Abstract
Prostate cancer is the second most common malignancy affecting men worldwide, and the commonest affecting men of African descent. Significant diagnostic and therapeutic advances have been made in the past decade. Improvements in the accuracy of prostate cancer diagnosis include the uptake of multi-parametric MRI and a shift towards targeted biopsy. We also now have more life-prolonging systemic and hormonal therapies for men with advanced disease at our disposal than ever before. However, the development of robust screening tools and targeted screening programs has not followed at the same pace. Evidence to support population-based screening remains unclear, with the use of PSA as a screening test limiting our ability to discriminate between clinically significant and insignificant disease. Prostate cancer has a large heritable component. Given that most men without risk factors have a low lifetime risk of developing lethal prostate cancer, much work is being done to further our knowledge of how we can best screen men in higher risk categories, such as those with a family history (FH) of the disease or those of African ancestry. These men have been reported to carry upwards of a two-fold increased risk of developing the disease at an earlier age, with evidence suggesting poorer survival outcomes. In men with a FH of prostate cancer, this is felt to be due to rare, high-penetrance mutations and the presence of multiple, common low penetrance alleles, with men carrying specific germline mutations in the BRCA and other DNA repair genes at particularly high risk. To date, large scale genome-wide association studies (GWAS) have led to the discovery of approximately 170 single nucleotide polymorphisms (SNPs) associated with prostate cancer risk, allowing over 30% of prostate cancer risk to be explained. Genomic tests, utilising somatic (prostate biopsy) tissue can also predict the risk of unfavourable pathology, biochemical recurrence and the likelihood of metastatic disease using gene expression. Targeted screening studies are currently under way in men with DNA repair mutations, men with a FH and those of Afro-Caribbean ethnicity which will greater inform our understanding of disease incidence and behaviour in these men, treatment outcomes and developing the most appropriate screening regime for such men. Incorporating a patient’s genetic mutation status into risk algorithms allows us an opportunity to develop targeted screening programs for men in whom early cancer detection and treatment will positively influence survival, and in the process offer male family members of affected men the chance to be counselled and screened accordingly.
Keywords: Prostate cancer, genetic screening, single nucleotide polymorphisms (SNPs)
Introduction
Prostate cancer remains one of the most commonly diagnosed cancers in men in the western world, with 1.1 million new cases annually and 307,000 deaths (1). It is the commonest cancer in UK with Caucasian males having a lifetime risk of 13.2–15% of developing the disease (2). However, not all men are at equal risk for developing the lethal form of the disease, and the vast majority will have unaffected overall survival (3). Both the ProtecT and PIVOT studies of PSA screened men demonstrated no difference in disease-specific or all-cause mortality irrespective if men were treated or just observed (4,5).
What we do know, is men with a family history (FH) of prostate cancer or those of Afro-Caribbean ancestry have a susceptibility to earlier onset and more aggressive disease making them an ideal group of men in whom to establish robust screening tools to improve survival by means of early diagnosis and treatment (6).
Genetics of prostate cancer in men with a FH and black men
Men with a FH of prostate cancer have a significantly higher lifetime risk of developing the disease, with a 2- to 8-fold increase reported (7) and worsening risk with the number of first degree relatives affected. A Swedish study reporting from a family-database of over 9 million people reported a standardized incidence ratio (SIR) of 23.72 for men whose father and sibling were affected (8). Further work in the same cohort established an SIR of 8.05 of developing prostate cancer before age 55, if a brother was affected before this age (9). Another group screened 34 first-degree relatives (sons/brothers) of 17 sets of (two) brothers with prostate cancer, using a combination of PSA, DRE and TRUS biopsy. Clinically significant, asymptomatic prostate cancer was found in 8 (24%) men with a reported RR of developing prostate cancer of 5–11 (10).
In a retrospective assessment of American men with a FH of prostate cancer undergoing prostate biopsies for either a raised PSA or abnormal DRE, it was found these men were significantly more likely to be diagnosed with both low-grade and high-grade prostate cancer (11).
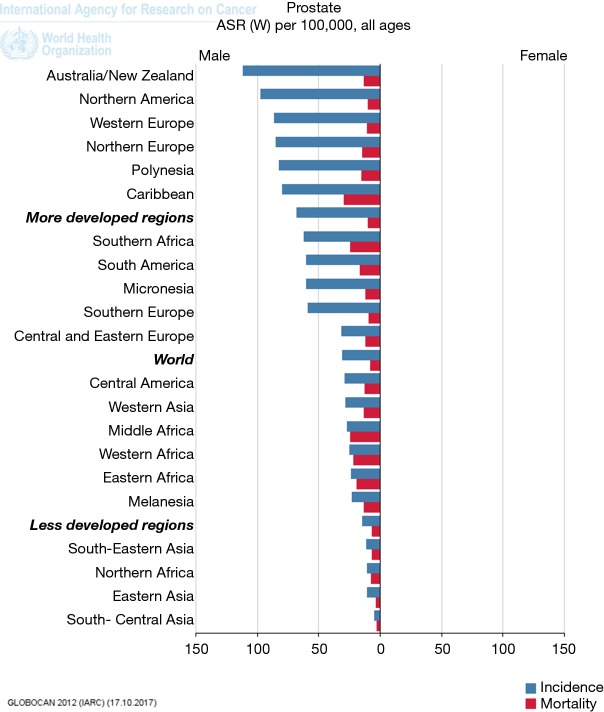
Retrospective studies of prostate cancer incidence in the USA and Africa reveal a higher disease occurrence in black men compared with white men (12). Age at disease onset is earlier, with tumours reported to be more aggressive (2). Mortality rates also significantly differ between African and Caribbean men compared with the UK, as depicted in GLOBOCANs mortality and incidence data (Figure 1).
Figure 1.
WHO Prostate cancer incidence and mortality figures by continent. Higher mortality noted in the Caribbean, Southern, Middle, Western and Eastern Africa, taken from Globocan (http://globocan.iarc.fr/Default.aspx) (1).
The PROCESS study examined 2,140 cases of prostate cancer in defined areas of London and Bristol with high proportions of black male residents, with black men being three times more likely to develop prostate cancer than white men but with no significant difference between black-African and black-Caribbean men (13). The most recent USA [2014] incidence and death rates per 100,000 population for black men with prostate cancer compared with white men, were 154.1 (86.9 in whites) and 38.0 (17.9 in whites) respectively (14).
Population screening for prostate cancer
Screening for prostate cancer would clearly aim to detect clinically important cancers, in parallel to not exposing men to the morbidity of unnecessary prostate biopsies and diagnosing clinically insignificant prostate cancer. Few other specialities have been as plagued with controversy as prostate cancer diagnostics, since the advent of PSA as tumour marker after its isolation from the serum of 219 patients with prostate cancer by Papsidero in 1981 (15). Work performed in the 1980s (16) produced evidence that although PSA is related to prostate cancer, its levels also rose in the presence of BPH rendering its relationship to prostate cancer diagnosis in Stamey’s own words as ‘tenuous at best’ (17).
In essence, PSA remains an imperfect screening tool in isolation for discriminating between a clinically significant cancer, and one which may have never affected a man during the course of his lifetime. The US Preventive Services Taskforce (USPSTF) report on this ‘pseudo-disease’ situation in their 2012 recommendation, citing the benefits of PSA screening as ‘small and potentially none, and the harms are moderate to substantial’ (18).
The UK national screening committee (NSC) last recommended in January 2014 there was not enough evidence to commence a national screening program, given significant limitations in PSA’s ability to perform as a robust screening tool. The harms of overdiagnosis and overtreatment were felt to clearly still outweigh the potential 21% reduced risk on mortality (19).
In a report to the NSC in March 2013, the University of Sheffield assessed the outcomes of four different screening options; a single screen at age 50, screening every 4 years age 50–74, screening every 2 years age 50–74 and screening annually aged 50–74. They estimated all repeat screening policy options were associated with 45–65% risk of overdetection of prostate cancer, with a rate of 30–40% for a single screen policy and in order to obtain 1 additional year of life, repeat screening policies are associated within the region of 22–32 years of additional prostate cancer management. Finally, they estimated an overall expected survival benefit of 2–4 days per person invited for a single screen aged 50, and 20–60 days for the repeat screen policies (20).
Pashayan et al. assessed the implications of using polygenic risk scoring (PRS) on reducing overdiagnosis. They constructed a PRS on 17,000 men aged 50–69 from three large studies [ProtecT, SEARCH and the UK Genetic Prostate Cancer Study (UKGPCS)] using 66 known SNPs, separating men with and without prostate cancer into risk quartiles. By introducing this method, they found a 56% reduction in overdiagnosis between the lowest risk quartile and the highest (21). In a separate study the same author examined 43,842 men within the ProtecT study alone, aged 50–69. A PSA threshold of ≥3.0 ng/mL−1 was used, with 3.5% of men being diagnosed with prostate cancer. It was estimated that 10–31% of these cases were overdiagnosed (22).
Moving towards developing specific risk-based strategies and targeting certain groups of men would allow us to expose less ‘low risk’ men to the harms of false-positive PSA testing and increase the usefulness of a screening program. The Rotterdam prostate cancer risk calculator developed by Roobol et al. describes a screening algorithm applied to 1,850 men within the Rotterdam section of the ERSPC. They calculated the probability of having a diagnosis of indolent prostate cancer detected on biopsy. Of the 1,850 men, 541 were diagnosed with prostate cancer on biopsy with an estimate of 44% of these likely to be indolent. Applying their algorithm, using a PSA biopsy threshold of ≥3.0 ng/mL and taking into account previous screening visits significantly reduced the number of unnecessary biopsies (23).
Evidence for targeted screening
We know certain groups of men have an elevated risk of an early onset of disease and for developing lethal prostate cancer. Men with a FH of prostate cancer and black men have repeatedly been shown to be at higher risk but a full explanation of the exact genetic mechanism eludes us still. It seems sensible therefore to investigate the best way of adopting screening programs in these specific groups of men, who are well placed to theoretically truly benefit from early cancer detection and treatment.
Interrogating the PLCO data, Liss et al. found that when they looked at all men within the study who had a FH of prostate cancer, those who were screened had a trend towards decreased prostate cancer specific mortality and time to death, with a significantly higher incidence of prostate cancer and mortality in all men with a FH compared to those without (6).
There is evidence to suggest genetic based scores improve prostate cancer detection and risk stratification. Using 14 known prostate cancer associated SNPs and the presence/absence of a FH of prostate cancer, Xu et al. built a risk prediction model. They concluded an OR of 4.92 for developing prostate cancer for men with a positive FH and ≥14 risk alleles for the Swedish cohort in their study (24) . Using data from the REDUCE trial (Reduction by Dutasteride of Prostate Cancer Events) which assessed the chemopreventive benefits of Dutasteride, Kader and colleagues analysed germline DNA from 1,654 controls. These men all had an initial negative prostate biopsy, with subsequent prostate biopsies at 2 and 4 years. They found adding a genetic score based on 33 risk SNPs with clinical variables was an independent prostate cancer risk predictor on repeat prostate biopsy, and demonstrated ability to reduce the number of repeat biopsies required (25).
High risk susceptibility protein-coding genes in prostate cancer
HOXB13
High-risk prostate cancer predisposition genes exist, with carriers of a rare missense mutation (G84E) of the HOXB13 gene having a 33% risk of developing prostate cancer, compared to a 12% risk of non-carriers when studied in a Swedish population. This mutation was present in 1.3% of population controls and >4% of cases (26). Further large-scale analysis of 4,000 prostate cancer cases in Finland for this specific mutation revealed a significantly higher carrier-rate amongst men with prostate cancer (3.5%) and those with a FH (8.4%) compared to controls (27). In a separate study of 5,083 unrelated European subjects who had prostate cancer, Ewing et al. found the carrier rate of the (G84E) mutation was increased by a factor of approximately 20. This mutation was significantly more common in men with disease at a young age and with a positive FH (1.4%), than those without (0.1%) (28) . This genetic mutation therefore seems particularly significant in young men with prostate cancer and with a strong FH in Finnish and Swedish populations.
DNA repair genes
Germline deleterious mutations in BRCA1/2 genes increase the risk of developing prostate cancer. Edwards et al. (29) reported the prevalence of a BRCA2 mutation to be 2.3% in young men presenting with PSA detected prostate cancer. The authors’ further analysis of 21 young men with prostate cancer who had BRCA2 mutations compared to approx. 1,500 controls demonstrated poorer overall survival (30) with an Icelandic study showing a mean survival of only 2.1 years in men with prostate cancer with the specific 999del5 BRCA2 mutation compared with non-carriers (31). Two further retrospective analyses found association between BRCA status and higher risk of disease recurrence, prostate cancer specific-mortality and high risk disease with a significant difference in CSS of 8.6 vs. 15.7 years for non-carriers (32,33).
Mutations of other genes involved in DNA repair such as ATM, CHEK2, MSH1, MLH1, MSH2, and MSH6 have also been associated with a high risk of developing prostate cancer, following analysis of a mainly white, European cohort of men in the UKGPCS study (34,35). In this study 7.3% of cases of prostate cancer patients with a positive FH were found to carry a germline mutation. The most frequent mutation was in BRCA2 (28.57% of all mutations), and importantly there was a significant association between genetic mutation carrier status with nodal and metastatic disease. A further review by Pritchard et al. of 692 men with metastatic castrate resistant prostate cancer revealed a germline DNA repair-gene mutation in 11.8% of all men, across 16 genes including BRCA1/2, ATM, CHEK2, PALB2 and RAD51D (36). A genetic mutation (657del5) in NBN has also been associated with a significantly higher risk of earlier onset disease, aggressive histology and reduced 5- and 10-year survival in a Polish cohort of approximately 3,800 men (34).
Germline single nucleotide polymorphisms (SNPs) in prostate cancer
Aside from high-penetrance mutations as described above, more common alleles (SNPs) associated with prostate cancer can occur in up to 5% of the population. These low-penetrance genetic variations can confer a low risk of prostate cancer risk if occurring alone, but result in an elevated and potentially clinically relevant risk when multiple SNPs occur together, strengthening their genetic effect. Large scale genome-wide association studies (GWAS), have led to the discovery of up to 170 SNPs associated with prostate cancer risk (37-40). Pending imminent publication of the latest susceptibility loci from the Oncoarray Initiative (41), 30% of the familial risk in prostate cancer can be explained based on previously published SNPs, with men in the top 1% of the risk profile having a 4.7-fold increase in risk of developing prostate cancer compared with controls (42).
Zheng & colleagues published their results examining the effect of the five commonest known SNPs associated with prostate cancer. They found their presence in combination with a FH accounted for 46% of the cases of prostate cancer in their cohort and conferred an odds ratio of 9.46 compared with men who had none of these factors, independent of PSA. Pending the results of current screening studies in high-risk men, it would appear that using genetic information with PSA and FH information could translate into a breakthrough in improving prostate cancer screening (40).
Precision oncology and genomic tests available for prostate cancer
Precision oncology and targeted cancer therapy based on somatic and germline mutations are expected to soon form a significant part of cancer care. Robinson et al. recently published a series of 500 adult patients with metastatic solid tumours, revealing 12.2% of cases carrying a pathogenic germline mutation, of which 75% were in DNA repair genes. This emphasises the clinical relevance of obtaining genomic information to map patients’ individual tumour genotype and phenotype to offer targeted oncological therapies (43).
Work from the Stand Up To Cancer (SU2C) Prostate Cancer Foundation Dream Team will discover and deliver precision therapy for advanced prostate cancer, targeting adaptive pathways in hormone-resistant disease and aiming to deliver individualised treatment to prostate cancer patients based on the genetic characteristics of their tumour (http://www.standuptocancer.org/dream_teams/view/targeting_adaptive_pathways_in_metastatic_crpc).
Mapping the mutational burden of both somatic tissue in prostate cancer and germline DNA in delivering specific therapeutic agents is already under way in the TOPARP and the BARCODE 2 studies (44,45). In 2015, Robinson et al. performed whole-exome and transcriptome sequencing of biopsies from 150 men with metastatic castrate-resistant prostate cancer (mCRPC). They showed that 89% of men had clinically actionable genetic mutations, and 8% had actionable germline alterations. Based on potential sensitivity to PARP inhibitors and the prevalence of BRCA/ATM mutations they also estimated 19.3% of men could benefit from these therapeutic options (46).
Predictive genomic tests
Genomic tests utilising prostate biopsy tissue such as OncotypeDx® (a 17-gene assay) can be used in men with low-intermediate risk disease on initial prostate biopsy considering active surveillance or radical treatment. The result is a ‘Genomic Prostate Score’ (GPS) of 1–100. Higher values correlate with a higher risk of adverse pathology at the time of prostatectomy, risk of 10-year prostate cancer-specific mortality and risk of developing metastases within 10 years (47).
The Decipher® test uses prostatectomy tissue to predict the risk of 5-year metastases and prostate cancer-specific mortality after radical treatment, using an oligonucleotide microarray to create a signature ‘genomic classifier’ score based on 22 genomic markers, ranging from 0–1. In addition to acting independently as a prediction tool, it also improves the performance of the CAPRA-S clinicopathological risk model. Its use mainly lies in helping determine which men with unfavourable radical prostatectomy pathology would best benefit from further adjuvant treatments. Decipher Biopsy® has also been developed to predict risk of metastases, high risk disease and prostate cancer-specific mortality from initial biopsies in men with localised disease considering active surveillance (48,49).
The Prolaris® test measures gene expression levels of 31 cell cycle genes, in RNA extracted from prostate tumour tissue (needle biopsies or prostatectomy specimens). It is designed to act as a decision aid in the setting of considering active surveillance versus radical treatment in men with low/intermediate risk prostate cancer. It estimates prostate cancer-specific mortality at 10 years and the risk of biochemical recurrence, expressed as a ‘CCP’ (cell-cycle progression) score.
ConfirmMDx is an epigenetic DNA methylation test performed on prostate biopsy tissue, acting as an independent predictor of the likelihood of a positive repeat biopsy after initial negative biopsy. SelectMDx, made by the same company is used in predicting high versus low-grade disease on first prostate biopsy. Unlike previously described tests, it uses urine collected after a DRE on which a reverse-transcriptome PCR assay is performed. This test aims to discriminate between men who may avoid a prostate biopsy based having on a low risk of high-grade prostate cancer, or men who would benefit from a biopsy in whom a high risk of harbouring high-grade disease is detected. The 4Kscore® goes not require genetic material but acts as a predictive blood test, consisting of a panel of four kallikrein markers including total PSA, and predicts the risk of Gleason ≥7 prostate cancer on initial prostate biopsy (50,51).
Prostate cancer consortia investigating genetic variants
Led by The Institute of Cancer Research, the UKGPCS is recruiting 26,000 men with a diagnosis of prostate cancer in order to map the genetic changes associated with the disease. This study has provided a wealth of data encompassing multiple new prostate cancer susceptibility loci (37,38,52).
Currently, the IMPACT study has enrolled over 3,000 men (cases and controls) to investigate the outcomes of targeted screening in men with BRCA1/2 germline mutations and Lynch Syndrome with annual PSA and a biopsy threshold of 3.0 ng/mL. Early results have suggested targeted screening in this population is beneficial (53). The GENPROS study will aim to assess the clinical outcomes of 1,000 men after prostate cancer treatment with mutations in cancer predisposition genes such as BRCA, HOXB13 and Lynch Syndrome.
Collaborative, international working has led to the creation of multiple consortia interested in pooling large numbers of cases with genetic information, allowing large scale GWAS. The Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium has brought together over 100 research groups investigating prostate cancer genetics, also forming part of the OncoArray Consortium.
The OncoArray Consortium designed a custom-made chip to perform large-scale germline genotyping of over 400,000 blood samples. This international effort will soon publish the most up to date developments in prostate cancer susceptibility loci, bringing the total number of known pathological SNPs to approximately 170.
Most GWAS studies establishing risk SNPs, risk-prediction models using genetic information and genomic test validation (i.e., OncotypeDx/Decipher and Prolaris) have used genetic information from predominantly Caucasian men. Information regarding specific risk alleles in men of African ethnicity is therefore grossly lacking, hampering the potential for ethnicity-specific genetic risk scoring. The Men of African Descent and Carcinoma of the Prostate Consortium (MADCaP) is crucially examining prostate cancer epidemiology, outcomes and aims to catalogue specific genetic mutations in this group of men across Africa and North America, with one participating centre in London (54).
The huge scale collection and validation of prostate cancer risk alleles as described, will allow an opportunity to more precisely define which men will fall into the highest risk group. In this vein, scope for further research exists in examining the feasibility and survival benefit of offering ‘early’ radical treatments for men with the highest risk profiles.
The PROFILE study
The PROFILE Feasibility study examined the role of upfront prostate biopsy regardless of PSA with a PRS in 100 men. They reported a cancer detection rate of 25%, with 48% of these being clinically significant cancers requiring radical treatment (55).
Presently, the PROFILE study is recruiting a total of 700 subjects investigating the role of targeted screening in men with a genetic susceptibility to prostate cancer. Germline analysis of likely over 100 SNPs will be used in conjunction with an outright MRI with prostate biopsy (regardless of PSA) in men aged 40–69 with either a FH of prostate cancer or black men. The aim is to recruit 350 men in each group, with men declining MRI/Biopsy undergoing 6-monthly PSA surveillance for a minimum of 5 years. This study will determine the association of genetic profiling with MRI/prostate biopsy result in men with a genetic susceptibility to prostate cancer undergoing targeted, intensive screening. Information of prostate cancer incidence, aggressiveness and incidence of abnormal MRI and its value in this cohort will also be assessed.
Conclusions
It has been established that a FH of prostate cancer or Afro-Caribbean ethnicity can predispose men to both earlier onset and aggressive disease, with the potential for poor outcomes. We know PSA is an imperfect test, and the basis for why these groups of men are ‘high risk’ is fundamentally genetic in origin. It therefore seems sensible to develop targeted screening strategies for these groups of men, using a combination of tools to improve cancer detection and influence the natural history of their disease.
Advances in the field of uro-oncology such as the diagnostic performance of multi-parametric MRI and genomic interrogation have led us to a position of potentially use these as screening tools in the right populations. In addition, by identifying the value of screening high-risk men and potentially extending a targeted national screening program to such groups, the burden of adverse effects associated with population screening could be minimised.
Acknowledgements
The PROFILE study is funded by Movember/PCUK.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Lloyd T, Hounsome L, Mehay A, et al. Lifetime risk of being diagnosed with, or dying from, prostate cancer by major ethnic group in England 2008-2010. BMC Med 2015;13:171. 10.1186/s12916-015-0405-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adolfsson J, Ronstrom L, Lowhagen T, et al. Deferred treatment of clinically localized low grade prostate cancer: the experience from a prospective series at the Karolinska Hospital. J Urol 1994;152:1757-60. 10.1016/S0022-5347(17)32379-0 [DOI] [PubMed] [Google Scholar]
- 4.Hamdy FC, Donovan JL, Lane JA, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med 2016;375:1415-24. 10.1056/NEJMoa1606220 [DOI] [PubMed] [Google Scholar]
- 5.Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 2012;367:203-13. 10.1056/NEJMoa1113162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liss MA, Chen H, Hemal S, et al. Impact of family history on prostate cancer mortality in white men undergoing prostate specific antigen based screening. J Urol 2015;193:75-9. 10.1016/j.juro.2014.07.085 [DOI] [PubMed] [Google Scholar]
- 7.Goldgar DE, Easton DF, Cannon-Albright LA, et al. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 1994;86:1600-8. 10.1093/jnci/86.21.1600 [DOI] [PubMed] [Google Scholar]
- 8.Dong C, Hemminki K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer 2001;92:144-50. [DOI] [PubMed] [Google Scholar]
- 9.Hemminki K, Czene K. Age specific and attributable risks of familial prostate carcinoma from the family-cancer database. Cancer 2002;95:1346-53. 10.1002/cncr.10819 [DOI] [PubMed] [Google Scholar]
- 10.McWhorter WP, Hernandez AD, Meikle AW, et al. A screening study of prostate cancer in high risk families. J Urol 1992;148:826-8. 10.1016/S0022-5347(17)36733-2 [DOI] [PubMed] [Google Scholar]
- 11.Elshafei A, Moussa AS, Hatem A, et al. Does positive family history of prostate cancer increase the risk of prostate cancer on initial prostate biopsy? Urology 2013;81:826-30. 10.1016/j.urology.2012.10.074 [DOI] [PubMed] [Google Scholar]
- 12.Angwafo FF. Migration and prostate cancer: an international perspective. J Natl Med Assoc 1998;90:S720-3. [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Shlomo Y, Evans S, Ibrahim F, et al. The risk of prostate cancer amongst black men in the United Kingdom: the PROCESS cohort study. Eur Urol 2008;53:99-105. 10.1016/j.eururo.2007.02.047 [DOI] [PubMed] [Google Scholar]
- 14.U.S. Cancer Statistics Working Group. United States Cancer Statistics: 1999-2014 Incidence and Mortality Web-based Report. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2017. Available online: www.cdc.gov/uscs
- 15.Papsidero LD, Wang MC, Valenzuela LA, et al. A prostate antigen in sera of prostatic cancer patients. Cancer Res 1980;40:2428-32. [PubMed] [Google Scholar]
- 16.Oesterling JE. Prostate specific antigen: a critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. J Urol 1991;145:907-23. 10.1016/S0022-5347(17)38491-4 [DOI] [PubMed] [Google Scholar]
- 17.Stamey TA, Caldwell M, McNeal JE, et al. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol 2004;172:1297-301. 10.1097/01.ju.0000139993.51181.5d [DOI] [PubMed] [Google Scholar]
- 18.Moyer VA, Force USPST. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2012;157:120-34. 10.7326/0003-4819-157-2-201207170-00459 [DOI] [PubMed] [Google Scholar]
- 19.UKNSC. Screening for Prostate Cancer Review - 2015 Update Medicine WIoP; 2015.
- 20.Hummel S, Chilcott J. Option appraisal: screening for prostate cancer model update: Report to the UK National Screening Committee March 2013. University of Sheffield, Research SoHaR; 2013.
- 21.Pashayan N, Duffy SW, Neal DE, et al. Implications of polygenic risk-stratified screening for prostate cancer on overdiagnosis. Genet Med 2015;17:789-95. 10.1038/gim.2014.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pashayan N, Duffy SW, Pharoah P, et al. Mean sojourn time, overdiagnosis, and reduction in advanced stage prostate cancer due to screening with PSA: implications of sojourn time on screening. Br J Cancer 2009;100:1198-204. 10.1038/sj.bjc.6604973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roobol MJ, Steyerberg EW, Kranse R, et al. A risk-based strategy improves prostate-specific antigen-driven detection of prostate cancer. Eur Urol 2010;57:79-85. 10.1016/j.eururo.2009.08.025 [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Sun J, Kader AK, et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate 2009;69:1565-72. 10.1002/pros.21002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kader AK, Sun J, Reck BH, et al. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. Eur Urol 2012;62:953-61. 10.1016/j.eururo.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson R, Aly M, Clements M, et al. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur Urol 2014;65:169-76. 10.1016/j.eururo.2012.07.027 [DOI] [PubMed] [Google Scholar]
- 27.Laitinen VH, Wahlfors T, Saaristo L, et al. HOXB13 G84E mutation in Finland: population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 2013;22:452-60. 10.1158/1055-9965.EPI-12-1000-T [DOI] [PubMed] [Google Scholar]
- 28.Ewing CM, Ray AM, Lange EM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med 2012;366:141-9. 10.1056/NEJMoa1110000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards SM, Kote-Jarai Z, Meitz J, et al. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet 2003;72:1-12. 10.1086/345310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards SM, Evans DG, Hope Q, et al. Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br J Cancer 2010;103:918-24. 10.1038/sj.bjc.6605822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tryggvadottir L, Vidarsdottir L, Thorgeirsson T, et al. Prostate cancer progression and survival in BRCA2 mutation carriers. J Natl Cancer Inst 2007;99:929-35. 10.1093/jnci/djm005 [DOI] [PubMed] [Google Scholar]
- 32.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748-57. 10.1200/JCO.2012.43.1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallagher DJ, Gaudet MM, Pal P, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 2010;16:2115-21. 10.1158/1078-0432.CCR-09-2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cybulski C, Wokolorczyk D, Kluzniak W, et al. An inherited NBN mutation is associated with poor prognosis prostate cancer. Br J Cancer 2013;108:461-8. 10.1038/bjc.2012.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leongamornlert D, Saunders E, Dadaev T, et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer 2014;110:1663-72. 10.1038/bjc.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 2008;40:316-21. 10.1038/ng.90 [DOI] [PubMed] [Google Scholar]
- 38.Kote-Jarai Z, Easton DF, Stanford JL, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev 2008;17:2052-61. 10.1158/1055-9965.EPI-08-0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macinnis RJ, Antoniou AC, Eeles RA, et al. A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol 2011;35:549-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med 2008;358:910-9. 10.1056/NEJMoa075819 [DOI] [PubMed] [Google Scholar]
- 41.Eeles R. Prostate cancer genome-wide association study from 89,000 men using the OncoArray chip to identify novel prostate cancer susceptibility loci. J Clin Oncol 2016;34:1525. [Google Scholar]
- 42.Eeles RA, Olama AA, Benlloch S, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 2013;45:385-91, 91e1-2. [DOI] [PMC free article] [PubMed]
- 43.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature 2017;548:297-303. 10.1038/nature23306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benafif S. The BARCODE 2 Study - The use of gentic profiling to guide prostate cancer treatment (BARCODE2). Available online: https://clinicaltrials.gov/ct2/show/NCT02955082
- 45.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373:1697-708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson D, Van Allen EM, Wu YM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162:454. 10.1016/j.cell.2015.06.053 [DOI] [PubMed] [Google Scholar]
- 47.Cullen J, Rosner IL, Brand TC, et al. A Biopsy-based 17-gene Genomic Prostate Score Predicts Recurrence After Radical Prostatectomy and Adverse Surgical Pathology in a Racially Diverse Population of Men with Clinically Low- and Intermediate-risk Prostate Cancer. Eur Urol 2015;68:123-31. 10.1016/j.eururo.2014.11.030 [DOI] [PubMed] [Google Scholar]
- 48.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol 2016;69:157-65. 10.1016/j.eururo.2015.05.042 [DOI] [PubMed] [Google Scholar]
- 49.Karnes RJ, Choeurng V, Ross AE, et al. Validation of a Genomic Risk Classifier to Predict Prostate Cancer-specific Mortality in Men with Adverse Pathologic Features. Eur Urol 2018;73:168-75. 10.1016/j.eururo.2017.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punnen S, Pavan N, Parekh DJ. Finding the Wolf in Sheep's Clothing: The 4Kscore Is a Novel Blood Test That Can Accurately Identify the Risk of Aggressive Prostate Cancer. Rev Urol 2015;17:3-13. [PMC free article] [PubMed] [Google Scholar]
- 51.Cucchiara V, Cooperberg MR, Dall’Era M, et al. Genomic Markers in Prostate Cancer Decision Making. Eur Urol 2017. [Epub ahead of print]. 10.1016/j.eururo.2017.10.036 [DOI] [PubMed] [Google Scholar]
- 52.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 2009;41:1116-21. 10.1038/ng.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitra AV, Bancroft EK, Barbachano Y, et al. Targeted prostate cancer screening in men with mutations in BRCA1 and BRCA2 detects aggressive prostate cancer: preliminary analysis of the results of the IMPACT study. BJU Int 2011;107:28-39. 10.1111/j.1464-410X.2010.09648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium launches genotyping effort. Cancer Discov 2013;3:1321-2. 10.1158/2159-8290.CD-NB2013-159 [DOI] [PubMed] [Google Scholar]
- 55.Castro E, Mikropoulos C, Bancroft EK, et al. The PROFILE Feasibility Study: Targeted Screening of Men With a Family History of Prostate Cancer. Oncologist 2016;21:716-22. 10.1634/theoncologist.2015-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]