fig. S4F). Interestingly, voluntary exercise in PKO mice still confers many benefits (Fig. S4F), suggesting that major adaptive functions of exercise, such as angiogenesis, mitochondrial biogenesis and oxidative remodeling can be elicited in absence of PGC1α/β as long as ERRγ-dependent signaling is intact. This suggests that ERRγ synthetic agonists could have substantial and predictable benefits in treatment of muscle disease when exercise (and/or PGC1α/β induction) is not possible or practical.
Differences in phenotype severity in previous PGC1α/β double-knockout models appear to correlate with the efficiencies of the muscle depletion, suggesting that the absolute levels of PGC1α/β are important (Zechner et al., 2010) (Rowe et al., 2013). Indeed, the severely compromised muscle phenotype described here is similar to that shown in Zechner et al., where both models efficiently deplete both PGC1α and β in muscle (Zechner et al., 2010). While injury is considered a product of exercise, PKO mice show evidence of severe muscle damage even under sedentary conditions, indicating a basal role for PGC1α/β in this process. Although the exact mechanism causing muscle damage in PKO mice is not clear, mitochondrial energy deficit and increased ROS production are likely involved (Powers et al., 2011). Notably, such damage is almost completely rescued by ERRγ overexpression, with mitochondrial ROS in HEPKO muscle fully restored to WT levels. This is associated with ERRγ-induced upregulation of antioxidant genes such as Sod2 and Gpx3 and suppression of developmental myosin heavy chain genes Myh3 and Myh8.
Previously, we have shown that the overexpression of ERRγ induces exercise-like oxidative muscle remodeling without engaging changes in the expression level or activity of PGC1α (Narkar et al., 2011). In this study, overexpression of ERRγ in PKO muscle reveals that ERRγ-activated target genes can achieve almost all aspects of oxidative muscle remodeling in the absence of PGC1α/β, including oxidative fiber-type transformation, angiogenesis, and increased mitochondrial energy metabolism. These data highly suggest that PGC1α/β are dispensable for ERRγ-induced oxidative muscle remodeling. Interestingly, both oxidative fiber-type determination and angiogenesis are minimally affected in PKO muscle, with no significant reduction in genes involved in these pathways, suggesting PGC1α/β are not required for their basal functions.
Our RNA-Seq and ChIP-Seq studies identify a network of 629 ERRγ directly controlled genes that are induced by its overexpression. These genes are highly enriched in all of the above-mentioned oxidative muscle remodeling processes, confirming the direct role of ERRγ in transcriptionally regulating oxidative muscle remodeling. PGC1α/β are transcriptional co-activators and require DNA-binding TFs to activate downstream genes. PGC1α is known to drive oxidative muscle remodeling by activating the same set of genes that are ERRγ direct targets (Lin et al., 2002) (Arany, 2008), indicating ERRα/γ as its primary partner TFs during PGC1α-induced oxidative muscle remodeling, although further studies are required to confirm this.
We have also demonstrated that exercise training alone is sufficient to significantly restore mitochondrial energetic dysfunctions and improve running performance in PKO mice (Fig. S4F). Surprisingly, these improvements are comparable to those achieved by ERRγ overexpression. This confirms that exercise has concurrent benefits on oxidative muscle remodeling that are PGC1α/β-independent. Notably, training combined with ERRγ overexpression further boosts mitochondrial energetic functions, resulting in 5-fold increase in running time indicating that ERRγ is itself not simply an exercise surrogate. In summary, these results suggest that while exercise activates PGC1α/β many of its benefits can be achieved independently with notable cross talk with ERRγ and its target genes (Fig. 4F).
METHODS
Mouse models
PKO (Cre+Pgc1afl/flPgc1bfl/fl) mice were generated by crossing Pgc1afl/fl (JAX 009666) with Pgc1bfl/fl ((Sonoda et al., 2007) (Wei et al., 2010)) and ACTA1-Cre (JAX 006139) mice. HE mice (ACTA1-Esrrg (Narkar et al., 2011)) were also crossed to Pgc1afl/fl and Pgc1bfl/fl mice to generate HE+Pgc1afl/flPgc1bfl/fl mice. Cre+Pgc1afl/flPgc1bfl/fl mice were then crossed to HE+Pgc1afl/flPgc1bfl/fl mice to generate WT (Cre−HE−Pgc1afl/flPgc1bfl/fl), HE (Cre−HE+Pgc1afl/flPgc1bfl/fl), PKO (Cre+HE−Pgc1afl/flPgc1bfl/fl), and HEPKO (Cre+HE+Pgc1afl/fl Pgc1bfl/fl) mice.
For exercise training, 2-month-old mice were transferred to cages with low-profile wireless running wheels (ENV-044, Med Associates Inc.) and housed for 2 months. Daily running activity was monitored and recorded with the wheel manager software.
Prior to exhaustion running, mice were pre-adapted to the treadmill for 5 minutes per day for 3 days at 5 meter/min. After 1 day recovery mice were subjected to the run-to-exhaustion test, which included running for 1 minute each at speeds of 5, 6, 7, 8 and 9 meter/min, 30 minutes at 10 meter/min, 2 minutes each at speeds of 11, 12, 13, 14 and 15 meter/min, and run at 15 meter/min till exhaustion. Blood lactate was measured in all mice using a Lactate Scout analyzer (EKF Diagnostic) with tail bleed upon exhaustion of the first mouse. Blood glucose was measured using a glucometer (Nova max) in individual mice upon exhaustion.
All animal protocols were reviewed and approved by the Institute of Animal Care and Use Committee (IACUC) of the Salk Institute, and studies were conducted in compliance with institutional and national guidelines.
Quantification and statistical analysis
ANOVA and post-hoc analysis were used to evaluate statistical significance in all studies.
Detailed experimental procedures are described in supplemental material.
Supplementary Material
Figure S1. ERRγ improves running defect and muscle damage in PGC1-null mice, related to Fig. 1.
Figure S2. ERRγ improves mitochondrial energetic defects in PGC1-null muscle, related to Fig. 2.
Figure S3. ERRγ induces oxidative muscle remodeling in the absence of PGC1, related to Fig. 3.
Figure S4. Exercise and ERRγ synergistically improve oxidative functions in PKO muscle, related to Fig. 4.
Table S1. Motif analysis for ERRγ ChIP-seq, related to Fig. 3.
Table S2. List of primer sequences, related to methods.
Document S1. Supplemental experimental procedures.
Acknowledgments
We thank C. Brondos and E. Ong for administrative assistance, Y. Dai and J. Nery for assistance with library preparation and sequencing, and H. Juguilon and J. Alvarez for technical assistance. This work was funded by grants from Office of Naval Research (ONR N00014-16-1-3159), NIH (DK057978, HL105278, HL088093, ES010337 and CA014195) and National Health and Medical Research Council of Australia Project Grants 512354 and 632886 (C.L. and M.D.), as well as The Leona M. and Harry B. Helmsley Charitable Trust (#2017PG-MED001), the Samuel Waxman Cancer Research Foundation, Ipsen/Biomeasure and the Glenn Foundation for Medical Research. R.M.E is an investigator of the Howard Hughes Medical Institute and March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute. Research reported in this publication was supported by the National Institute Of Environmental Health Sciences of the National Institutes of Health under Award Number P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was sponsored by the Department of the Navy, Office of Naval Research through grant N00014-16-1-3159. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Office of Naval Research.
Footnotes
Author Contributions
W.F., M.D., and R.M.E. designed the study. W.F., N. He, C.S.L., Z.W., N. Hah, W.W., and M.H. conducted all experiments. C.L. and R.T.Y. analyzed genomic data. W.F., A.R.A., M.D., and R.M.E. drafted and revised the manuscript.
Accession Numbers
RNA-Seq and ChIP-Seq data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database, Accession # SRP110311.
References
- Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Current opinion in genetics & development. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmann C, Tang Y, Bush Z, Rowe GC. Adult expression of PGC-1alpha and -1beta in skeletal muscle is not required for endurance exercise-induced enhancement of exercise capacity. American journal of physiology Endocrinology and metabolism. 2016;311:E928–E938. doi: 10.1152/ajpendo.00209.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current opinion in lipidology. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Estrella NL, Desjardins CA, Nocco SE, Clark AL, Maksimenko Y, Naya FJ. MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. The Journal of biological chemistry. 2015;290:1256–1268. doi: 10.1074/jbc.M114.589838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Atkins AR, Yu RT, Downes M, Evans RM. Road to exercise mimetics: targeting nuclear receptors in skeletal muscle. Journal of molecular endocrinology. 2013;51:T87–T100. doi: 10.1530/JME-13-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Lin CS, Potluri P, Procaccio V, Wallace DC. mtDNA lineage analysis of mouse L-cell lines reveals the accumulation of multiple mtDNA mutants and intermolecular recombination. Genes & development. 2012;26:384–394. doi: 10.1101/gad.175802.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Waizenegger W, Lin CS, Sorrentino V, He MX, Wall CE, Li H, Liddle C, Yu RT, Atkins AR, et al. PPARdelta Promotes Running Endurance by Preserving Glucose. Cell Metab. 2017;25:1186–1193. e1184. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z, Rumsey J, Hazen BC, Lai L, Leone TC, Vega RB, Xie H, Conley KE, Auwerx J, Smith SR, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. The Journal of clinical investigation. 2013;123:2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng T, Li P, Okutsu M, Yin X, Kwek J, Zhang M, Yan Z. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. American journal of physiology Cell physiology. 2010;298:C572–579. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. The Journal of biological chemistry. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. PGC-1 coactivators and the regulation of skeletal muscle fiber-type determination. Cell Metab. 2011;13:351. doi: 10.1016/j.cmet.2011.03.008. author reply 352. [DOI] [PubMed] [Google Scholar]
- Holloszy JO. Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. The Journal of biological chemistry. 1967;242:2278–2282. [PubMed] [Google Scholar]
- Holloszy JO, Booth FW. Biochemical adaptations to endurance exercise in muscle. Annual review of physiology. 1976;38:273–291. doi: 10.1146/annurev.ph.38.030176.001421. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metabolism. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Ji LL, Kavazis AN, Jackson MJ. Reactive oxygen species: impact on skeletal muscle. Comprehensive Physiology. 2011;1:941–969. doi: 10.1002/cphy.c100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, Beaulieu V, Gao J, Turner G, Markovits J. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. The Journal of biological chemistry. 2010;285:22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robergs RA, Ghiasvand F, Parker D. Biochemistry of exercise-induced metabolic acidosis. American journal of physiology Regulatory, integrative and comparative physiology. 2004;287:R502–516. doi: 10.1152/ajpregu.00114.2004. [DOI] [PubMed] [Google Scholar]
- Rowe GC, Patten IS, Zsengeller ZK, El-Khoury R, Okutsu M, Bampoh S, Koulisis N, Farrell C, Hirshman MF, Yan Z, et al. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell reports. 2013;3:1449–1456. doi: 10.1016/j.celrep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, Mobberley M, Ryder TA, Rowlerson A, Scott J, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 2007;6:236–245. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summermatter S, Santos G, Perez-Schindler J, Handschin C. Skeletal muscle PGC-1alpha controls whole-body lactate homeostasis through estrogen-related receptor alpha-dependent activation of LDH B and repression of LDH A. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8738–8743. doi: 10.1073/pnas.1212976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. The FEBS journal. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Wang X, Yang M, Smith LC, Dechow PC, Sonoda J, Evans RM, Wan Y. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11:503–516. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, Heligon C, Barish GD, Desvergne B, Evans RM, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner C, Lai L, Zechner JF, Geng T, Yan Z, Rumsey JW, Collia D, Chen Z, Wozniak DF, Leone TC, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metabolism. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner C, Leone TC, Kelly DP. Response to Handschin and Spiegelman. Cell Metabolism. 2011;12:352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ERRγ improves running defect and muscle damage in PGC1-null mice, related to Fig. 1.
Figure S2. ERRγ improves mitochondrial energetic defects in PGC1-null muscle, related to Fig. 2.
Figure S3. ERRγ induces oxidative muscle remodeling in the absence of PGC1, related to Fig. 3.
Figure S4. Exercise and ERRγ synergistically improve oxidative functions in PKO muscle, related to Fig. 4.
Table S1. Motif analysis for ERRγ ChIP-seq, related to Fig. 3.
Table S2. List of primer sequences, related to methods.
Document S1. Supplemental experimental procedures.