Abstract
The human cytomegalovirus protein US11 induces the dislocation of MHC class I heavy chains from the endoplasmic reticulum (ER) into the cytosol for degradation by the proteasome. With the use of a fractionated, permeabilized cell system, we find that US11 activity is needed only in the cell membranes and that additional cytosolic factors are required for heavy chain dislocation. We identify ubiquitin as one of the required cytosolic factors. Cytosol depleted of ubiquitin does not support heavy chain dislocation from the ER, and activity can be restored by adding back purified ubiquitin. Methylated-ubiquitin or a ubiquitin mutant lacking all lysine residues does not substitute for wild-type ubiquitin, suggesting that polyubiquitination is required for US11-dependent dislocation. We propose a new function for ubiquitin in which polyubiquitination prevents the lumenal domain of the MHC class I heavy chain from moving back into the ER lumen. A similar mechanism may be operating in the dislocation of misfolded proteins from the ER in the cellular quality control pathway.
INTRODUCTION
The MHC class I complex binds intracellularly derived peptides and presents them at the cell surface to the cytotoxic T cells of the immune system. The MHC class I heavy chain contains the peptide-binding site and is a type I transmembrane protein with a large luminal/extracellular domain and a short cytosolic tail. Human class I heavy chains have a molecular mass of 43 kDa and contain a single N-linked glycan. Human cytomegalovirus (HCMV) evades detection by the immune system by targeting class I heavy chains for destruction soon after they have been synthesized. To do this, HCMV seems to co-opt the quality control process by which the cell normally disposes of misfolded or misassembled secretory proteins in the endoplasmic reticulum (ER) (Wiertz et al., 1996a,b; Shamu et al., 1999). Like proteins targeted for destruction by the ER quality control pathway, class I heavy chains in HCMV-infected cells are reverse-translocated, or dislocated, across the ER membrane into the cytosol, where they undergo proteasome-dependent degradation. Proteins are probably dislocated across the membrane via the Sec61 complex, which forms the channel through which they originally entered the ER (Wiertz et al., 1996; Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999).
There are two HCMV proteins responsible for targeting MHC class I heavy chains for destruction, US11 and US2 (Jones et al., 1995). Either protein, when stably expressed in human astrocytoma cell lines, causes constitutive destruction of class I heavy chains. The process is very rapid: the half-life of the heavy chains is <3 min. In the presence of proteasome inhibitors, degradation is inhibited, but the heavy chains are still quickly dislocated into the cytosol, where they are deglycosylated by the activity of an N-glycanase and are found as soluble species (Wiertz et al., 1996a,b). US11 and US2 are small (<30 kDa), transmembrane glycoproteins that are localized to the ER. Both bind to class I heavy chains (Wiertz et al., 1996a; Story et al., 1999), but the mechanism by which they act to effect heavy chain dislocation is unknown.
The actual dislocation process might involve the function of ubiquitin. In human cells expressing US11, class I heavy chains are ubiquitinated before they are degraded (Shamu et al., 1999). Because ubiquitinated heavy chains are associated with membranes, ubiquitination may occur early during dislocation. However, it has not yet been shown that ubiquitination is actually required for US11-dependent dislocation and degradation. Moreover, it is unclear whether ubiquitination would simply signal degradation of the heavy chain after its dislocation into the cytosol, act as a targeting signal during the dislocation process (by analogy with ubiquitin's role as a signal during endocytosis [Hicke, 1999]), or possibly function in a novel way.
A robust system that can be dissected biochemically is required to understand in detail the mechanisms underlying protein dislocation across the ER membrane. We have previously reported the development of a permeabilized cell system for dislocation that allows cytosolic factors to be manipulated (Shamu et al., 1999). The system recapitulates important features of US11-dependent dislocation and degradation of MHC class I heavy chains found in living cells. Most importantly, the half-life of MHC heavy chains in permeabilized US11-expressing cells is ∼10 min, much shorter than the half-lives of misfolded proteins degraded by the standard ER quality control pathway (Finger et al., 1993; Yuk and Lodish, 1993; Ward et al., 1995; Biederer et al., 1996; Yu et al., 1997). With the use of this permeabilized cell system, we show here that US11 activity is required in the cell membranes and that cellular proteins from the cytosol are required for heavy chain dislocation. We also show that ubiquitin-depleted cytosol does not support heavy chain dislocation across the ER membrane. Purified wild-type ubiquitin rescues the activity of ubiquitin-depleted cytosols, but ubiquitin unable to form polyubiquitin chains does not. Thus, polyubiquitination is required for the US11-dependent dislocation of class I heavy chains, and not just for their degradation by proteasomes. A similar mechanism may apply for degradation of ER proteins in the cellular quality control pathway.
MATERIALS AND METHODS
Cells and Cell Culture
Control and US11-expressing U373MG human astrocytoma cells (Jones et al., 1995) were cultured as described previously (Wiertz et al., 1996a).
Antibodies
Anti-heavy chain (αHC), anti-US11, and anti-Ub (αUb) antibodies were described previously (Shamu et al., 1999). Anti-hsc70 mouse monoclonal antibodies (1B5) were purchased from Stressgen (Victoria, British Columbia, Canada). Antibodies directed against the α1 (iota/p27) subunit of the 20S proteasome were purchased from ICN Pharmaceuticals (Aurora, OH). Antibodies against the S7 (Mss1) ATPase subunit of the 19S proteasome cap were purchased from Affiniti Research Products (Mamhead, United Kingdom).
Preparation of Bovine Liver Cytosol
Bovine livers were obtained immediately after slaughter. The tissue was kept in ice-cold homogenization buffer (50 mM HEPES pH 7.5, 80 mM KCl, 15 mM NaCl, 3 mM MgCl2, 250 mM sucrose, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride; 0.2 mM spermine, 0.5 mM spermidine) and cut into small pieces to remove blood vessels and connective tissue. The resulting 500 g of liver was washed with 2 liters of homogenization buffer to remove as much blood as possible. Homogenization buffer (∼750 ml), containing extra protease inhibitors (10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastatinal, 1 μg/ml pepstatin), was added. The liver was crudely homogenized in a Polytron blender and then further homogenized with the use of a Potter homogenizer, rotating at ∼1000 rpm. The homogenate was centrifuged at 9000 rpm in a Beckman JA-10 rotor for 15 min. The supernatant was filtered through eight layers of cheesecloth, recentrifuged in the JA-10 rotor at 9000 rpm for 30 min, and filtered through cheesecloth a second time. The clarified supernatant was then centrifuged in a Beckman Ti45 rotor at 45,000 rpm for 3 h. The cytosol supernatant was removed carefully, avoiding the top fat layer and the loose membrane pellet. The protein concentration of this liver cytosol was 20–30 mg/ml, as measured with the use of a Micro BCA Protein Assay (Pierce, Rockford, IL).
Permeabilized Cells and Heavy Chain Dislocation Assays
MHC class I heavy chain dislocation assays were carried out on membrane pellets isolated from permeabilized astrocytoma cells. Cells were labeled intact for 3 min with [35S]methionine and cysteine as described previously (Shamu et al., 1999). They were washed once with ice-cold phosphate-buffered saline, containing 0.9 mM CaCl2, and permeabilized on ice for 10 min in permeabilization buffer (PB; 25 mM HEPES pH 7.3, 115 mM potassium acetate, 5 mM sodium acetate, 2.5 mM MgCl2, 0.5 mM EGTA) containing 0.025% digitonin, an ATP-regenerating system, and protease inhibitors (Shamu et al., 1999). Cytosol was squeezed out of the permeabilized cells by centrifuging in a microfuge at 14,000 rpm at 4°C for 10 min. This “squeezed-out” astrocytoma cytosol was 3–5 mg/ml of protein and contained proteins with a wide range of molecular masses (Figure 1A). Cytosol made in this way from control astrocytoma cells was used in the experiment shown in Figure 1C. The permeabilized cell membrane pellets were washed once with ice-cold PB containing the ATP-regenerating system and protease inhibitors but no digitonin and then resuspended in ice-cold permeabilization buffer (containing an ATP-regenerating system and protease inhibitors but no digitonin) or in cytosol. Dislocation assays were started by incubating the resuspended membranes at 37°C. Samples were taken at various time points and lysates were made as described below.
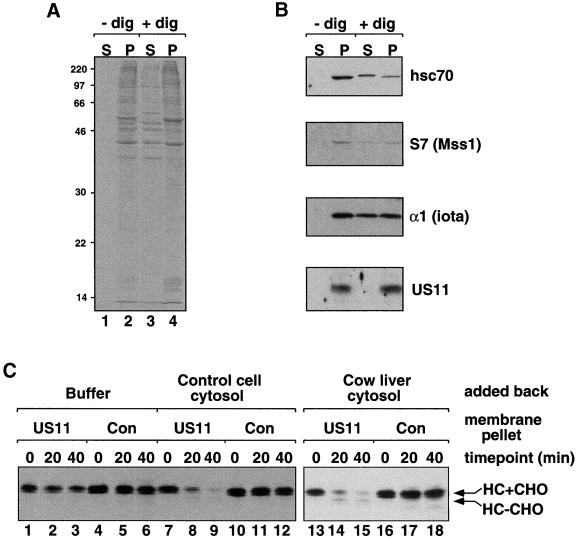
Figure 1.
A variety of different mammalian cytosols support US11-dependent dislocation and degradation of class I heavy chain. Astrocytoma cells expressing US11 were permeabilized (+dig) or mock-permeabilized (−dig) according to the procedure described in MATERIALS AND METHODS. The squeezed-out cytosol fractions (S) and the membrane pellet fractions (P) were run on SDS gels and either Coomassie stained (A) or processed for immunoblotting (B). Migration of molecular mass markers is indicated at the left of the Coomassie-stained gel. The blots were probed with antibodies directed against cytosolic Hsc70 protein and the ER membrane protein US11, as well as with antibodies against the proteasome subunits α1 (iota) and S7(Mss1). (C) MHC class I heavy chain dislocation assays were carried out with the use of membrane pellets from [35S]methionine-labeled permeabilized US11-expressing or control cells. The membranes were resuspended in buffer (lanes 1–6), in cytosol squeezed out from control astrocytoma cells (lanes 7–12), or in cow liver cytosol (lanes 13–18). The bands corresponding to glycosylated heavy chain (HC+CHO) and deglycosylated heavy chain (HC-CHO) are labeled.
In several experiments, time points from the dislocation assays were processed further before lysate preparation. Fractionation of samples from dislocation assays was carried out in the experiments shown in Figures 3B and 4. At the 30-min time point, two samples were taken from each dislocation assay. Lysis buffer was added immediately to 1 sample (total). The second sample was centrifuged in a microfuge at 14,000 rpm at 4°C for 10 min. The supernatant was removed and saved. The pelleted fraction was resuspended in PB containing the ATP-regenerating system and protease inhibitors but no digitonin. Lysates of each fraction (total, supernatant, and pellet) were made and immunoprecipitations were carried out, as described below. Proteolysis protection experiments shown in Figure 3C were carried out on samples from the dislocation assays after a protocol described previously (Shamu et al., 1999).
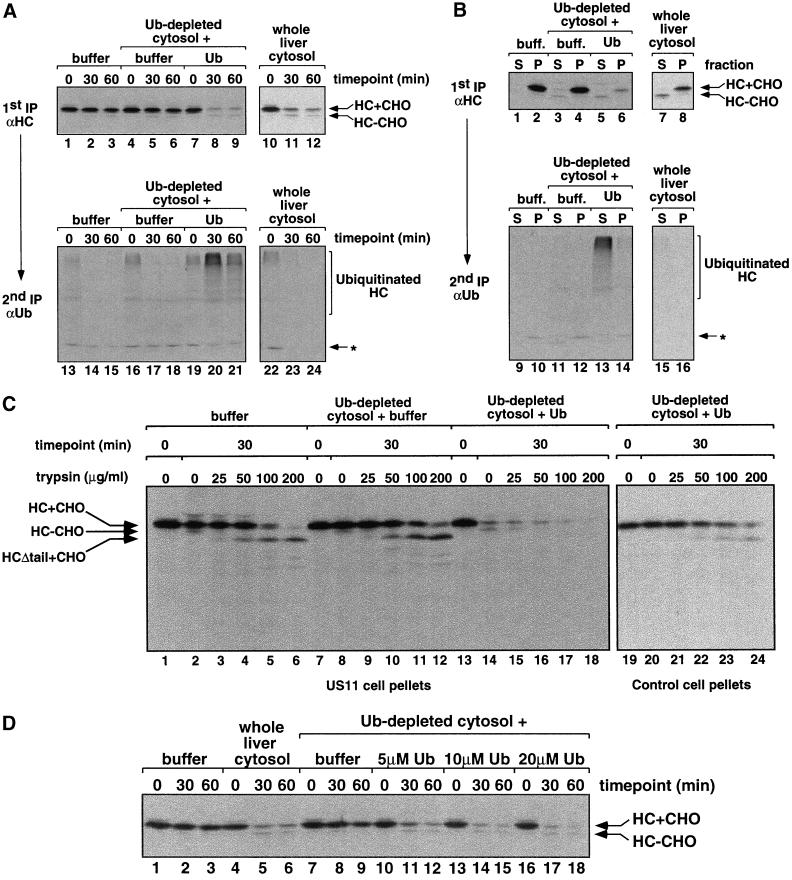
Figure 3.
Ubiquitin is required for the dislocation and degradation of MHC class I heavy chain. (A) Membrane pellets were prepared from [35S]methionine-labeled permeabilized US11 cells, resuspended as indicated in buffer or cytosol mix. The cytosols used were either whole liver cytosol, Ub-depleted cytosol with permeabilization buffer added, or Ub-depleted cytosol with 20 μM bovine ubiquitin added. Heavy chain dislocation assays were carried out as described in MATERIALS AND METHODS. A first immunoprecipitation was carried out with αHC serum followed by incubation with Staph A. Bound material was eluted with SDS and one-third of each sample was analyzed directly by SDS-PAGE and autoradiography (lanes 1–12). The remaining two-thirds of each sample was diluted into NP-40 buffer and reimmunoprecipitated with αUb before SDS-PAGE (lanes 13–24). Background bands that precipitate with Staph A alone are identified by the asterisk (Shamu et al., 1999). Exposure time of the gels for lanes 1–12 is 16 h and for lanes 13–24 is 3 wk. (B) Samples taken at the 30-min time point from the experiment described in A were fractionated by centrifugation before lysate preparation into supernatant (S) and pellet (P) fractions (see MATERIALS AND METHODS). Immunoprecipitations with αHC and αUb serum were carried out as described in A. Exposure time of the gels for lanes 1–8 is 24 h and for lanes 9–16 is 3 wk. (C) The heavy chain in US11 cell pellets is protected from trypsin proteolysis unless complete liver cytosol is added. [35S]methionine-labeled membrane pellets were prepared from permeabilized US11 and control cells, resuspended in buffer (with 1 μM ubiquitin aldehyde) or the indicated cytosol mix, and dislocation assays were carried out. At 0 and 30 min, samples from the dislocation assays were treated on ice with trypsin at the final concentrations indicated. Denaturing SDS lysates were made and MHC class I heavy chains were immunoprecipitated with αHC serum. Where added in the cytosol mix (lanes 13–24), bovine ubiquitin was at 20 μM. Proteolysis produced a glycosylated heavy chain species that lacks its cytosolic tail (HCΔtail+CHO). (D) Heavy chain dislocation assays were carried out as described above with the use of permeabilized US11 cell membranes resuspended in buffer, whole liver cytosol, or Ub-depleted cytosol supplemented with permeabilization buffer or increasing concentrations of bovine ubiquitin.
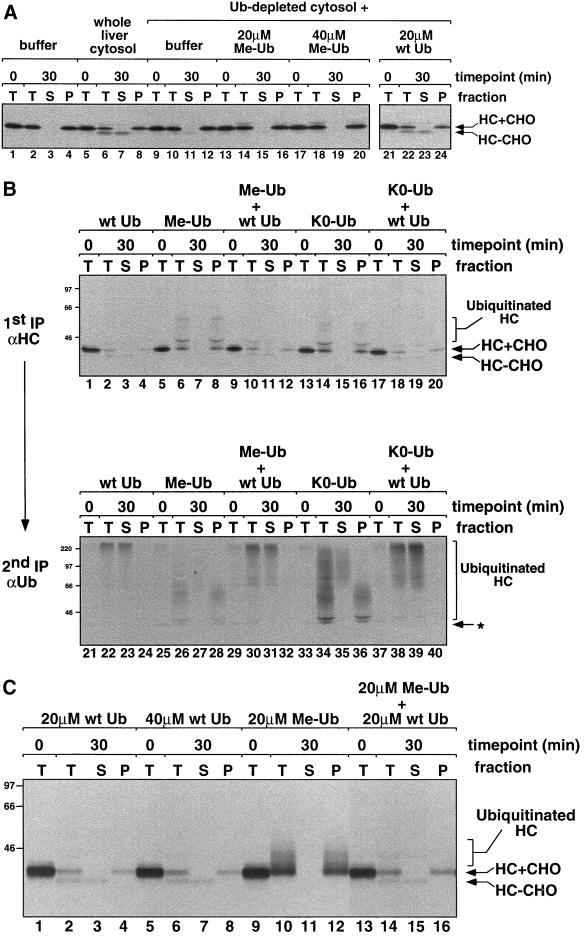
Figure 4.
Polyubiquitination is required for heavy chain dislocation and degradation. (A) Heavy chain dislocation assays were carried out with the use of permeabilized US11 cell pellets resuspended in buffer alone (lanes 1–4), in whole liver cytosol (lanes 5–8), or in Ub-depleted cytosol supplemented with permeabilization buffer, 20 μM Me-Ub, 40 μM Me-Ub, or 20 μM wild-type (wt) bovine ubiquitin (lanes 9–24). Fractionation of the 30-min time points before lysate preparation was carried out as described in MATERIALS AND METHODS. In this experiment, additional Uba1p E1 enzyme and ubiquitin aldehyde were not added to the Ub-depleted cytosol after it was thawed for use in the dislocation assay (see MATERIALS AND METHODS). (B) Heavy chain dislocation assays were carried out with the use of permeabilized US11 cell pellets resuspended in Ub-depleted cytosol supplemented with 20 μM wt Ub, Me-Ub, or K0- Ub (which lacks all lysine residues), or a mixture of 20 μM each wt Ub and Me-Ub or K0-Ub. Note that additional Uba1p E1 enzyme and ubiquitin aldehyde were added to the Ub-depleted cytosol after it was thawed for use in this experiment. Immunoprecipitations with αHC and αUb serum were carried out as described in Figure 3A. The low molecular weight ubiquitinated heavy chains seen in the presence of Me-Ub or K0-Ub, running between 46 and 66 kDa, appear to be less efficiently immunoprecipitated with the αUb serum than the more highly polyubiquitinated heavy chains. Exposure time of the gel for lanes 1–20 is 20 h and for lanes 21–40 is 3 wk. (C) A portion of each αHC immunoprecipitation from the experiment shown in lanes 1–20 of B was eluted from the Staph A with SDS and DTT, diluted into NP-40 buffer, and reimmunoprecipitated with αHC serum to confirm that the immunoprecipitated bands were indeed heavy chains. The samples were run on very long polyacrylamide gels in wide lanes to enhance separation of the different heavy chain species.
Lysate Preparation and Immunoprecipitation
Nondenaturing NP-40 lysates were made from samples taken during dislocation assays as described previously (Shamu et al., 1999), except that SDS and DTT were added to all lysates after the clarifying spin to final concentrations of 0.1% and 0.2 mM, respectively. Denaturing SDS lysates, made after the proteolysis protection experiment (Figure 3C), were as described previously (Shamu et al., 1999). All immune complexes were recovered by precipitation with fixed Staphylococcus aureus (Staph A) bacteria. Fluorography of gels was carried out as described by Ploegh (1995).
Ubiquitin Reagents
Bovine ubiquitin was purchased from Sigma (St. Louis, MO). The bovine ubiquitin was methylated (Me-Ub) according to the protocol described by Hersko and Heller (1985). Ubiquitin with all lysine residues replaced by arginine (K0-Ub) was purified in recombinant form from bacteria as described previously (You et al., 1999).
Ubiquitin was modified with the fluorescent probe Oregon Green by derivatizing an engineered ubiquitin in which the amino acid sequence MCHHHHHH has been fused to the N terminus of human ubiquitin protein. The engineered ubiquitin fusion was obtained by expression in bacteria and purified by metal-affinity chromatography. Labeling was carried out by adding 0.8 mg of 2′,7′-difluorofluorescein (Oregon Green) iodoacetamide “mixed isomer” (Molecular Probes, Eugene, OR) in 50 μl of dimethyl sulfoxide to a 1-ml solution, containing 2 mg of MCH6-ubiquitin in 0.05 M HEPES pH 7.0. The reaction was incubated at room temperature for 4 h. At the end of the incubation, protein was separated from other reaction components by filtration on a P10 column (Pierce). Analysis of the labeled protein by SDS-PAGE revealed a small percentage of labeled protein that migrated with slower mobility than ubiquitin. These contaminants were removed by size exclusion chromatography on Superdex G75 16/60 (Amersham Pharmacia Biotech, Arlington Heights, IL). The purified labeled protein was stored in −70°C until use. Incorporation of the fluorophore Oregon Green into MCH6-ubiquitin was indicated by the presence of fluorescence in the purified protein, with excitation and emission maxima at 487 and 515 nm, respectively. Under our reaction conditions, incorporation of label was estimated to be 0.7 mol/mol of protein, calculated by assuming a molar extinction coefficient of 7.1 × 104 cm−1M−1 at 491 nm for the fluorescent label and protein determination by Bradford analysis.
The active site serine mutant of E214K was obtained by replacing the C88 codon TGT in the human Ubc2B gene (GenBank accession NM_003337) with TCC to code for serine. The mutated sequence was inserted between the BamHI and EcoRI sites in the vector pGEX-4T2 (Amersham Pharmacia Biotech) to express the mutant glutathione (GST)-tagged SerE214K in bacteria. The fusion protein was purified by glutathione-affinity chromatography. The concentration of SerE214K was calculated by active-site titration with Oregon Green-labeled ubiquitin. The Saccharomyces cerevisiae ubiquitin-activating enzyme Uba1p was purified from yeast cells that harbor a plasmid that encodes a polyHis-tagged UBA1 gene (kindly provided by Jurgen Dohmen, Heinrich-Heine-Universitat, Dusseldorf, Germany). The enzyme was purified by metal-chelation chromatography, followed by ubiquitin-affinity chromatography. E1 activity was tested by its ability to form thioester bonds with ubiquitin.
Depleting Ubiquitin from Liver Cytosol
Ubiquitin was depleted from cow liver cytosol with the use of the recombinant GST-tagged ubiquitin-conjugating enzyme GST-SerE214K, as described above. The depletion mixture contained liver cytosol, 16 μM SerE214K, 0.2 μg/ml Uba1p, and an ATP-regenerating system (Feldman et al., 1997), and was incubated at 37°C for 10–15 min. Ubiquitin aldehyde (Calbiochem, San Diego, CA), an inhibitor of deubiquitinating enzymes, was then added to a final concentration of 1 μM and the depletion mixture was incubated in batch with glutathione Sepharose beads (Amersham Pharmacia Biotech) at 4°C for 15 min. The beads, bound to ubiquitin-conjugated SerE214K, were pelleted by centrifugation. The resulting supernatant was collected as the ubiquitin-depleted (Ub-depleted) cytosol and was stored in aliquots at −80°C. When thawed for dislocation assays, additional ATP-regenerating system components were added to the Ub-depleted cytosol along with the buffer or purified ubiquitin protein. In all but the experiment shown in Figure 4A, Uba1p (0.26 μg/ml), and ubiquitin aldehyde (1 μM) were also added to the thawed Ub-depleted cytosol before use.
RESULTS
US11-dependent Dislocation of MHC Class I Heavy Chain from ER Requires Cytosolic Factors
To study the mechanism of US11-dependent class I heavy chain movement from the ER to the cytosol, we used a permeabilized cell system. Astrocytoma cells were labeled with [35S]methionine, permeabilized with the use of low concentrations of the mild detergent digitonin, as described previously (Shamu et al., 1999), and then centrifuged in a microfuge. Centrifugation separated permeabilized cells into a membrane pellet and a cytosol fraction (Figure 1A). The cell membrane pellet contained, among many other proteins, US11 (Figure 1B) and the dislocation substrate MHC class I heavy chain integrated into the ER membrane (see below). The cytosol fraction contained a large number of proteins with a wide range of molecular masses, suggesting that it comprises a significant portion of the cytosol (Figure 1A, lane 3). However, some of the cytosolic chaperone Hsc70 and the 26S proteasome fractionated with the membrane pellet, as determined by immunoblot with antibodies to Hsc70 and to the proteasome subunits α1 (iota), α2 (C3), and S7 (Mss1) (Figure 1B; our unpublished data). Thus, permeabilization and centrifugation results in release of a substantial fraction, but not of all, cytosolic proteins.
We next asked whether the proteins present in the membrane pellet were sufficient to promote US11-dependent dislocation and degradation of heavy chain. [35S]methionine-labeled membranes from US11-expressing and control cells were resuspended in buffer or in a variety of different mammalian cytosols. The extent of heavy chain dislocation and degradation was assayed for each condition by incubating the resuspended membranes at 37°C and observing the fate of the heavy chain over time by immunopreciptation. We found that the heavy chain was not dislocated or degraded efficiently from US11-containing membranes resuspended in buffer alone (Figure 1C, lanes 1–3). In contrast, heavy chain dislocation and degradation were observed when the membranes were resuspended in mammalian cytosol from control astrocytoma cells (lanes 7–9), cow liver (lanes 13–15), or cow brain (our unpublished data). Importantly, none of the conditions supported degradation of heavy chain from control cell membranes not containing US11. Moreover, in the absence of cytosol, heavy chains remained integrated in the ER membrane during the course of the dislocation assay (see below). Cytosol from astrocytoma cells, cow liver, and cow brain had approximately the same dislocation and degradation activity, although more of a lower molecular weight heavy chain species accumulated when US11 cell pellets were incubated in liver cytosol (Figure 1C, lanes 14 and 15). This species is deglycosylated heavy chain (our unpublished data). Because deglycosylated heavy chain accumulates in US11-expressing astrocytoma cells only in the presence of proteasome inhibitors (Wiertz et al., 1996b), its accumulation under these conditions suggests that the activity of the proteasomes in the liver cytosol was not well preserved during cytosol preparation.
Taken together, these results demonstrate that the proteins present in US11 cell membrane pellets are not sufficient to promote dislocation and degradation of MHC class I heavy chain and that cytosolic factors are required. These factors are inactivated by a 5-min incubation at 65°C and are larger than 3.5 kDa, as determined by dialysis (our unpublished data). Moreover, because cytosols from cells lacking US11 support efficient dislocation and degradation of heavy chain, US11 is required in the membrane fraction only.
Ubiquitin Is Required for Class I Heavy Chain Dislocation
US11-dependent heavy chain degradation is accompanied by the formation of ubiquitinated heavy chains (Shamu et al., 1999). To test whether ubiquitination is essential for dislocation and degradation, we depleted ubiquitin from liver cytosol. Liver cytosol was incubated with a GST-tagged mutant ubiquitin-conjugating enzyme, SerE214K, in which the active site cysteine has been mutated to serine. SerE214K covalently binds ubiquitin at a rate that is only ∼3 times slower than that of wild-type E214K enzyme (Chau, unpublished observations). However, instead of forming a thioester bond with ubiquitin, SerE214K forms a stable ester bond that prevents the subsequent transfer of ubiquitin to E3 enzymes or to target proteins. Thus, SerE214K-ubiquitin conjugates accumulate over time and ubiquitin can be removed from the cytosol by precipitating SerE214K with glutathione beads (see MATERIALS AND METHODS for details). With the use of this method, we were able to reproducibly deplete >90% of ubiquitin from cytosol (Figure 2).
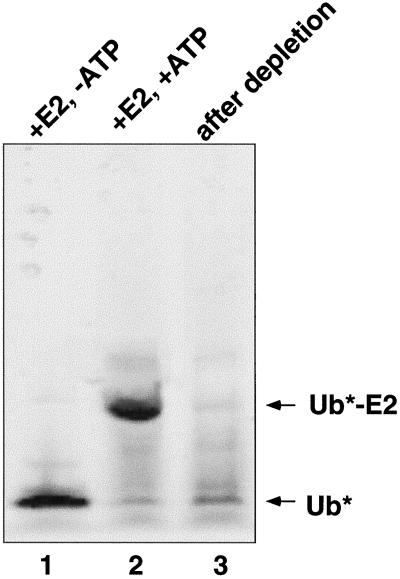
Figure 2.
Monitoring the extent of ubiquitin depletion. Oregon Green-labeled ubiquitin (Ub*) was added to liver cytosol to a final concentration of 1 μM before the depletion procedure. Samples were taken at various stages during the procedure, run on a nonreducing 12% Tricine-SDS polyacrylamide gel, and visualized with the use of a FluorImager (Molecular Dynamics, Sunnyvale, CA). Lane 1, cow liver cytosol with Oregon Green Ub and GST-tagged SerE214K(E2). Lane 2, same as lane 1, except 1 mM ATP was also added and the sample was incubated at 37°C for 10 min. Lane 3, same as lane 2, except the sample was further treated with glutathione Sepharose beads to remove GST-tagged SerE214K. We were able to reproducibly deplete >90% of the Oregon Green ubiquitin from the liver cytosol. We could also monitor ubiquitin depletion by Western blot, with the use of αUb antibodies, but this method was much less sensitive than the Oregon Green ubiquitin method (our unpublished data).
[35S]methionine-labeled membranes from US11-expressing cells were resuspended in buffer, in whole liver cytosol, or in Ub-depleted cytosol and dislocation assays were carried out as described above. We found that heavy chains were not significantly degraded in the Ub-depleted cytosol (Figure 3A, compare lanes 4–6 with lanes 10–12), showing that ubiquitin is required for degradation. Interestingly, incubation with Ub-depleted cytosol did not result in the accumulation of deglycosylated heavy chains, suggesting that dislocation was also impaired. This was confirmed by fractionation experiments, which showed that, in assays with Ub-depleted cytosol, the heavy chain remained associated with the membrane fraction (Figure 3B, lanes 3 and 4). Furthermore, in the absence of ubiquitin, the heavy chain luminal domain remained largely protected from protease digestion (Figure 3C, lanes 7–12). Thirty minutes into the dislocation assay, only the cytosolic tail of the heavy chain was susceptible to trypsin digestion (lanes 10–12), indicating that the heavy chain remains integrated in the ER membrane in the absence of cytosolic ubiquitin. This dislocation defect is due solely to the absence of ubiquitin and not due to the removal of unknown, coprecipitating factors, because the addition of purified bovine ubiquitin (Figure 3A, lanes 7–9; B, lanes 5 and 6; and C, lanes 13–18) or bacterially expressed ubiquitin (our unpublished data) fully restored the ability of the extracts to support heavy chain dislocation and degradation. We found that 5–10 μM bovine ubiquitin was sufficient to restore full activity to the Ub-depleted cytosol (Figure 3D). This is the same range in which other in vitro ubiquitination reactions are optimal (Podust et al., 2000).
To confirm that depletion of ubiquitin affected polyubiquitination of heavy chains, we subjected the class I heavy chains immunoprecipitated with the αHC serum to a second round of immunoprecipitation with anti-ubiquitin serum (αUb) (Figure 3A, bottom panels). In ubiquitin-depleted cytosol, very few high molecular weight bands, representing polyubiquitinated heavy chains, were seen (lanes 16–18). When ubiquitin was added back, polyubiquitinated heavy chains were prominent at the 30-min time point and decreased thereafter (lanes 19–21). The polyubiquitinated chains were found largely in the cytosol fraction (Figure 3B, lane 13). It should be noted that, in these samples, ubiquitin aldehyde was present during the dislocation assay to inhibit deubiquitination. When the inhibitor was absent, polyubiquitinated chains were less abundant and rapidly degraded (Figure 3A, lanes 22–24). Overall, these experiments clearly demonstrate a requirement for ubiquitin in US11-dependent heavy chain dislocation.
Polyubiquitination Is Required for Heavy Chain Dislocation
We next tested whether formation of polyubiquitin chains is required for heavy chain dislocation or if monoubiquitination is sufficient. Polyubiquitin chains are attached to lysine residues in ubiquitinated protein substrates and are extended by isopeptide bond formation between a ε-amino group of a lysine in one ubiquitin molecule and the C-terminal glycine in another. Ubiquitin that lacks available lysines, through methylation (Me-Ub; Hershko and Heller, 1985) or mutation (K0-Ub; Pickart, 2000), does not form polyubiquitin chains. When Me-Ub or K0-Ub were added back to Ub-depleted cytosol, they were much less active than wild-type ubiquitin in supporting dislocation and degradation of class I heavy chain from US11 membranes (Figure 4A, compare lanes 13–16 with lanes 21–24; Figure 4B, compare lanes 1–4 with lanes 5–8 and 13–16). Quantitation of the heavy chain in these experiments shows some degradation in the presence of Me-Ub or K0-Ub, but clearly less than in the presence of wild-type Ub (our unpublished data). US11-dependent dislocation was restored to assays containing Me-Ub or K0-Ub when an equal amount of wild-type ubiquitin was added (Figure 4B, lanes 9–12 and 17–20), demonstrating that potential impurities in the Me-Ub and K0-Ub preparations are not poisoning the reactions.
We know that our preparations of Me-Ub and K0-Ub are capable of being coupled as monoubiquitin adducts to substrates because they conjugate to S. cerevisiae Cdc34p in vitro (our unpublished data). Moreover, when the dislocation assays were carried out in the presence of higher concentrations of ubiquitin aldehyde, heavy chain species running between 43 and 66 kDa were seen in samples where only Me-Ub or K0-Ub was added. These can be reimmunoprecipitated with antiubiquitin antibodies (Figure 4B, lanes 28 and 36) and antiheavy chain antibodies (Figure 4C, lanes 10 and 12). Thus, they are likely heavy chains that have been mono-ubiquitinated on multiple lysine residues or that bear very short polyubiquitin chains capped by Me-Ub or K0-Ub. Interestingly, these low molecular weight ubiquitinated heavy chains fractionate with the cell membrane pellets, whereas more highly ubiquitinated heavy chains present in the same samples are found in the soluble, cytosolic fractions (Figure 4, B and C). This observation supports a model for heavy chain dislocation in which ubiquitination of heavy chain occurs early, while the heavy chain is still associated with the ER membrane (Shamu et al., 1999). Furthermore, these results suggest that polyubiquitination is required for US11-dependent heavy chain dislocation.
DISCUSSION
Our results have implications for the function of US11 in the specific pathway of MHC class I degradation, as well as more general implications for the process of protein movement from the ER into the cytosol. To identify and characterize factors that are required for the US11-dependent dislocation and degradation of MHC class I heavy chain, we have fractionated a permeabilized cell system into cytosolic and membrane components. We find that cytosolic proteins are essential for dislocation and that US11 is required only in the membrane. US11 probably functions to initiate heavy chain dislocation, feeding heavy chain into the cellular ER degradation pathway at one of its early steps. Because US11-dependent heavy chain degradation is much faster than degradation of misfolded proteins that accumulate in the ER, this would suggest that the initial dislocation step is usually rate-limiting.
We have identified ubiquitin as one of the cytosolic proteins required for US11-dependent heavy chain dislocation. Previous experiments in other systems suggested that ubiquitination is required for the dislocation and degradation of misfolded/unassembled ER proteins. However, these studies were performed in vivo, by using either loss-of-function or dominant negative mutants in the components of the ubiquitination machinery (Ward et al., 1995; Biederer et al., 1997; de Virgilio et al., 1998; Yu and Kopito, 1999). The effects of such mutants are probably pleiotropic and therefore difficult to interpret. We have used a new method, the use of the modified conjugating enzyme SerE214K, to deplete ubiquitin from the cytosol. This in vitro method is less likely to give rise to indirect effects because it allows for a positive control, the readdition of ubiquitin to the depleted extract, and is performed on a short time scale. Moreover, the in vitro approach has allowed us to demonstrate that the attachment of only one or a few ubiquitin molecules is not sufficient for heavy chain dislocation and that polyubiquitination is necessary. A requirement for polyubiquitination in the dislocation of proteins from the ER has been claimed previously (Yu and Kopito, 1999), but had not been directly demonstrated.
US11-dependent dislocation of heavy chain differs from recently characterized endocytic pathways in which modification with a single ubiquitin molecule is sufficient to target substrate proteins for movement between cellular compartments (Shih et al., 2000). In the absence of ubiquitin, the heavy chain remains tightly associated with, and probably integrated in, the cell membranes, as shown by fractionation and protease protection experiments. The polyubiquitination step required for US11-dependent heavy chain degradation must therefore occur very early in the process, before the heavy chain is released from the ER membrane and before it is deglycosylated.
The simplest interpretation of our data is that polyubiquitination of the class I heavy chain itself is required for its dislocation. However, it could be that the modification of a different protein is actually important. If we assume that modification of the heavy chain is essential, the required sites of ubiquitination are probably not in the cytosolic tail, because a mutant heavy chain lacking lysines in the tail is still dislocated and degraded (Shamu et al., 1999). Thus, the key ubiquitination step would have to occur after at least part of the lumenal domain of the heavy chain has been dislocated from the ER. The relevant ubiquitinating enzymes may be bound to the ER membrane, as are the ubiquitin-conjugating enzymes Ubc6p and Ubc7p, and the ubiquitin ligase-containing Hrd1p and Hrd3p, all of which are involved in ER-associated degradation in yeast (Sommer and Jentsch, 1993; Hiller et al., 1996; Biederer et al., 1997; Gardner et al., 2000; Bays et al., 2001).
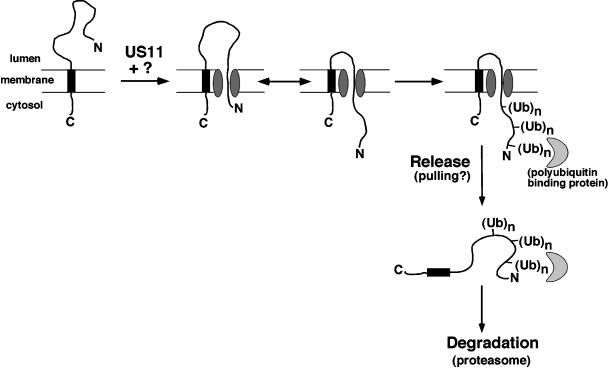
Figure 5 illustrates one model for the mechanism of heavy chain dislocation. In this model, dislocation is initiated, by an as-yet-uncharacterized mechanism, involving proteins in the ER lumen that include US11. This leads to exposure of a lumenal portion of the heavy chain to the cytosol. The partially dislocated heavy chain is free to slide backwards and forwards through the dislocation channel. Polyubiquitination of the heavy chain acts as part of a molecular ratchet, preventing portions of heavy chain that have already been dislocated onto the cytosol from slipping back into the ER lumen. Polyubiquitin chains would be ideal in such a ratcheting role, not only would they be covalently attached to the heavy chain substrate but also they would be too massive to fit through the channel in the ER membrane. In addition, the polyubiquitin chain may be recognized by a binding partner that may also prevent back-sliding and could additionally act in the next step of dislocation (see below). Although a single ubiquitin molecule may simply be too small for ratcheting, a single polyubiquitin adduct may be sufficiently large to keep a portion of the heavy chain on the cytosolic side of the membrane. This may explain why a substrate thought to be ubiquitinated only at its N terminus can still be efficiently dislocated (Yu and Kopito, 1999). An interesting feature of our model is that it proposes a new function for ubiquitin, distinct from its previously identified functions as a signal for degradation or endocytosis.
Figure 5.
Tentative model for US11-dependent heavy chain dislocation. For energetic reasons, we favor the possibility that the heavy chain N terminus exits the ER first, leaving the transmembrane domain in the ER membrane until the last stage of dislocation. The lumenal domain would then be able to slide back and forth through the channel. Polyubiquitination, either alone or in conjunction with a polyubiquitin-binding protein, would prevent back-sliding into the ER (ratcheting). Release of the heavy chain from the membrane probably involves an active pulling mechanism, possibly again involving a polyubiquitin-binding protein.
The final step in retrograde translocation is the release of the heavy chain from the ER membrane. Although the proposed ratcheting mechanism allows for the retention of the lumenal domain of heavy chain in the cytosol, it is likely that a pulling force would be required to fully extract the protein from the membrane. Indeed, ubiquitin is not the sole cytosolic factor required for release of the heavy chain from the ER membrane into the cytosol. When ubiquitin and the purified ubiquitin-activating enzyme Uba1p were added together in buffer to US11 cell membranes, heavy chain dislocation was not observed (our unpublished data). One possibility is that an additional, cytosolic polyubiquitin-binding protein or protein complex is required, which would actively pull the ubiquitinated heavy chain out of the ER membrane. An obvious candidate for this role is the 19S regulatory subunit of the proteasome, acting either on its own, or as part of the 26S proteasome complex (Yu et al., 1997; Mayer et al., 1998). Components of the 19S subunit include ATPases as well as polyubiquitin binding proteins and one of its well-known functions is to feed polyubiquitin-tagged proteins into the proteasome 20S core.
To further elucidate the mechanism of US11-dependent heavy chain dislocation, including the role of ubiquitin, the additional cytosolic and membrane components required for the process must be identified. The fractionated permeabilized cell system described here should provide a useful experimental starting point.
ACKNOWLEDGMENTS
We thank T. Gladysheva (Millenium Pharmaceuticals, Cambridge, MA) for the MCHHHHHH ubiquitin construct used for Oregon Green labeling; Lars Dreier and Pascal Stein for bovine liver cytosol; Cecile Pickart for the K0-Ub expression system; Melissa Rolls and Peter Sorger for comments on the manuscript; and Domenic Tortorella, Margo Furman, Maurits Kleijnen, Benedikt Kessler, and Seth Sadis for reagents, protocols, and helpful discussions. C.E.S. was supported by a Special Fellowship from the Leukemia and Lymphoma Society. T.A.R. is an Investigator of the Howard Hughes Medical Institute. V.C. was supported by National Institutes of Health Grant GM-62194.
Abbreviations used:
- HC
MHC class I heavy chain
- HCMV
human cytomegalovirus
- MHC
major histocompatibility complex
- PB
permeabilization buffer
- Staph A
fixed Staphylococcus aureus
- Ub
ubiquitin
REFERENCES
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985;128:1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hicke L. Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Jones TR, Hanson LK, Sun L, Slater JS, Stenberg RM, Campbell AE. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Braun T, Jentsch S. Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. Current Protocols in Protein Science (eds. J.E. Coligan, B.M. Dunn, H.L. Ploegh, D.W. Speicher, and P.T. Wingfield) New York: John Wiley & Sons; 1995. One-dimensional isoelectric focusing of proteins in slab gels; pp. 10.12.11–10.12.18. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownwell JE, Gladysheva TB, Lu R-S, Wang C, Coggins MB, Pierce JW, Chau V. A Nedd8 conjugation pathway is essential for proteolyic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Story CM, Rapoport TA, Ploegh HL. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J Cell Biol. 1999;147:45–58. doi: 10.1083/jcb.147.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Sloper-Mold KE, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Story CM, Furman MH, Ploegh HL. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc Natl Acad Sci USA. 1999;96:8516–8521. doi: 10.1073/pnas.96.15.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996a;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996b;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- You J, Cohen RE, Pickart CM. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli. Biotechniques. 1999;27:950–954. doi: 10.2144/99275st01. [DOI] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- Yuk MH, Lodish HF. Two pathways for the degradation of the H2 subunit of the asialoglycoprotein receptor in the endoplasmic reticulum. J Cell Biol. 1993;123:1735–1749. doi: 10.1083/jcb.123.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Schekman R. The engagement of Sec61p in the ER dislocation process. Mol Cell. 1999;4:925–934. doi: 10.1016/s1097-2765(00)80222-1. [DOI] [PubMed] [Google Scholar]