Abstract
Active surveillance (AS) is an important treatment modality aiming to reduce the overtreatment of patients with prostate cancer (PCa) who have a low risk of disease reclassification. After enrolling in AS patients are actively monitored using different diagnostic tests (e.g., prostate specific-antigen, digital rectal exams (DREs), medical imaging, and prostate biopsies). Biopsy is the most burdensome test. We aimed to review schedules for monitoring men on AS. We compare fixed versus risk based dynamic monitoring, where biopsies are scheduled during follow-up based on dynamic risk predictions. Several prediction models and scheduling techniques have been published. All proposed risk prediction models need further external validation. We conclude that risk based, dynamic monitoring is a promising new strategy to further reduce overtreatment in PCa patients.
Keywords: Prostate cancer (PCa), active surveillance (AS), prediction model, review
Introduction
Screening for prostate cancer (PCa) leads to a reduction in PCa-related mortality as shown in the ERSPC trial (1). However, screening also results in relatively large number of men being diagnosed with Gleason grade 3+3=6 cancer (2). These men typically have a very low risk of dying due to PCa, and hence many of these cancers may be considered overdiagnosed. In these men active surveillance (AS) is typically proposed as a treatment option (3). AS consists of closely monitoring the disease status of the patient using clinical work up and repeat biopsies to detect a possible disease reclassification. If a disease reclassification is observed, curative treatment may often be initiated. Several simulation studies have shown benefit of AS compared to immediate treatment for some patient groups (4-6).
Commonly used surveillance tools in AS are prostate-specific antigen (PSA), digital rectal exam (DRE) and follow-up biopsies (7). Follow-up biopsies are burdensome diagnostic tests which have the additional risk of infection and/or bleeding (8). Therefore it would be desirable to minimize the number of biopsies. On the other hand, biopsies are currently the reference standard in detecting disease reclassification in AS. Omitting biopsies may lead to too late detection of disease reclassification, potentially foregoing the window of opportunity for curative treatment.
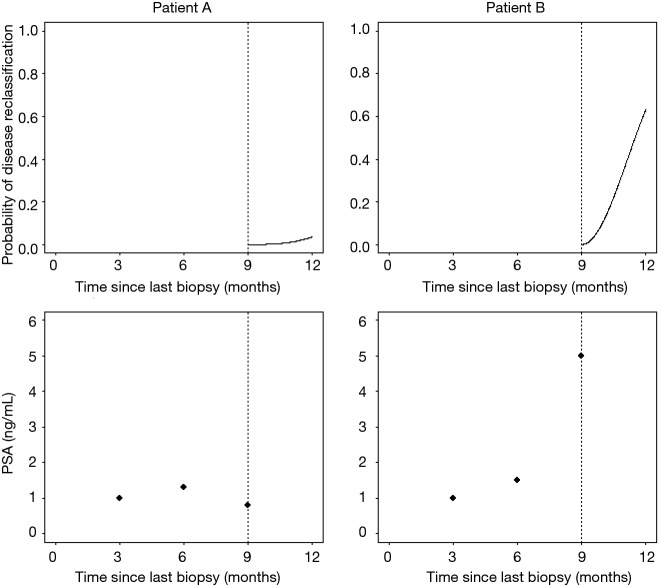
A promising alternative to fixed protocols for AS is risk based, dynamic monitoring. Consider for instance that we have two hypothetical patients who have been followed for nine months with PSA measurements every three months and are scheduled to receive a biopsy at 12 months (Figure 1). Patient A shows a stable PSA around 1 ng/mL and consequently has a very low risk of showing disease reclassification at 12 months. Patient B shows a fast rising PSA and has a relatively high risk of disease reclassification. For patient A it may be preferable to defer the biopsy to a later time point in AS, while patient B might opt for an earlier biopsy. The use of prediction models makes it possible to give risk estimates based on information collected during AS and potentially allow for such individualized schedules.
Figure 1.
Hypothetical PSA profile and risk of progression for two hypothetical patients. PSA, prostate-specific antigen.
Our aim was to compare current fixed schedules with risk-based, dynamic monitoring and to discuss proposed prediction models which may be used in risk-based follow-up during AS.
Current surveillance schedules and stopping criteria
A recent review compared guidelines on AS schedules developed by different medical associations (7). Moreover, the Movember AS database comprises patients from all major studies on AS from all over the world. An overview of patient characteristics in the GAP3 database and on overview of selection criteria, monitoring tools and surveillance schedules has been published recently (9).
Here we present a short overview of currently recommended monitoring tools and surveillance schedules mentioned in different clinical guidelines as developed by medical associations and research cohorts. The previous reviews identified several surveillance methods during AS, namely serum PSA measurements, DRE, repeat biopsy sampling, PSA kinetics, and MRI (Table 1).
Table 1. Summary of monitoring tools, timing of monitoring tools and criteria for switching to definitive therapy.
| Category | Monitoring tools | Triggers for definitive therapy |
|---|---|---|
| Follow-up monitoring and triggers for definitive therapy recommendations in guidelines | ||
| Guideline | ||
| EAU (10) | Serum PSA, DRE and rebiopsy mentioned | NR*** |
| NCCN (11) | Serum PSA ≤6 months, unless clinically indicated, DRE ≤12 months unless clinically indicated, rebiopsy ≤12 months unless clinically indicated or at 6 months if the initial biopsy sample had <10 cores, MRI mentioned | Rebiopsy sample GS contains grade 4 or 5 disease, increased number of tumor-positive cores or increased extent of cancer per core |
| NICE (12) | Serum PSA, 3–4 months year 1 post-diagnosis, every 3–6 months year 2–4 and then every 6 months, PSA kinetics measured throughout AS, DRE every 6–12 months year 1–4 and then annually, rebiopsy 12 months after diagnosis or if concern exists about clinical or PSA changes, MRI at enrollment f not done, before or in the presence of concern about clinical or PSA changes. | Switching recommended if disease progression observed, also taking into account patient’s life expectancy, preferences and comorbidities |
| GSU (13) | Serum PSA every 3 months year 0–3 post-diagnosis, every 6 months thereafter, PSA kinetics mentioned, DRE every 3 months year 0–3 post-diagnosis then every 6 months, if PSA stable rebiopsy every 12–18 months within 3 years post-diagnosis, then every 3 years | PSA >10 ng/mL, PSA-DT <3 years, tumor stage > cT2a, repeat biopsy GS >6, >2 tumor-positive cores, >50% cancer per biopsy core |
| KCE (14) | Serum PSA every 6 months, DRE every 6 months, rebiopsy within 1-year and repeated thereafter, annual MRI | PSA>10 ng/mL, PSA-DT <3 years, clinical change detected during DRE, or suspicious lesions on MRI |
| FCCG (15) | Serum PSA, DRE and rebiopsy mentioned | PSA-DT <3 years, repeat biopsy GS >6, >2 tumor-positive cores, or disease is reclassified as clinically relevant |
| SCAN (16) | Serum PSA every 3 months year 1 post-diagnosis, every 6 months thereafter, PSA-DT after 5 PSA measurements, DRE ≤6 months, repeat biopsy within 6 months post diagnosis then years 1, 4, 7, and 10 | PSA-DT <3, progression of palpable T2-stage disease on DRE or palpable lesions appearing. GS 4 or 5 at rebiopsy >50% spread of cancer in any one core, >50% of core samples, or bilateral disease |
| CCNS (17) | Serum PSA every 6 months, DRE every 6 months, rebiopsy at 6 months if initial biopsy <10 cores or findings discordant with clinical findings, within 18 months otherwise, periodically thereafter | Switch recommended at disease progression |
| I+CS (18) | Serum PSA every 3 months 0–3 years post-diagnosis, every 6 months thereafter, PSAV with at least 5 PSA measurements, DRE every 3 months within year 0–2 post-diagnosis, every 6 months thereafter, rebiopsy year 1, 4, and 7 post-diagnosis | PSAV >1 ng/mL, DRE, rebiopsy GS, number of tumor-positive cores, maximum extent of cancer per core all mentioned |
| AHS (19) | Serum PSA every 3–6 months, PSA kinetics mentioned, DRE annually, rebiopsy year 1–2 post-diagnosis, every 2–3 years thereafter or as clinically indicated | PSA-DT <3 years, increase clinical stage from baseline at DRE, Gleason pattern 4 or higher, >50% of cancer in any one core, patient preferences |
| CCO (20) | Serum PSA every 3–6 months, DRE annually, rebiopsy 6–12 months post-diagnosis, every 3–5 years thereafter, MRI mentioned | Repeat biopsy GS ≥7 with pattern 4 accounting for >10% of the total tumor and/or significant increase in tumor volume |
| PCT (21) | Serum-PSA every 6 months, or every 3 months when concern of progression, DRE mentioned, PSA kinetics mentioned, repeat biopsy within 12 months of initial biopsy or when clinically indicated | NR |
| Follow-up monitoring recommendations of cohorts in the GAP3 database | ||
| Center* | ||
| USA | ||
| JHU | Serum PSA every 6 months, DRE every 6 months, rebiopsy every 12 months | |
| UCSF | Serum PSA every 3 months, DRE every 6 months, rebiopsy every 12–24 months | |
| MSKCC | Serum PSA every 6 months, DRE every 6 months, rebiopsy every 3 years, mpMRI every 18 months | |
| MDACC | Serum PSA every 6 months, DRE every 12 months, rebiopsy every 12–24 months | |
| EU | Serum PSA every 6 months, DRE every 12 months, rebiopsy every 12 months, mpMRI annually for the first 3 years then final after 5 years |
|
| MUSIC | Serum PSA every 3–6 months, DRE every 12 months, rebiopsy every other year, mpMRI every other year and confirmatory test in first 3–4 months | |
| Canada | ||
| UOFC | Serum PSA every 6 months, DRE every 6 months, rebiopsy at year 1 then every 2 years, mpMRI when PSA >10 ng/mL | |
| UOFT | Serum PSA every 3 months until 2 years, then every 6 months, PSA kinetics every 12 months, DRE every 6 months, rebiopsy at year 1, 4, 7, 10, and 15, mpMRI every 12 months | |
| United Kingdom | ||
| GSTT | Serum PSA every 6 months, PSA kinetics every 12 months, DRE every 12 months, mpMRI every 12 months | |
| UCL | Serum PSA 3–4 monthly in year 1 then every 6 months, rebiopsy for men with change in MRI and uncertainty about switch to definitive therapy, mpMRI at baseline and 12 months the dependent on risk factors including MRI, PSA density and GS | |
| Camb | Serum PSA every 3 months, rebiopsy within 1 year post-diagnosis, then at 36 and 60 months and if mpMRI shows a change, mpMRI every 12 months | |
| Europe | ||
| KSB | Serum PSA every 6 months, DRE every 6 months, rebiopsy every 24 months | |
| UCD | NR | |
| SU | Serum PSA every 3–6 months, DRE every 6–12 months, rebiopsy every 2–3 years | |
| Lille-U | Serum PSA every 6 months, DRE every 12 months, rebiopsy at month 12, mpMRI at month 12 | |
| SUS | Serum PSA every 3 months, DRE every 6 months, rebiopsy after month 12, 48, and 84 | |
| INT-MILAN (SAINT) | Serum PSA every 3 months, PSA kinetics after first year every 3 months, DRE every 6 months, rebiopsy every 12 months first 2 years, then every 24 months | |
| INT-MILAN (PRIAS) | Serum PSA every 3 months, PSA kinetics after first year every 3 months, DRE every 6 months, rebiopsy after 12, 48, and 84 months | |
| IVO | Serum PSA every 6 months, DRE every 6 months, rebiopsy at month 6 and 24 then every 3 years, mpMRI before every biopsy | |
| Australasia | ||
| MEASCAP | Serum PSA every 3 months, DRE every 6 months, rebiopsy month 12, 48 and 84 | |
| SGH | Serum PSA every 3–6 months first 2 years then every 6–12 months, PSA kinetics every 12 months, DRE every 12 months, rebiopsy every 12 months, mpMRI every 12 months | |
| YUHS | Serum PSA every 3 months, rebiopsy considered when change on mpMRI is detected, mpMRI every 12 months | |
| KU | Serum PSA every 2 months for 6 months then every 3 months, DRE every 12 months, rebiopsy every 12 months | |
| Other | ||
| PRIAS** | Serum PSA every 3 months, PSA kinetics every 6 months, DRE every 6 months, rebiopsy after 12, 48, and 84 months | |
*, Institute abbreviations: JHU, Johns Hopkins University, Baltimore, USA; UCSF, University of California, San Francisco, San Francisco, USA; MSKCC, Memorial Sloan Kettering Cancer Center, New York, USA; MDACC, MD Anderson Cancer Centre, Houston, USA; EU, Emory University School of Medicine, Winship Cancer Institute, Atlanta, USA; MUSIC, University of Michigan and Michigan Urological Surgery Improvement Collaborative, Michigan, USA; UOFC, University of Calgary, Southern Alberta Institute of Urology, Calgary, Canada; UOFT, University of Toronto, Sunnybrook Health Sciences Centre, Toronto, Canada; GSTT, Guy’s and St Thomas’ NHS Foundation Trust, London, United Kingdom; UCL, University College London & University College London Hospitals Trust, London, United Kingdom; Camb, Cambridge University Hospitals NHS Trust, Cambridge, United Kingdom; KBS, Kantonsspital Baden, Baden, Switzerland; UCD, University College Dublin, Dublin, Ireland; SU, Sahlgrenska University Hospital, Göteborg, Sweden; Lille-U, Lille University Hospital Center, Lille, France; SUS, Skåne University Hospital, Malmö, Sweden; INT-MILAN, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy; IVO, Instituto Valenciano de Oncología, Valencia, Spain; MEASCAP, Monash University and Epworth HealthCare, Melbourne, Australia; YUHS, Gangnam Severance Hospital, Yonsei University Health System, Seoul, Republic of Korea; KU, Kagawa University Faculty of Medicine, Kagawa, Japan. **, PRIAS (Prostate Cancer Research International Active Surveillance): includes Erasmus Medical Center, Rotterdam, the Netherlands; Helsinki University Central Hospital, Helsinki, Finland; University of British Columbia, BC Cancer Agency, Vancouver, Canada). ***, NR, not reported.
Serum PSA measurements
All guidelines that mention follow-up criteria include recommendations to use serum PSA measurements as a monitoring tool during follow-up. The NCCN, KCE, CCNS and PCT guidelines mention that PSA measurements should be implemented with intervals of at most 6 months. Additionally the PCT guidelines recommend doing a PSA measurement every three months whenever there is a concern over PSA progression. Three guidelines recommend to do PSA measurements every 3–6 months. Four guidelines (NICE, GSU, SCAN, I+CS) use intervals based on the duration of being on AS, typically they recommend PSA testing every 3 months when starting AS and every 6 months after 1–2 years on AS. Most cohorts included in Movember AS database recommend to perform PSA testing every 3–6 months, four cohorts recommend the frequency of PSA testing to depend on the duration in AS.
Digital rectal examination
All guidelines also recommend the use of DRE as a monitoring tool. The majority of the guidelines recommend to perform a DRE biannually or annually. One guideline recommend a DRE every three months at the start of AS and every six months after 2 years on AS (I+CS) or when the serum PSA is stable in the first two years (GSU).
DRE is less frequently used as a monitoring tool in the studies in the Movember AS database, due to the use of multi-parametric magnetic resonance imaging (mpMRI). When DRE is used as a monitoring tool it is typically performed every 6 or 12 months.
Repeat biopsy scheduling
Repeat TRUS guided biopsy scheduling is recommended by all protocols. Repeat biopsy scheduling is included to periodically evaluate if disease reclassification has occurred, which might influence the prognosis of the patient and is the trigger to switch from AS to curative therapy. There is substantial variation in biopsy schedules between the different guidelines and studies. Most guidelines recommend an early repeat biopsy within 1.5 years after starting AS. Hereafter, guidelines show substantial variation recommending yearly repeat biopsies (NCCN) or an early rebiopsy and 2–3 years intervals between later repeat biopsies (GSU, SCAN, AHS, CCO). While other guidelines mention only an early rebiopsy and make no specific recommendations for later repeat biopsies (KCE, CCNS, NCCS). One guideline recommends a rebiopsy after 1 year and only later repeat biopsies when concern rises about clinical and/or serum PSA changes (NICE).
Substantial variation in the frequency and timing of the repeat biopsies is also present in the cohorts in the Movember AS database. Some cohorts perform a repeat biopsy every 1–2 years, and some cohorts perform an early rebiopsy (typically after 1 year of starting AS). Two cohorts do not routinely perform repeat biopsy, but only if changes on mpMRI lead to uncertainty about the decision to switch to definitive therapy (UCL) or whenever a change on mpMRI is observed (YUHS).
PSA kinetics
Some guidelines recommend the use of PSA kinetics during follow-up. Remarkably, none of the clinical guidelines mention specific cut-offs to use with PSA kinetics. The PRIAS study recommend to perform a repeat biopsy whenever the PSA-DT was less than 10 years.
MRI
Four guidelines mention MRI as a possible tool to be used in AS. Most suggest that MRI might be used when clinical parameters such as PSA, DRE, and PSA kinetics signal disease reclassification but results from the repeat biopsy are discordant.
Some cohorts in the Movember AS database use the mpMRI routinely during follow-up. UCL and YUHS only perform repeat biopsies whenever changes on mpMRI are observed. Whenever mpMRI is used it is typically performed annually. One cohort performed an mpMRI before every repeat biopsy (IVO).
Summary of triggers to switch to definitive therapy
Most guidelines recommend to switch to definitive treatment whenever an increase in Gleason grade is detected during AS or if an increase in the number of tumor positive cores is detected (>2 of a recommended 10 cores) or an increase in the extent of cancer in biopsy cores (>50% of cancer per tumor-positive core). Clinical progression detected during DRE (although not clearly defined), PSA-DT <3 years are also mentioned as criteria in five guidelines (GSU, KCE, FCCG, SCAN, AHS). Three guidelines explicitly mentioned patient preference as a trigger to switch treatment. The NICE guidelines also mention that comorbidities and life expectancy should be taken into account when considering the switch from AS to definitive therapy. Triggers for switching to definitive therapy were not mentioned in the overview of cohorts in the Movember AS database. Currently the guidelines do not mention the option to switch from AS to watchful waiting. Watchful waiting is similar but distinct to AS, the main difference is that in watchful waiting no curative treatment is initiated when disease reclassification is detected. Watchful waiting may be a viable alternative for patients with low life-expectancy and/or comorbidities.
Summary of protocolized surveillance
After initiating AS for a patient, most guidelines and individual study protocols recommend the use of serum PSA measurements, DRE and repeat biopsies as tools to detect progression of the PCa. However there is a wide variation in the timing and frequency of the measurements of these quantities. Some variation in the definition of progression of PCa exists; all guidelines and studies consider an increase in Gleason grade as progression. However, progression due to increase of the tumor volume is operationalized differently between studies and guidelines. MRI and PSA kinetics are mentioned less frequently as monitoring tools in the guideline, and need further investigation how to best use these.
Risk-based dynamic monitoring
Performing a prostate biopsy is a medical burdensome procedure. Better scheduling of biopsies might allow for less unnecessary procedures and the same benefits of AS. To be able to implement risk-based dynamic monitoring schedule a prediction model is required that dynamically incorporates all relevant information of the patients and provides an estimate of the risk of disease reclassification. A major challenge is to incorporate the serial measurements of the monitoring tools into the prediction model. Three such models have been developed recently: the Canary PASS model, Johns Hopkins model, and the PRIAS model (Table 2).
Table 2. Characteristics of prediction models developed to estimate the risk of disease reclassification during AS.
| Prediction model | Statistical technique | Development population | Endpoint |
|---|---|---|---|
| Canary-PASS | Generalized estimating equations (GEE) | 859 patients from the Canary-PASS cohort | GS ≥7 or ≥34% of tumor-positive cores |
| Johns Hopkins | Bayesian hierarchical latent class model | 964 patients from Johns Hopkins | GS ≥7 at radical prostatectomy |
| PRIAS | Joint model* | 5,624 patients from the PRIAS project | Time until GS ≥7 at biopsy |
*, a combination of a mixed effect model and a survival model. AS, active surveillance.
Canary-PASS risk calculator
The Canary PASS is an ongoing multicenter prospective and observational AS study (22,23). Progression was defined as an increase in GS from ≤6 to ≥7 or more than 34% of positive biopsy cores at biopsy. Using a cohort of 859 patients, of whom 354 (41%) eventually showed disease reclassification, a prediction model was developed using logistic regression with generalized estimating equations. Reclassification rates varied between 11–29% at the different biopsy rounds. Predictors included in the prediction model were age at biopsy, time since last biopsy, last PSA measurement, percentage of tumor-positive cores on the last biopsy and the number of tumor-negative biopsies while on AS. The discriminative ability of the prediction model was assessed using the area under the receiver-operating characteristic (AUC) and was equal to 0.74.
Johns Hopkins post radical prostatectomy prediction model
The Johns Hopkins Active Surveillance study is a cohort of men who have low-risk PCa according to the Epstein criteria (24). A prediction model based on a Bayesian hierarchical latent class model was developed to predict the GS after a radical prostatectomy (25). The development cohort consisted of 964 patients, of whom 195 (20%) experienced reclassification after a biopsy and 191 patients received radical prostatectomy after opting for active treatment. Of these 191 patients, 79 patients had a GS ≥7 at radical prostatectomy. The developed prediction model included the covariates patient age, prostate volume, PSA, time since diagnosis, number of previous biopsies, and all previous biopsy results. The AUC of the resulting model for predicting the GS after RP was 0.74 and the model showed reasonable calibration at internal validation.
PRIAS
The PRIAS study is the world’s largest ongoing web-based AS study (26). Based on 5,264 patients a Bayesian joint model was developed to study the association between the evolution of PSA measurements and the risk of Gleason grade reclassification (27,28). PSA velocity was found to be more strongly associated with the risk of Gleason grade reclassification than PSA. Using this model, individual predictions could be derived for each patient for progression from GS6 to GS ≥7. These predictions are updated as new PSA measurements and biopsy results become available. Time-varying AUCs at year 1, 2 and 3 were only moderate (0.61, 0.66, and 0.60 respectively, for a time window of 1 year) (29).
Protocol based monitoring versus risk-based monitoring
A number of studies have aimed to compare the impact of different fixed follow-up schedules on patient outcomes. Using data from the Johns Hopkins cohort, delay in detection of disease reclassification and the reduction in the number of biopsies was compared if the PRIAS protocol was applied in these patients (30). It was found that using the PRIAS protocol would lead to a median delay in detection time of 1.9 years.
Several simulation studies have been performed aiming to estimate the trade-off between the reduction in number of biopsies taken and the delay in detection of disease reclassification (31-33). Although all studies used different modelling strategies, two studies used data from the Johns Hopkins cohort to calibrate their models (33,34). Both these studies concluded that switching from yearly biopsies to less frequent biopsies would lead to a relatively modest increase in delay of detection of the disease reclassification. The third simulation study used data from Johns Hopkins, Canary Prostate Active Surveillance Study, University of California, San Francisco, and University of Toronto. A delay in detection of disease reclassification of 3-5 months was estimated when switching from yearly to biennial biopsies.
Another study used simulations to compare the consequences of protocol based screening versus risk-based dynamic monitoring using patient data from PRIAS (28). The time to Gleason score upgrading of the PCa was estimated using joint models, taking into account the results from PSA tests and previous biopsy results. A risk-based dynamic monitoring approach was compared to the Johns Hopkins follow-up schedule and the PRIAS schedule. It was found that the risk based dynamic monitoring approach could potentially reduce the average number of biopsies taken during follow-up from 5 to 2, at the price of increasing the time between disease reclassification and the detection of upgrading from 6–8 months to approximately 14 months. No major difference in the number of biopsy taken and the delay in detection of the disease reclassification was found between the Johns Hopkins and PRIAS schedule.
Discussion
Currently few models are available to predict the risk of disease reclassification reliably during follow-up in AS. Whenever models have been developed thorough external validation of these models is lacking. Several simulation studies have been performed comparing more strict and more relaxed follow-up schedules. These have illustrated the trade-off between the number of biopsies taken and the delay in detection of disease progression. These simulation studies suggested that a yearly schedule could be relaxed to a biennial or triennial follow-up schedule without a large increase in delay of PCa detection, although a formal risk-benefit analysis is needed for firmer conclusions.
A recent microsimulation study investigated the optimal age at which AS should be stopped. After such an age, further biopsies would provide no further benefit, indicating that a switch to watchful waiting is warranted (31). The optimal age at which to stop AS was strongly dependent on the disease risk and life expectancy of a patient. Most guidelines and currently proposed prediction models do not consider life expectancy explicitly in recommendations for switching from AS to definitive therapy. Life-expectancy may modeled using a competing risk analysis, where death due to other causes is a competing risk to disease progression.
The majority of the cohorts containing AS patients are observational cohorts. Developing prediction models using observational data runs the risk of introducing biases from the dataset into the prediction model. In the context of AS cohorts this is present in the form of ascertainment bias, patients who are at high risk of progression receive more biopsies and hence are detected earlier. For instance, some guidelines recommend to do a biopsy if the PSA-DT is too short. Most statistical methods provide unbiased estimates as long as all relevant patient and tumor characteristics are entered in the prediction model that is used in the decision to take a biopsy. Having such a statistically correct model is a rather strong assumption.
The vast majority of AS schedules use PSA and DRE measurements as a surveillance tool. There is an increasing interest in the use of mpMRI in AS as a tool to reduce the number of biopsies (35). Additionally, various blood biomarkers are investigated as monitoring tools in AS (36,37). These markers can potentially improve the previously developed prediction models and may further reduce the number of biopsies required to detect a disease reclassification and/or the delay in the detection of disease reclassification. External validation across a range of setting is required before clinical application of such models is considered (38,39).
In conclusion, risk-based dynamic monitoring is a promising alternative to a fixed follow-up schedule for all patients in AS. Simulation studies have shown that the number of biopsies required to detect disease reclassification could be reduced at the price a limited delay in detection of the disease reclassification. External validation of the proposed prediction models is currently lacking and is essential before implementing these dynamic prediction models into clinical practice.
Acknowledgements
Funding: A Tomer and D Rizopoulos would like to acknowledge support by the Netherlands Organization for Scientific Research’s VIDI grant nr. 016.146.301, and Erasmus MC funding.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014;384:2027-35. 10.1016/S0140-6736(14)60525-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeb S, Bjurlin MA, Nicholson J, et al. Overdiagnosis and overtreatment of prostate cancer. Eur Urol 2014;65:1046-55. 10.1016/j.eururo.2013.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klotz L. Active surveillance for low-risk prostate cancer. F1000 Med Rep 2012;4:16. 10.3410/M4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Carvalho TM, Heijnsdijk EA, de Koning HJ. Estimating the individual benefit of immediate treatment or active surveillance for prostate cancer after screen- detection in older (65+) men. Int J Cancer 2016;138:2522-8. 10.1002/ijc.29976 [DOI] [PubMed] [Google Scholar]
- 5.Hayes JH, Ollendorf DA, Pearson SD, et al. Active surveillance compared with initial treatment for men with low- risk prostate cancer: a decision analysis. JAMA 2010;304:2373-80. 10.1001/jama.2010.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeb S, Zhou Q, Siebert U, et al. Active Surveillance Versus Watchful Waiting for Localized Prostate Cancer: A Model to Inform Decisions. Eur Urol 2017;72:899-907. 10.1016/j.eururo.2017.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruinsma SM, Bangma CH, Carroll PR, et al. Movember GAP3 consortium Active surveillance for prostate cancer: a narrative review of clinical guidelines. Nature reviews. Urology 2016;13:151-67. 10.1038/nrurol.2015.313 [DOI] [PubMed] [Google Scholar]
- 8.Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64:876-92. 10.1016/j.eururo.2013.05.049 [DOI] [PubMed] [Google Scholar]
- 9.Bruinsma SM, Zhang L, Roobol MJ, et al. The Movember Foundation’s GAP3 cohort: A profile of the largest global prostate cancer active surveillance database to date. BJU Int 2017. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 10.European Association of Urology (EAU). Guidelines on prostate cancer, 2014.
- 11.The National Comprehensive Cancer Network (NCCN) NCCN Clinical Practice Guidelines in Oncology: Prostate cancer. 2014. [DOI] [PubMed] [Google Scholar]
- 12.The National Institute for Health and Clinical Excellence (NICE). Prostate cancer: diagnosis and management. 2014. [PubMed]
- 13.German Society of Urology (GSU). Interdisziplinäre Leitlinie der Qualität S3 zur Früherkennung, Diagnose und Therapie der verschiedenen Stadien des Prostatakarzinoms. 2014.
- 14.Belgian Healthcare Knowledge Centre (KCE). A National Clinical Practice Guideline on the Management of Localised Prostate Cancer. 2013.
- 15.The Finnish Medical Society Duodecim (FCCG) Prostate cancer. 2014. [Google Scholar]
- 16.South East Scotland Cancer Network (SCAN). SCAN Guideline for Active Surveillance (Deferred Radical Treatment) of Early, Low-risk, Prostate Cancer. 2015.
- 17.Cancer Care Nova Scotia (CCNS). Guidelines for the Management of Prostate Cancer. 2006.
- 18.Aragon Institute of Health Sciences (I+CS). Clinical Practice Guideline for Prostate Cancer Treatment. 2008.
- 19.Alberta Health Services (AHS) Alberta Health Services Clinical Practice Guideline: Prostate Cancer. 2014. [Google Scholar]
- 20.Morash C, Tey R, Agbassi C, et al. Active surveillance for the management of localized prostate cancer: Guideline recommendations. Can Urol Assoc J 2015;9:171-8. 10.5489/cuaj.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prostate Cancer Taskforce (PCT). Diagnosis and management of prostate cancer in New Zealand men: recommendations from the Prostate Cancer Taskforce. The New Zealand Ministry of Health 2013. [Google Scholar]
- 22.Newcomb LF, Brooks JD, Carroll PR, et al. Canary Prostate Active Surveillance Study: design of a multi- institutional active surveillance cohort and biorepository. Urology 2010;75:407-13. 10.1016/j.urology.2009.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ankerst DP, Xia J, Thompson IM, et al. Precision Medicine in Active Surveillance for Prostate Cancer: Development of the Canary-Early Detection Research Network Active Surveillance Biopsy Risk Calculator. Eur Urol 2015;68:1083-8. 10.1016/j.eururo.2015.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coley RY, Zeger SL, Mamawala M, et al. Prediction of the Pathologic Gleason Score to Inform a Personalized Management Program for Prostate Cancer. Eur Urol 2017;72:135-41. 10.1016/j.eururo.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 25.Coley RY, Fisher AJ, Mamawala M, et al. A Bayesian hierarchical model for prediction of latent health states from multiple data sources with application to active surveillance of prostate cancer. Biometrics 2017;73:625-34. 10.1111/biom.12577 [DOI] [PubMed] [Google Scholar]
- 26.Bokhorst LP, Valdagni R, Rannikko A, et al. A Decade of Active Surveillance in the PRIAS Study: An Update and Evaluation of the Criteria Used to Recommend a Switch to Active Treatment. Eur Urol 2016;70:954-60. 10.1016/j.eururo.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 27.Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: an overview. Statistica Sinica 2004;14:809-34. [Google Scholar]
- 28.Tomer A, Nieboer D, Roobol MJ, et al. Personalized Schedules for Surveillance of Low Risk Prostate Cancer Patients. [DOI] [PMC free article] [PubMed]
- 29.Rizopoulos D, Molenberghs G, Lesaffre EMEH. Dynamic predictions with time- dependent covariates in survival analysis using joint modeling and landmarking. Biom J 2017;59:1261-76. 10.1002/bimj.201600238 [DOI] [PubMed] [Google Scholar]
- 30.Kates M, Tosoian JJ, Trock BJ, et al. Indications for intervention during active surveillance of prostate cancer: a comparison of the Johns Hopkins and Prostate Cancer Research International Active Surveillance (PRIAS) protocols. BJU Int 2015;115:216-22. 10.1111/bju.12828 [DOI] [PubMed] [Google Scholar]
- 31.de Carvalho TM, Heijnsdijk EA, de Koning HJ. When should active surveillance for prostate cancer stop if no progression is detected? Prostate 2017;77:962-9. 10.1002/pros.23352 [DOI] [PubMed] [Google Scholar]
- 32.Inoue LY, Lin DW, Newcomb LF, et al. Comparative Analysis of Biopsy Upgrading in Four Prostate Cancer Active Surveillance Cohorts. Ann Intern Med 2018;168:1-9. 10.7326/M17-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnett CL, Auffenberg GB, Cheng Z, et al. Optimizing active surveillance strategies to balance the competing goals of early detection of grade progression and minimizing harm from biopsies. Cancer 2017. [Epub ahead of print]. 10.1002/cncr.31101 [DOI] [PubMed] [Google Scholar]
- 34.de Carvalho TM, Heijnsdijk EA, de Koning HJ. Estimating the risks and benefits of active surveillance protocols for prostate cancer: a microsimulation study. BJU Int 2017;119:560-6. 10.1111/bju.13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberts AR, Roobol MJ, Drost FH, et al. Risk-stratification based on magnetic resonance imaging and prostate- specific antigen density may reduce unnecessary follow-up biopsy procedures in men on active surveillance for low-risk prostate cancer. BJU Int 2017;120:511-9. 10.1111/bju.13836 [DOI] [PubMed] [Google Scholar]
- 36.Lin DW, Newcomb LF, Brown MD, et al. Evaluating the Four Kallikrein Panel of the 4Kscore for Prediction of High-grade Prostate Cancer in Men in the Canary Prostate Active Surveillance Study. Eur Urol 2017;72:448-54. 10.1016/j.eururo.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tosoian JJ, Patel HD, Mamawala M, et al. Longitudinal assessment of urinary PCA3 for predicting prostate cancer grade reclassification in favorable-risk men during active surveillance. Prostate Cancer Prostatic Dis 2017;20:339-42. 10.1038/pcan.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. New York: Springer, 2009. [Google Scholar]
- 39.Steyerberg EW, Harrell FE. Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol 2016;69:245-7. 10.1016/j.jclinepi.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]