Abstract
The ubiquitin-proteasome pathway acts as a regulator of the endocytosis of selected membrane proteins. Recent evidence suggests that it may also function in the intracellular trafficking of membrane proteins. In this study, several models were used to address the role of the ubiquitin-proteasome pathway in sorting of internalized proteins to the lysosome. We found that lysosomal degradation of ligands, which remain bound to their receptors within the endocytic pathway, is blocked in the presence of specific proteasome inhibitors. In contrast, a ligand that dissociates from its receptor upon endosome acidification is degraded under the same conditions. Quantitative electron microscopy showed that neither the uptake nor the overall distribution of the endocytic marker bovine serum albumin-gold is substantially altered in the presence of a proteasome inhibitor. The data suggest that the ubiquitin-proteasome pathway is involved in an endosomal sorting step of selected membrane proteins to lysosomes, thereby providing a mechanism for regulated degradation.
INTRODUCTION
After internalization from the plasma membrane, molecules are rapidly delivered to early endosomes, also known as sorting endosomes. Most of the soluble content of sorting endosomes is delivered to lysosomes for degradation, whereas the majority of membrane-bound proteins recycle back to the plasma membrane. Recycling receptors such as the transferrin receptor and the low-density lipoprotein (LDL) receptor are segregated into tubular membrane extensions of the sorting endosome and recycle with >99% efficiency, thereby avoiding proteolysis (reviewed in Trowbridge et al., 1993; Mellman, 1996). Recycling receptors are reused many times and are important for nutrient delivery and scavenging of nonfunctional proteins such as protease/protease inhibitor complexes and altered glycoproteins. On the other hand, signal-transducing membrane receptors such as the epidermal growth factor receptor (EGFR) and the growth hormone receptor (GHR) are transported together with their ligand into lysosomes for degradation, a process often referred to as signal down-regulation. Membrane proteins destined for lysosomal degradation are segregated into intraendosomal vesicles, which results in the formation of late endosomes or multivesicular bodies (MVBs), thus providing a mechanism to remove this class of proteins from the limiting membrane of the endosome (Felder et al., 1990). Down-regulation of growth factor receptors is important for cellular regulation; disrupted internalization or degradation often results in the loss of cell growth control (reviewed in Lemmon and Traub, 2000).
The ubiquitin-proteasome pathway controls a multitude of regulatory processes via ubiquitin-mediated degradation of essential cytosolic and nuclear proteins. The pathway comprises ubiquitin, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin-ligase (E3), and a multisubunit protease, the 26S proteasome. The enzymes E1, E2, and E3 act in concert and accomplish the covalent attachment of multiple ubiquitin molecules to the specific target proteins. Polyubiquitinated proteins are then degraded by the 26S proteasome (reviewed in Hershko and Ciechanover, 1998). In addition to this well-known role in recognition by the proteasome, ubiquitination is also involved in endocytosis and down-regulation of membrane receptors, transporters, and channels (reviewed in Hicke, 1999; Strous and Govers, 1999). In yeast it was shown that a single ubiquitin moiety is sufficient to mediate internalization of an activated receptor that lacks all cytoplasmic tail sequences (Roth and Davis, 2000; Shih et al., 2000). The uracil permease Fur4p undergoes ubiquitin-dependent internalization and vacuolar degradation with lysine-63 in ubiquitin serving as a critical residue for ubiquitin chain addition (Galan and Haguenauer-Tsapis, 1997). Recent evidence suggests that the ubiquitin-proteasome pathway may also regulate protein sorting after the initial internalization step, at the level of the endosome. The tyrosine kinase adaptor protein c-Cbl mediates EGFR ubiquitination and its subsequent lysosomal and/or proteasomal degradation. c-Cbl does not accelerate internalization of the EGFR but may function at the endosome to facilitate sorting of the receptor into the MVB, thereby attenuating kinase signaling (Levkowitz et al., 1998). In yeast, the F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome. Members of the F-box family of proteins have been shown to mediate ubiquitination of substrate proteins as components of SKP1/cullin/F-box ubiquitin ligase complexes (reviewed in Deshaies, 1999). Degradation of the α-factor receptor and uracil permease is inhibited at a postinternalization step in Rcy1Δ mutant cells (Wiederkehr et al., 2000).
The GHR is a mammalian plasma membrane protein whose internalization is mediated by the ubiquitin-proteasome pathway (Strous et al., 1996). A 10 amino acid motif within the GHR cytosolic tail (the UbE motif; DSWVEFIELD) is involved in both receptor ubiquitination and endocytosis (Govers et al., 1999). Mutation of residue Phe-327 within this motif to alanine abolished receptor ubiquitination and ligand internalization and degradation (Govers et al., 1997). GHR ubiquitination occurs at the cell surface and coincides with the recruitment of the receptor to clathrin-coated membrane areas (van Kerkhof et al., 2001). Growth hormone (GH)-induced internalization of the full-length GHR is inhibited in the presence of specific proteasome inhibitors, whereas a receptor truncated at position 369 enters the cells unaffected (van Kerkhof et al., 2000). In addition to the ubiquitin-dependent endocytosis signal, the cytosolic tail of the GHR contains a di-leucine motif. On truncation of the GHR at amino acid residue 349, this di-leucine motif becomes functional and mediates ubiquitin system-independent internalization (Govers et al., 1998). We used this feature to study the involvement of the ubiquitin-proteasome pathway in lysosomal targeting. For the GHR truncated at amino acid 349, both the UbE-motif and proteasomal activity are required for endosomal sorting of the GH–GHR complex to the lysosome. In addition, we show that proteasome inhibitors block the degradation of nerve growth factor (NGF), the ligand for the receptor tyrosine kinase TrkA, at a postinternalization step. Transport of the general endocytic marker bovine serum albumin (BSA)-gold and acid-labile ligands was not blocked under these conditions. Together, these data reveal an important role for the ubiquitin-proteasome pathway in endosomal sorting of membrane proteins for lysosomal degradation.
MATERIALS AND METHODS
Materials and Antibodies
The polyclonal antibody generated against amino acid residues 271–318 of the cytosolic tail of the GHR (anti-T) was described previously (van Kerkhof et al., 2000). Human GH was a gift of Eli Lilly (Indianapolis, IN). MG-132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal) and clasto-lactacystin β-lactone were purchased from Calbiochem-Novabiochem (San Diego, CA) and transferrin (Tf) was purchased from Sigma (St. Louis, MO). Human recombinant RAP was expressed in a glutathione (GST) expression vector and isolated as described previously (Li et al., 2000). Murine NGF (2.5S) was obtained from Promega (Leiden, The Netherlands).
Plasmids, Cell Culture, and Transfection
Full-length rabbit GHR cDNA in pCB6 was described (Strous et al., 1996). The truncated GHR cDNAs GHR(349) and GHR(399) were subcloned into the CMV-NEO expression plasmid pcDNA3.1 (Invitrogen, Gröningen, The Netherlands) as previously described (Govers et al., 1998). cDNA of mutants GHR(349)(F327A) and GHR(399)(K271-362R) were constructed as previously described (Govers et al., 1999). Rat TrkA cDNA was kindly provided by Dr. D. Holtzman (Washington University School of Medicine, St. Louis, MO) and subcloned from pDM115 into the CMV-NEO expression plasmid pcDNA3.1. The construction of mLRP4T100, the membrane-containing minireceptor of LDL receptor-related protein (LRP) was previously described (Li et al., 2000). The Chinese hamster cell line ts20, bearing a thermolabile ubiquitin-activating enzyme E1, was used in this study (Kulka et al., 1988). cDNA constructs were transfected into the ts20 cells with the use of the calcium phosphate transfection procedure. For all constructs stably expressing clonal cell lines were obtained. The ts20 cells were grown at 30°C in minimum essential medium α (MEMα) supplemented with 10% fetal calf serum, 4.5 g/l glucose, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.45 mg/ml geneticin. For experiments cells were grown in 60-mm dishes in the absence of geneticin to a confluence of ∼75% and 10 mM sodium butyrate was added overnight to increase GHR expression (Strous et al., 1996). The LRP-null Chinese hamster ovary (CHO) cell line stably transfected with the mLRP4T100 cDNA was cultured at 37°C in Ham's F12 medium (Li et al., 2000).
Ligand Binding, Internalization, and Degradation
125I-human GH and 125I-RAP were prepared with the use of chloramine T (Strous et al., 1996). 125I-NGF was prepared with the use of lactoperoxidase (Sutter et al., 1979). For internalization studies, cells were grown in 12-well plates, washed with MEMα supplemented with 20 mM HEPES pH 7.4 and 0.1% BSA, and incubated in a water bath. 125I-GH (8 nM) or 125I-NGF (1 nM) was bound on ice for 2 h, the cells were washed free of unbound ligand, and incubated for 0–30 min at 30°C. Membrane-associated ligand was removed by acid wash (0.15 M NaCl, 50 mM glycine, 0.1% BSA pH 2.5) on ice. Internalized ligand was determined by measuring the radioactivity after solubilization of the acid-treated cells in 1 N NaOH with the use of a LKB gamma counter. For degradation studies, cells were incubated with 125I-GH (8 nM) or 125I-RAP (5 nM) for 6 min at 30°C. The medium was aspirated and the cells were washed and incubated in medium without ligand. At the indicated times, the medium was collected and precipitated with 1 volume of ice-cold 20% trichloroacetic acid (TCA) for 30 min on ice. Acid-soluble radioactivity was determined in the supernatant after centrifugation and was used as a measurement for degraded ligand. Membrane associated and internalized ligands were determined as described above. Nonspecific degradation was determined in the presence of excess unlabeled ligand and subtracted.
Transferrin Recycling
Tf was saturated with Fe3+ and labeled with 125I with the use of iodo-beads (Pierce, Rockford, IL) according to standard procedures. The ts20 cells were grown in 6-cm dishes and depleted from serum by 60-min incubation in MEMα supplemented with 20 mM HEPES pH 7.4 and 0.1% BSA at 30°C. 125I-Tf was added at 2 μg/ml and cells were incubated in the presence of ligand for 30 min. The medium was aspirated and cells were washed on ice for 5 min with buffer pH 5 [20 mM 2-(N-morpholino)ethanesulfonic acid pH 5.0, 130 mM NaCl, 50 μM desferal, 2 mM CaCl2, 0.1% BSA] followed by 10 min on ice with buffer pH 7.4 (MEMα supplemented with 20 mM HEPES pH 7.4 and 0.1% BSA). Then cells were incubated in MEMα supplemented with 20 mM HEPES pH 7.4 and 0.1% BSA containing 50 μM desferal at 30°C. At the indicated time points, 200-μl samples were taken and the amount of released 125I-Tf was measured with the use of a LKB gamma counter. Background label was determined in the presence of 200 μg/ml of unlabeled Tf and subtracted. TCA-precipitation of the medium verified that the released 125I-Tf was not degraded.
Metabolic Labeling
Cells were grown in 60-mm dishes and incubated in methionine- and cysteine-free MEM. Then [35S]methionine (3.7 MBq/ml Tran35S Label, 40 Tbq/mmol; ICN, Costa Mesa, CA) was added and the incubation was continued at 30°C in a CO2 incubator. The radioactivity was replaced with medium containing 100 μM unlabeled methionine, 0.1% BSA, and 16 nM GH and chased for 0–60 min. Cells were lysed and samples were immunoprecipitated (see below). Radioactivity was determined with the use of a Storm imaging system (Molecular Dynamics, Sunnyvale, CA) and quantified with Molecular Dynamics Image QuaNT software, version 4.2a.
Cell Lysis and Immunoprecipitation
Immunoprecipitations were performed as described previously (Strous et al., 1996). For GHR immunoprecipitations, cells were lysed on ice in 0.3 ml of lysis buffer containing 1% Triton X-100, 1 mM EDTA in phosphate-buffered saline (PBS), containing 50 mM NaF, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μM MG-132, and 1 mM phenylmethylsulfonyl fluoride. Immunoprecipitation of the supernatant was carried out in 1% Triton X-100, 0.5% SDS, 0.25% sodium deoxycholate, 0.5% BSA in PBS with the various inhibitors. The lysates were incubated with the indicated antibodies for 2 h on ice and immune complexes were isolated with the use of protein A-agarose beads (Repligen, Cambridge, MA). The immunoprecipitates were washed twice with the same buffer and twice with 10-fold diluted PBS. Immune complexes were subjected to SDS-PAGE and immunoblotting as described (Govers et al., 1997).
Electron Microscopy
BSA was coupled to 5-nm gold particles and dialyzed overnight against PBS at 4°C. Cells were incubated for 1 h in MEMα + 0.1% BSA in the presence or absence of 20 μM MG-132. After addition of BSA-gold at a final optical density of 5 at 520 nm, cells were further incubated for 1 h to label the entire endocytic pathway. After washing, cells were fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M phosphate buffer pH 7.4 for 30 min on ice, followed by 3 h at room temperature. Further processing for ultrathin-cryosectioning was done as described previously (Slot et al., 1991). To pick up ultrathin cryosections, a 1:1 mixture of 2.3 M sucrose and 1.8% methylcellulose was used (Liou et al., 1996).
Semiquantitative Analysis of BSA-Gold Distribution
To establish the distribution of internalized BSA-gold, per condition 50 cell profiles with a visible nucleus were analyzed at magnification of 25,000×. The number of gold particles located over a specific compartment was expressed as a percentage of total gold. The various endocytic compartments were distinguished by morphological criteria, which were deduced as a general concept from studies on the endocytic pathway of a large variety of cells (Klumperman et al., 1991, 1993; Kleijmeer et al., 1997; De Wit et al., 1999). Primary endocytic vesicles and tubules were recognized by size (80–90 nm) and electron lucent lumen. Recycling vesicles and tubules had an electron dense lumen and were 60 nm in diameter. Early or sorting endosomes were recognized as elongated, irregular-shaped vacuoles with an electron lucent content and few internal vesicles. Late endosomes or MVBs were characterized by their numerous internal vesicles, whereas lysosomes were defined by their electron dense content with the occasional appearance of membrane sheets. Clathrin-coated pits were defined as invaginations of the plasma membrane positive for clathrin, which was recognized by its electron dense appearance. Clathrin-coated vesicles near the plasma membrane were counted as a separate category, but part of these might in fact be connected to the plasma membrane, out of the plane of sectioning. Noncoated, flask-like, and sometimes branched invaginations were designated as caveolae. Membranes located in the vicinity of the trans-side of a Golgi stack were assigned as trans-Golgi area.
RESULTS
GHR Amino Acid Phenylalanine 327 Is Required for Both Receptor and Ligand Degradation
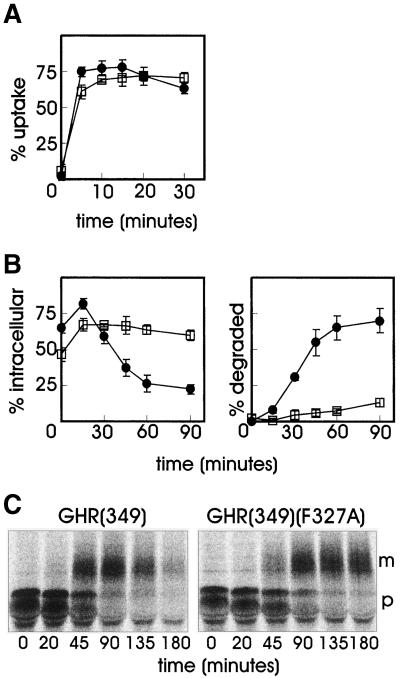
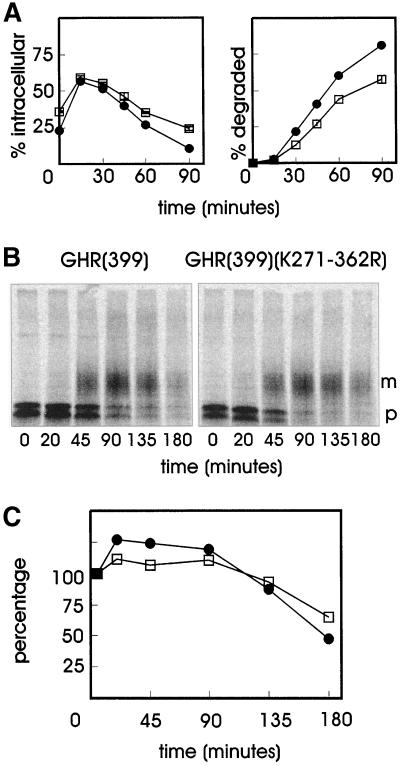
GHR endocytosis is mediated by the ubiquitin-proteasome pathway via a 10 amino acid internalization motif (UbE motif; DSWVEFIELD) (Strous et al., 1996; Govers et al., 1999). Previously, we identified a di-leucine motif in the cytosolic tail of the receptor, which upon truncation at amino acid 349 [GHR(349), Figure 1], is activated and mediates internalization in an ubiquitin system-independent manner (Govers et al., 1998). Mutation of phenylalanine residue 327 to alanine in the UbE motif abolished ubiquitination of both full-length and truncated GHR (Govers et al., 1999) but did not influence the internalization of the truncated GHR(349) (Figure 2A). To determine whether the truncated receptor can direct its ligand to the degradation pathway, cells were incubated with 125I-GH and chased for various time points in the absence of ligand after which the amount of TCA soluble radioactivity in the medium was analyzed, indicating 125I-GH degradation. In the GHR(349) transfected cells, 50–75% of the ligand was found intracellularly within the first 15 min (Figure 2B, left). Thereafter, the amount of intracellular ligand decreased rapidly with a concomitant increase of degraded ligand as acid-soluble radioactivity in the medium (Figure 2B, right). Endocytosis of ligand by the GHR(349)(F327A) mutant (Figure 1) was comparable to GHR(349) during the first 15 min. However, upon prolonged chase times, only a minor decrease in intracellular ligand was measured and almost no degraded ligand was detected in the medium, which is in striking contrast to what was observed with GHR(349). Thus, mutation of phenylalanine 327 in the UbE motif of the truncated GHR(349) has no effect on GH internalization but interferes significantly with its degradation.
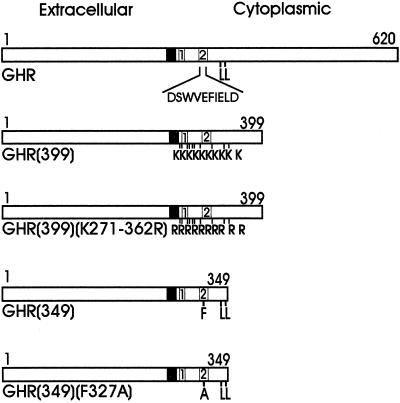
Figure 1.
Schematic representation of the wild-type and mutant GHRs. For the GHR mutants, truncated at residue 399, all cytoplasmic lysines and lysine mutations are indicated. For GHR mutants truncated at residue 349, Phe327 or Ala327 and Leu347, Leu348 are indicated. The black square represents the transmembrane domain. The numbered boxes represent box 1 and box 2, corresponding to the conserved homology domains within members of the cytokine receptor family. DSWVEFIELD indicates the position of the UbE motif, which is important for ubiquitination and endocytosis.
Figure 2.
Phe327 is required for degradation of the truncated GHR(349). (A) GHR(349) or GHR(349)(F327A) expressing ts20 cells were incubated for 2 h on ice with 8 nM 125I-GH. Unbound label was removed and the cells were incubated at 30°C for the indicated times. The amounts of internalized 125I-GH are plotted as a percentage of the cell-associated radioactivity after binding on ice. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. ●, GHR(349); □, GHR(349)(F327A). (B) GHR(349) or GHR(349)(F327A) expressing ts20 cells were incubated for 6 min at 30°C with 8 nM 125I-GH. The ligand was removed and the cells were incubated for the indicated times at 30°C. At each time point, the amount of cell surface, internalized, and degraded ligand was determined as explained in MATERIALS AND METHODS. The amount of 125I-GH is plotted as a percentage of total radioactivity. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. (Left) Intracellular 125I-GH. (Right) Degraded 125I-GH in the medium. ●, GHR(349); □, GHR(349)(F327A). (C) GHR(349) or GHR(349)(F327A) expressing ts20 cells were labeled with [35S]methionine for 20 min at 30°C and chased in the absence of radioactivity for the indicated times. GHR was immunoprecipitated with anti-GHR antibody (anti-T). p, precursor GHR (50 kDa); m, mature GHR (70 kDa).
Next, we analyzed the turnover of the truncated receptor itself with the use of pulse-chase labeling with [35S]methionine. The receptor is synthesized as a glycoprotein precursor (Figure 2C, p) and converted in the Golgi apparatus to the complex glycosylated mature form (Figure 2C, m). For the truncated GHR(349) we observed a rapid disappearance of the mature form after prolonged chase times in the presence of GH, indicating a fast degradation of the receptor. Although the maturation of the mutated GHR(349)(F327A) was somewhat delayed compared with GHR(349), quantification showed that the half-life of the mature mutant receptor was approximately fivefold prolonged. Together, these results show that residue phenylalanine 327 in the truncated GHR(349) is required for efficient degradation of both ligand and receptor. As the degradation of the ligand occurs in lysosomes (Murphy and Lazarus, 1984; Yamada et al., 1987), we conclude that the GHR UbE motif is required for endosome-to-lysosome sorting of both receptor and ligand.
Proteasomal Activity Is Required for Degradation of a Truncated GHR and Its Ligand
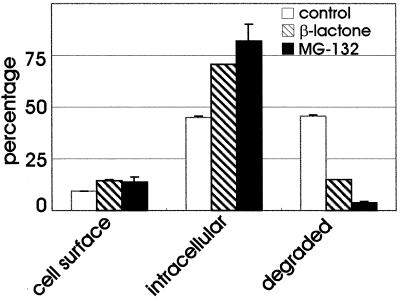
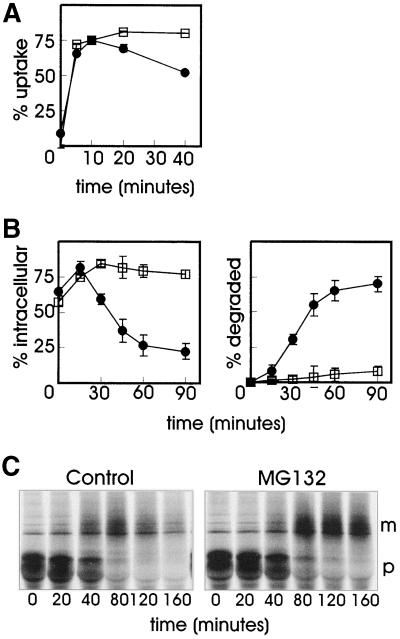
Because the UbE-motif is involved in GHR ubiquitination, we investigated the role of the ubiquitin-proteasome pathway in the degradation of GH, with the use of proteasome inhibitors. The peptide aldehyde MG-132 is a substrate analog and a reversible inhibitor of the chymotrypsin-like activity of the proteasome. Because peptide aldehydes might inhibit certain lysosomal cysteine proteases and the calpains, it is important to show that similar biological effects occur with other proteasome inhibitors. Lactacystin and its derivative clasto-lactacystin β-lactone are structurally different from the peptide aldehyde and act as a pseudo substrate that becomes irreversibly linked to the active site threonine of the proteasome β-subunits (Rock et al., 1994; Lee and Goldberg, 1998). Lactacystin shows high specificity for the proteasome but can also inhibit cathepsin A (Ostrowska et al., 1997). GHR(349)-transfected cells were used to compare the effect of two specific proteasome inhibitors, MG-132 and clasto-lactacystin β-lactone (Craiu et al., 1997), on 125I-GH degradation. After a short incubation with 125I-GH, cells were chased for 45 min, whereafter the amounts of intracellular and degraded ligand in the medium were determined (Figure 3). In untreated cells (control), 45% of the ligand was found intracellular and about the same amount was degraded. In cells treated with either of the proteasome inhibitors, the amount of intracellular GH was markedly increased compared with the control cells, however, degradation was almost completely inhibited. Because the two proteasome inhibitors showed a similar effect on ligand internalization and degradation, a more detailed analysis was performed with only MG-132 (Figure 4). In Figure 4A the effect of MG-132 on the uptake of 125I-GH after binding on ice was determined. Both in control and MG-132-treated GHR(349) cells, ∼75% of the ligand was internalized after 10 min, indicating that the proteasome inhibitor has no effect on endocytosis. For the control cells it has been shown (Figure 2) that internalized GH is rapidly degraded, starting after ∼15 min of chase. However, in the presence of proteasome inhibitor, the degradation of GH was almost completely inhibited (Figure 4B). To monitor the effect of the proteasome inhibitor on the fate of the truncated receptor, a pulse-chase labeling experiment with [35S]methionine was performed. As seen in Figure 4C, the mature form of the receptor was rapidly degraded upon prolonged chase times. In the presence of MG-132, the maturation of the truncated GHR(349) was unaffected while degradation was almost completely inhibited, which is in line with the effect of the proteasome inhibitor on the degradation of GH. These results clearly indicate an involvement of the ubiquitin-proteasome pathway in the degradation of the truncated GHR(349) and its ligand at a postinternalization step.
Figure 3.
Effect of proteasome inhibitors on GH degradation. GHR(349) expressing ts20 cells were incubated with 20 μM clasto-lactacystin β-lactone (2 h), 20 μM MG-132 (1 h), or solvent alone (control) then 8 nM 125I-GH was added and the incubation was continued for 6 min at 30°C. The ligand was removed and the cells were incubated for 45 min at 30°C. The amount of cell surface, internalized and degraded ligand was determined as explained in MATERIALS AND METHODS. The amount of 125I-GH is plotted as a percentage of total radioactivity. Each bar represents the mean value of one experiment performed in duplicate ± SD.
Figure 4.
MG-132 inhibits the degradation of the truncated GHR(349) and its ligand. (A) GHR(349)-expressing ts20 cells were incubated for 1 h with 20 μM MG-132 or solvent and put on ice for 2 h with 8 nM 125I-GH. Unbound label was removed and the cells were incubated at 30°C in the absence or presence of MG-132 as indicated. The amount of internalized 125I-GH is plotted as a percentage of the cell-associated radioactivity after the 2-h binding on ice. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. ●, control; □, MG-132. (B) GHR(349) expressing ts20 cells were incubated for 1 h at 30°C with 20 μM MG-132 or solvent (control) then 8 nM 125I-GH was added and the incubation was continued for 6 min. The ligand was removed and the cells were incubated at 30°C as indicated. At each time point the amount of cell surface, internalized, and degraded ligand was determined as explained in MATERIALS AND METHODS. The amount of 125I-GH is plotted as a percentage of total radioactivity. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. (Left) Intracellular 125I-GH. (Right) Degraded 125I-GH in the medium. ●, control; □, MG-132. (C) GHR(349) expressing ts20 cells were labeled with [35S]methionine for 20 min after preincubation for 1 h with or without 20 μM MG-132. Cells were chased in the absence of radioactivity for the indicated times. GHR was immunoprecipitated with anti-GHR antibody (anti-T). p, precursor GHR (50 kDa); m, mature GHR (70 kDa).
Lysine Residues in Cytoplasmic Domain of GHR Are Not Required for Degradation
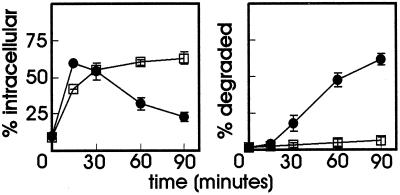
The preceding experiments indicate that the ubiquitin-proteasome pathway is involved in directing the GHR and its ligand to the degradative pathway. Next, we addressed the question whether ubiquitination of the receptor itself is required for this sorting step. Previously, with the use of a GHR truncated at amino acid 399 in which all the 10 cytoplasmic lysine residues were mutated to arginine [GHR(399)(K271-362R); Figure 1], we have shown that GHR ubiquitination is not required for internalization at the plasma membrane. By replacing phenylalanine 327 for alanine in this mutant, we could show that the internalization is controlled by the UbE motif (Govers et al., 1999). Here, we have used cell lines stably expressing GHR(399) or truncation mutant GHR(399)(K271-362R) to measure the degradation of 125I-GH. Both cell lines showed comparable amounts of intracellular ligand after incubation with 125I-GH, again indicating that there is no effect on internalization if the GHR tail lacks attachment sites for ubiquitin (Figure 5A, left). After prolonged incubation, the percentage of intracellular ligand decreased, due to the degradation of the 125I-GH, as can be seen in the Figure 5A, right. Degradation of GH in the case of the lysine-less GHR was only slightly less efficient compared with the lysine-containing truncation, strongly indicating that ubiquitination of the GHR itself is not required for its degradation. The degradation of the truncated receptors was monitored with the use of a pulse-chase labeling with [35S]methionine (Figure 5B). As shown by immunoprecipitation of the GHR, the signal for the mature form decreased rapidly upon prolonged chase times, for both GHR(399) and GHR(399)(K271-362R) (Figure 5C). From these experiments we conclude that ubiquitination of the GHR itself is not required for the ubiquitin-proteasome pathway-dependent sorting to the degradative pathway.
Figure 5.
Degradation of GHR and ligand is independent of lysine residues. (A) GHR(399)- or GHR(399)(K271-362R)-expressing ts20 cells were incubated for 6 min at 30°C with 8 nM 125I-GH. The ligand was removed and the cells were incubated for the indicated times at 30°C. At each time point the amount of cell surface, internalized, and degraded ligand was determined as explained in MATERIALS AND METHODS. The amount of 125I-GH is plotted as a percentage of total radioactivity. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. (Left) Intracellular 125I-GH. (Right) Degraded 125I-GH in the medium. ●, GHR(399); □ GHR(399)(K271-362R). (B) Cells were labeled with [35S]methionine for 20 min at 30°C and chased in the absence of radioactivity for the indicated times. GHR was immunoprecipitated with anti-GHR antibody (anti-T). p, precursor GHR (60 kDa); m, mature GHR (80 kDa). (C) Amounts of radioactivity of the total lanes were determined with the use of Image Quant software and were expressed as percentages of the radioactivity incorporated in the lanes at 0 min. ●, GHR(399); □, GHR(399)(K271-362R).
Proteasome Inhibitors Inhibit Degradation of TrkA-bound NGF at Level of Endosomes
Internalization and degradation of the GHR depends on the UbE motif in its cytoplasmic tail. Next, we addressed the question whether the degradation of a receptor, which is sorted into the degradative pathway but does not contain an obvious UbE motif, is also regulated by the ubiquitin-proteasome pathway. Similar to other receptor tyrosine kinases, TrkA, the receptor tyrosine kinase for the neurothrophin NGF, dimerizes upon ligand binding, which in turn results in an activation of the intracellular kinase domain and rapid internalization (Grimes et al., 1997). With the use of Chinese hamster ts20 cells stably transfected with TrkA we found that the ubiquitin-proteasome pathway is not involved in the endocytosis of this receptor (Alves dos Santos and Strous, unpublished results). We used these cells to determine the effect of proteasome inhibitors on the degradation of NGF. As seen in Figure 6, left, the internalization of the ligand was not inhibited in the presence of MG-132. After prolonged incubation, the intracellular amount of 125I-NGF decreased in the control cells and could be detected as TCA-soluble radioactivity in the medium (Figure 6, right). In the MG-132-treated cells, the label remained intracellular during the chase with no measurable degradation of NGF. This result indicates an involvement of the ubiquitin-proteasome pathway in lysosomal transport of NGF bound to TrkA.
Figure 6.
Effect of MG-132 on degradation of 125I-NGF. TrkA-expressing ts20 cells were incubated for 2 h with 20 μM MG-132 or solvent and put on ice for 1 h with 1 nM 125I-NGF. Unbound label was removed and the cells were incubated at 30°C in the absence or presence of MG-132 as indicated. The amounts of 125I-NGF are plotted as a percentage of the cell-associated radioactivity after 2-h binding on ice. (Left) Intracellular NGF. (Right) Degraded NGF. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. ●, control; □, MG-132.
Dissociating Ligands and an Endocytic Marker Are Transported to Lysosomes in Presence of Proteasome Inhibitors
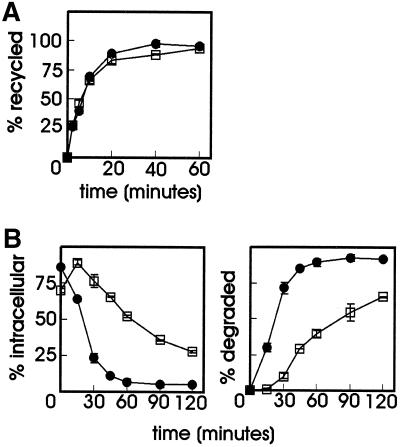
Most of the soluble content of sorting endosomes is delivered to lysosomes for degradation, while the majority of membrane proteins recycle from the endocytic pathway back to the plasma membrane. To address the question whether proteasome inhibitors interfere with general mechanisms in membrane transport, we examined their effect on the recycling pathway and on the degradation of a ligand that dissociates from its receptor upon endosome acidification. To this end, ts20 cells were loaded with 125I-Tf in the absence or presence of MG-132 after which the amount of recycling was measured by determining the release of internalized Tf into the medium. As can be seen in Figure 7A, recycling of Tf was unaffected in the presence of the proteasome inhibitor.
Figure 7.
Effect of MG-132 on transferrin recycling and degradation of 125I-RAP. (A) ts20 cells were serum depleted for 1 h with or without 20 μM MG-132 and then loaded with 2 μg/ml 125I-Tf for 30 min at 30°C. Cells were chilled on ice, plasma membrane-bound 125I-Tf was removed, and the cells were incubated at 30°C in medium containing 50 μM desferal in the absence or presence of MG-132. The release of 125I-Tf was determined and expressed as a percentage of the total amount of radioactivity loaded in the cells. ●, control; □, MG-132. (B) LRP-null CHO cells stably transfected with mLRP4T100 were incubated for 1 h at 37°C with or without 20 μM MG-132 before 5 nM 125I-RAP was added. The incubation was continued for 6 min, after which the unbound radioactivity was removed and the cells were incubated at 37°C in the absence of ligand with or without MG-132 for the time points indicated. At each time point the amount of cell surface, internalized, and degraded ligand was determined as explained in MATERIALS AND METHODS. The amount of 125I-RAP is plotted as a percentage of total radioactivity. Each point in the graph represents the mean value of two experiments performed in duplicate ± SD. (Left) Intracellular 125I-RAP. (Right) Degraded 125I-RAP in the medium. ●, control; □ MG-132.
The LRP is an endocytic receptor that belongs to the LDL receptor gene family (Herz et al., 1988). Ligand interactions with LRP can be antagonized by a 39-kDa receptor-associated protein (RAP). The recombinant form of RAP has been used extensively in the study of ligand–receptor interactions. Endocytosis of cell-surface bound RAP is rapid and the internalized ligand is delivered to lysosomes while the receptor recycles to the cell surface (Iadonato et al., 1993; Czekay et al., 1997). In this study we used an LRP-null CHO cell line, stably transfected with the LRP minireceptor mLRP4T100, which mimics the function and trafficking of LRP (Li et al., 2000). Cells were incubated for a short time with 125I-RAP in the presence or absence of MG-132. After prolonged incubation in the absence of labeled ligand, the amount of internalized and degraded 125I-RAP was determined as described in MATERIALS AND METHODS (Figure 7B). As seen in the left panel, most of the ligand was already intracellular after 6 min of incubation, both in the absence or presence of MG-132. The amount of intracellular ligand decreased rapidly, accompanied by increased levels of TCA-soluble radioactivity in the medium (right). Degradation of RAP was less efficient in MG-132-treated cells, an effect that was also found in ts20 cells transfected with mLRP4T100 (van Kerkhof, unpublished results). MG-132 inhibited RAP degradation threefold, whereas both GH and NGF degradation was 16–17-fold decreased. Even although MG-132 affects RAP degradation, the effect differs in magnitude from the effect on GH and NGF degradation: once internalized, all RAP molecules are completely degraded, be it somewhat later (Figure 7B). This is not the case for NGF, neither for GH, if taken up via GHR(349)(F327A) or in the presence of MG-132: they are probably forced into the recycling pathway, escaping degradation. Additional evidence for this was provided by experiments with GHR(349)(F327A)-expressing cells, which were continuously incubated with 125I-GH: cell-associated radioactivity increased linearly with the GHR rate of synthesis, whereas no soluble radioactivity was released from the cells (van Kerkhof, unpublished results). Why MG-132 had a moderate affect on RAP degradation is unclear at the moment. One reason might be that the inhibitor slows down lysosomal degradation.
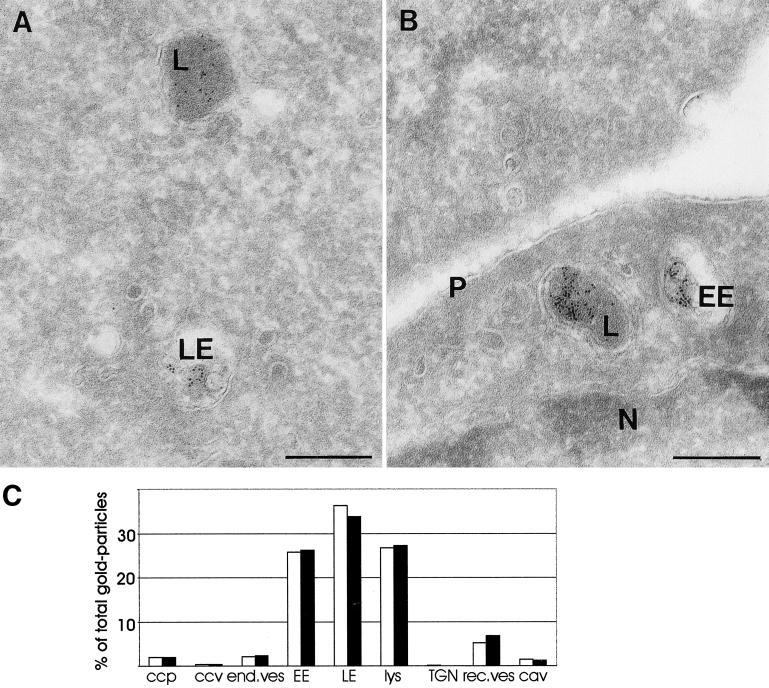
To investigate a possible general effect of proteasome inhibitors on the lysosomal pathway, we visualized transport along the endocytic tract with internalized BSA-gold (Slot et al., 1988). After 1-h uptake, significant amounts of BSA-gold reached the lysosomes in both control (Figure 8A) and MG-132-treated cells (Figure 8B). Quantitative analysis of the relative distribution of BSA-gold over the distinct endocytic compartments (for detailed definitions, see MATERIALS AND METHODS) revealed nearly identical distribution patterns (Figure 8C). Of particular interest in this respect, in control cells 36 and 26% of total gold was found in late endosomes and lysosomes, respectively, whereas in MG-132-treated cells these figures amounted to 34 and 27%. These data are a strong indication that transport of BSA-gold to lysosomes is unaltered in the presence of proteasome inhibitors. The low amount of BSA-gold in TGN and recycling vesicles and tubules is in agreement with the notion that this marker mainly follows the degradative pathway. Together, the data demonstrate that transport of an endocytic marker is unaltered and that degradation of a dissociating ligand is only slightly inhibited after incubation with MG-132, and indicate that proteasome inhibitors block a late step in lysosomal transport of selected membrane proteins but not in transport of soluble proteins.
Figure 8.
Transport of BSA-gold is not altered by a proteasome inhibitor. (A and B) Ultrathin cryosections of wild-type GHR-expressing ts20 cells, which were incubated for 1 h with BSA coupled to 5-nm gold. (A) In control cells, BSA-gold was found in the entire endocytic tract. Here localization in a late endosome (LE) and a lysosome (L) is shown. (B) In the presence of MG-132 transport of BSA-gold in the endosomal-lysosomal pathway is not affected, as illustrated by the presence of BSA-gold in early endosomes (EE) as well as in lysosomes. N, nucleus; p, plasma membrane. Bars, 200 nm. (C) Relative distribution of internalized BSA-gold particles over distinct endocytic compartments was established as explained in MATERIALS AND METHODS and expressed as percentage of the total number of gold-particles counted (3803 and 3580 in control and MG-132-treated cells, respectively). ccp, clathrin-coated pits; ccv, clathrin-coated vesicles; end.ves, primary endocytic vesicles and tubules; EE, early endosome; LE, late endosome; Lys, lysosome; TGN, trans-Golgi network; rec ves, recycling vesicle; cav, caveolae. □, control; ▪, MG-132.
DISCUSSION
Accumulating evidence suggests a role for ubiquitination in regulating protein sorting in the endosomal system. In this study, we used several models to address the role of the ubiquitin-proteasome pathway in sorting of internalized proteins at the level of the endosome. First, degradation of GH bound to GHR, a receptor that is ubiquitinated dependent on its UbE motif, was compared with degradation of NGF bound to TrkA, a receptor without an UbE motif. Both receptors remain bound to their ligand in endosomes. Proteasome inhibitors completely blocked the degradation of GH and NGF. In contrast, degradation of RAP, which needs to dissociate from its recycling receptor for targeting to lysosomes, was only slightly inhibited in the presence of the proteasome inhibitor, and transport of an general endocytic marker to the lysosome was unaffected. Together, the data suggest that proteasome inhibitors block the sorting of a select set of membrane proteins to the degradative pathway without interfering with transport of soluble proteins.
The GHR UbE motif is important for ubiquitination of the full-length receptor and its truncations. The truncated GHR(349) is internalized from the plasma membrane independent of the ubiquitin system by a C-terminally located di-leucine motif. Here, we corroborated the finding that mutation of residue phenylalanine 327 in the UbE motif of GHR(349) did not interfere with initial internalization from the plasma membrane and provide novel evidence that this mutation abolished subsequent degradation of both receptor and ligand. Therefore, we conclude that the UbE motif is not only involved in endocytosis of the GHR but also in its subsequent sorting to the lysosomes. The data also indicate that the di-leucine motif, which is involved in endocytosis of this GHR truncation, does not mediate the sorting to the degradative pathway. The finding that one motif can mediate different transport steps is in itself not new. For tyrosine-based motifs it has been reported, that they can function at different sorting steps (Peters et al., 1990; Prill et al., 1993) and also the di-leucine motif can act as an internalization motif and in lysosomal targeting (reviewed in Hunziker and Geuze, 1996). The role of the ubiquitin-proteasome pathway in intracellular sorting to the lysosome was corroborated by the use of specific proteasome inhibitors. Because the use of pharmacological inhibitors could be a potential problem with regard to specificity, we used two different inhibitors:β-lactone and MG-132; both were able to inhibit degradation of GH and GHR to the same extent (75–90%, Figure 3; Van Kerkhof et al., 2000). Although these inhibitors are structurally different, it cannot be excluded that part of the inhibition is due to an inhibitory effect on lysosomal hydrolases. The GH–GHR complex dissociates at a pH below 3, indicating that receptor and ligand remain associated in the endosomal system. A common localization throughout the endocytic pathway was shown for EGF and EGFR and similarly a pool of intact platelet-derived growth factor (PDGF)–PDGFR and NGF–TrkA receptor complexes could be detected in endosomes (Sorkin and Waters, 1993). We show that the degradation of NGF, internalized via its receptor, TrkA, is inhibited in the presence of proteasome inhibitors. Proteasome inhibitors were also shown to inhibit the degradation of the PDGFR (Mori et al., 1995) and the Met tyrosine kinase receptor (Jeffers et al., 1997). Furthermore, it was shown that proteasome inhibitors inhibit the down-regulation of the EGFR (Levkowitz et al., 1998) and the lysosomal degradation of interleukin-2 internalized by the interleukin-2 receptor (Yu and Malek, 2001), suggesting a possible role for the proteasome in regulating trafficking to the lysosome.
Do proteasome inhibitors interfere with trafficking to the lysosome or with the activity of lysosomal enzymes? Degradation of GH and NGF, which remain associated with their receptor, was inhibited 16-fold, whereas degradation of RAP, which dissociates from its receptor, was inhibited only threefold in the presence of MG-132. This difference in magnitude, together with the observation that mutation of the UbE motif causes comparable inhibition of GH degradation in the absence of MG-132, suggests a role of the ubiquitin-proteasome pathway in the delivery of GH and NGF to the lysosome. The most likely scenario is that inhibition of the ubiquitin-proteasome pathway induces recycling of the ligand–receptor complex: 1) More GH is associated with the cell surface after incubation with proteasome inhibitors (Figure 3.). 2) Proteasome inhibitors do not interfere with transferrin recycling (Figure 7A). 3) Morphological quantification data, with the use of the GHR(349)(F327A) mutant, showed colocalization of GH with transferrin receptor in recycling vesicles (Sachse and Klumperman, unpublished results). 4) Continuous uptake of 125I-GH in this mutant resulted in accumulation of radioactivity, both intracellular and at the cell surface, suggesting recycling of the mutant receptor–ligand complex. Why is the degradation of RAP inhibited while the morphological data show that transport of the endocytic marker BSA-gold is unaltered in the presence of MG-132? One possible explanation is that the proteasome inhibitor inhibits lysosomal hydrolases to some extent, which would result in a delayed degradation. Another possibility is that proteasome inhibitors interfere with the dissociation of receptor and ligand in the sorting endosome, resulting in recycling of RAP together with the receptor. This should result in more receptor and ligand at the cell surface, although the complex will be reinternalized very efficiently once it reaches the plasma membrane. Consistent with this, we detect more RAP (9 versus 4%) at the cell surface in the presence of proteasome inhibitors.
What is the molecular mechanism that regulates endosomal sorting to the lysosome? For a number of ligand-stimulated receptor tyrosine kinases, the tyrosine kinase adaptor c-Cbl is involved. Overexpression of c-Cbl increases ligand-induced ubiquitination and down-regulation of EGFR, PDGFR, and colony stimulating factor-1 receptor (Levkowitz et al., 1998; Miyake et al., 1998; Lee et al., 1999). For the EGFR, down-regulation depends on its intrinsic kinase activity and involves the trans-phosphorylation of the c-Cbl adaptor. For the GHR, the mechanism is unknown but must be different, because although the receptor is tyrosine phosphorylated upon addition of GH, no phosphorylation of c-Cbl could be detected (van Kerkhof, unpublished results). Possibly, the GHR UbE motif may serve, directly or via adaptor proteins, as an anchoring site for the ubiquitinating enzymes, leading to coated pit localization, internalization, and subsequent endosomal sorting. It was recently suggested that the ubiquitination state of proteins at the late endosome might help concentrate them in regions that will invaginate and form the internal vesicles (Amerik et al., 2000). The results with the truncated GHR without intracellular attachment sites for ubiquitin [GHR(399)(K271-362R)] show that ubiquitination of the receptor itself is not required for lysosomal sorting. Which proteins do need to be ubiquitinated is unclear at present, possibly it is the receptor that brings the ubiquitination machinery in proximity to the sorting machinery that is regulated by ubiquitination. Dynamic ubiquitination/deubiquitination of components of the endocytic machinery could play a role in the subsequent transport steps along the endocytic pathway. Ligand-induced ubiquitination was shown for Eps15, a clathrin-coated pit-associated protein required for EGFR uptake (van Delft et al., 1997) and genetic data implicate the product of the Drosophila liquid facets gene, epsin, which binds to Eps15, as a target for the fat facets deubiquitinating enzyme (Cadavid et al., 2000). From recent data it appeared that the yeast Doa4 deubiquitinating enzyme localizes reversibly with the late endosome/prevacuolar compartment along with a group of proteins essential for targeting of membrane proteins to the vacuole and it was proposed that Doa4 is responsible for deubiquitination events at the late endosome (Amerik et al., 2000).
An important question that remains is what is the target protein for the proteasome? It is not clear whether the down-regulated receptors are degraded in lysosomes, or by the proteasome, or both. However, because GH and NGF are also not degraded in the presence of proteasome inhibitors, this would favor a model in which one of the components of the endocytic sorting machinery is the target for the proteasome. Inhibition of degradation of (part of) this component would then lead to inhibition of assembling the sorting machinery and possibly result in recycling of the receptor–ligand complex. In the case of GHR sorting, a direct role for both the ubiquitin system and the proteasome is anticipated, although the target of the proteasome is probably not the GHR cytosolic tail. It remains to be seen whether receptors, in which c-Cbl plays a role in lysosomal sorting, are direct targets of the proteasome. It is also possible that there is no direct role for the proteasome. Use of proteasome inhibitors bears the risk of exhausting the cells of free ubiquitin (Swaminathan et al., 1999), which might lead to reduced ubiquitination of the target protein and reduced endosomal sorting. Recently, ubiquitin was implicated in retrovirus assembly and budding, a process in which the host machinery for endocytosis or MVB formation could play a role (Patnaik et al., 2000; Schubert et al., 2000). The late domain in the retroviral Gag protein was shown to recruit ubiquitin ligases to the site of viral assembly, and it was suggested that the engagement of the ubiquitin conjugation machinery plays a crucial role in the release of retroviruses. Proteasome inhibitors caused defects in virus budding, possibly through depletion of the pool of free ubiquitin, suggesting that there is no direct role for the proteasome.
In conclusion, the results of this study point to a specific role of the ubiquitin-proteasome pathway in the regulated sorting of specific sets of membrane proteins. Based on our observation that ubiquitination of the GHR itself is not required for this sorting, we speculate that a specific membrane protein recruits a ubiquitin ligase, which then directly or via ubiquitination of target proteins recruits the sorting machinery to accomplish its subsequent degradation.
ACKNOWLEDGMENTS
We thank Rene Scriwanek and Marc van Peski for excellent preparations of EM photographs, and Erica Vallon for carefully reading the manuscript. We thank Willem Stoorvogel, Jürgen Gent, Julia Schantl, and Toine ten Broeke for stimulating discussions; Dr. D. Holzman for kindly providing the rat TrkA cDNA; and Ellen van Dam for help with transferrin recycling experiments. This work was supported by a grant from the Netherlands Organization for Scientific Research (NWO-902-23-192), a European Union Network Grant (ERBFMRXCT96-0026), and by grants from the National Institutes of Health (HL-59150 and NS-37525).
Abbreviations used:
- EGFR
epidermal growth factor receptor
- GH
growth hormone
- GHR
growth hormone receptor
- LDL
low density lipoprotein
- LRP
LDL receptor-related protein
- MVB
multivesicular body
- NGF
nerve growth factor
- PDGFR
platelet-derived growth factor receptor
- RAP
receptor-associated protein
- Tf
transferrin
- TrkA
receptor tyrosine kinase activated by NGF
- UbE
ubiquitin-dependent endocytosis
REFERENCES
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadavid AL, Ginzel A, Fischer JA. The function of the Drosophila Fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- Czekay RP, Orlando RA, Woodward L, Lundstrom M, Farquhar MG. Endocytic trafficking of megalin/RAP complexes: dissociation of the complexes in late endosomes. Mol Biol Cell. 1997;8:517–532. doi: 10.1091/mbc.8.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- De Wit H, Lichtenstein Y, Geuze HY, Kelly RB, Vandersluijs P, Klumperman J. Synaptic vesicles form by budding from tubular extensions of sorting endosomes in PC12 cells. Mol Biol Cell. 1999;10:4163–4176. doi: 10.1091/mbc.10.12.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S, Miller K, Moehren G, Ullrich A, Schlessinger J, Hopkins CR. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 1990;61:623–634. doi: 10.1016/0092-8674(90)90474-s. [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers R, ten Broeke T, van Kerkhof P, Schwartz AL, Strous GJ. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 1999;18:28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers R, van Kerkhof P, Schwartz AL, Strous GJ. Linkage of the ubiquitin-conjugating system and the endocytic pathway in ligand-induced internalization of the growth hormone receptor. EMBO J. 1997;16:4851–4858. doi: 10.1093/emboj/16.16.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers R, van Kerkhof P, Schwartz AL, Strous GJ. Di-leucine-mediated internalization of ligand by a truncated growth hormone receptor is independent of the ubiquitin conjugation system. J Biol Chem. 1998;273:16426–16433. doi: 10.1074/jbc.273.26.16426. [DOI] [PubMed] [Google Scholar]
- Grimes ML, Beattie E, Mobley WC. A signaling organelle containing the nerve growth factor-activated receptor tyrosine kinase, trka. Proc Natl Acad Sci USA. 1997;94:9909–9914. doi: 10.1073/pnas.94.18.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kD liver membrane protein closely related to the LDL-receptor suggest a physiological role as a lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- Hunziker W, Geuze HJ. Intracellular trafficking of lysosomal membrane proteins. Bioessays. 1996;18:379–389. doi: 10.1002/bies.950180508. [DOI] [PubMed] [Google Scholar]
- Iadonato SP, Bu G, Maksymovitch EA, Schwartz AL. Interaction of a 39 kDa protein with the low-density-lipoprotein-receptor-related protein (LRP) on rat hepatoma cells. Biochem J. 1993;296:867–875. doi: 10.1042/bj2960867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers M, Taylor GA, Weidner KM, Omura S, Vandewoude GF. Degradation of the met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol Cell Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijmeer MJ, Morkowski S, Griffith JM, Rudensky AY, Geuze HJ. Major histocompatibility complex class II compartments in human and mouse B lymphoblasts represent conventional endocytic compartments. J Cell Biol. 1997;139:639–649. doi: 10.1083/jcb.139.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Boekestijn JC, Mulder AM, Fransen JA, Ginsel LA. Intracellular localization and endocytosis of brush border enzymes in the enterocyte-like cell line Caco-2. Eur J Cell Biol. 1991;54:76–84. [PubMed] [Google Scholar]
- Klumperman J, Hille A, Veenendaal T, Oorschot V, Stoorvogel W, von Figura K, Geuze HJ. Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J Cell Biol. 1993;121:997–1010. doi: 10.1083/jcb.121.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J Biol Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER. The cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Paz Marzolo M, van Kerkhof P, Strous GJ, Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. J Biol Chem. 2000;275:17187–17194. doi: 10.1074/jbc.M000490200. [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. HistochemCell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Miyake S, Lupher ML, Jr, Druker B, Band H. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc Natl Acad Sci USA. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Tanaka K, Omura S, Saito Y. Degradation process of ligand-stimulated platelet-derived growth factor beta-receptor involves ubiquitin-proteasome proteolytic pathway. J Biol Chem. 1995;270:29447–29452. doi: 10.1074/jbc.270.49.29447. [DOI] [PubMed] [Google Scholar]
- Murphy LJ, Lazarus L. The mouse fibroblast growth hormone receptor: ligand processing and receptor modulation and turnover. Endocrinology. 1984;115:1625–1632. doi: 10.1210/endo-115-4-1625. [DOI] [PubMed] [Google Scholar]
- Ostrowska H, Wojcik C, Omura S, Worowski K. Lactacystin, a specific inhibitor of the proteasome, inhibits human platelet lysosomal cathepsin A-like enzyme. Biochem Biophys Res Commun. 1997;234:729–732. doi: 10.1006/bbrc.1997.6434. [DOI] [PubMed] [Google Scholar]
- Patnaik A, Chau V, Wills JW. Ubiquitin is part of the retrovirus budding machinery. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Braun M, Weber B, Wendland M, Schmidt B, Pohlmann R, Waheed A, von Figura K. Targeting of a lysosomal membrane protein: a tyrosine-containing endocytosis signal in the cytoplasmic tail of lysosomal acid phosphatase is necessary and sufficient for targeting to lysosomes. EMBO J. 1990;11:3497–3506. doi: 10.1002/j.1460-2075.1990.tb07558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill V, Lehmann L, von Figura K, Peters C. The cytoplasmic tail of lysosomal acid phosphatase contains overlapping but distinct signals for basolateral sorting and rapid internalization in polarized MDCK cells. EMBO J. 1993;12:2181–2193. doi: 10.1002/j.1460-2075.1993.tb05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Roth AF, Davis NG. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275:8143–8153. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
- Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Sloper-Mold KE, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ, Weerkamp AH. Localization of macromolecular components by application of the immunogold technique on cryosectioned bacteria. Methods Microbiol. 1988;20:211–236. [Google Scholar]
- Sorkin A, Waters CM. Endocytosis of growth factor receptors. Bioessays. 1993;15:375–382. doi: 10.1002/bies.950150603. [DOI] [PubMed] [Google Scholar]
- Strous GJ, Govers R. The ubiquitin-proteasome system and endocytosis. J Cell Sci. 1999;112:1417–1423. doi: 10.1242/jcs.112.10.1417. [DOI] [PubMed] [Google Scholar]
- Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Sutter A, Riopelle RJ, Harris-Warrick RM, Shooter EM. Nerve growth factor receptors. Characterization of two distinct classes of binding sites on chick embryo sensory ganglia cells. J Biol Chem. 1979;254:5972–5982. [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge IS, Collawn JF, Hopkins CR. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- van Delft S, Govers R, Strous GJ, Verkleij AJ, Henegouwen PMPVE. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272:14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- van Kerkhof P, Govers R, Alves Dos Santos CM, Strous GJ. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J Biol Chem. 2000;275:1575–1580. doi: 10.1074/jbc.275.3.1575. [DOI] [PubMed] [Google Scholar]
- van Kerkhof P, Sachse M, Klumperman J, Strous GJ. Growth hormone receptor ubiquitination coincides with recruitment to clathrin-coated membrane domains. J Biol Chem. 2001;276:3778–3784. doi: 10.1074/jbc.M007326200. [DOI] [PubMed] [Google Scholar]
- Wiederkehr A, Avaro S, Prescianottobaschong C, Haguenauer-Tsapis R, Riezman H. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lipson KE, Donner DB. Structure and proteolysis of the growth hormone receptor on rat hepatocytes. Biochemistry. 1987;26:4438–4443. doi: 10.1021/bi00388a037. [DOI] [PubMed] [Google Scholar]
- Yu A, Malek T. The proteasome regulates receptor-mediated endocytosis of interleukin-2. J Biol Chem. 2001;276:381–385. doi: 10.1074/jbc.M007991200. [DOI] [PubMed] [Google Scholar]