Abstract
Graves’ disease (GD) is a common autoimmune disorder with a genetic predisposition. Owing to the biological effect of tumor necrosis factor-α (TNF-α) on the thyroid gland and its gene location, TNF-α should be able to influence an individual’s susceptibility to GD. In the present study, we conduct a meta-analysis of rs1800629 and rs361525 in TNF-α gene from all eligible case–control studies to assess the associations amongst reported TNF-α gene with GD. A total of ten case–control studies involving 2790 GD patients and 3472 healthy controls were included. The results showed that a significant association was characterized between the rs1800629 polymorphism and GD in the homozygous model (AA compared with GG: odds ratio (OR) = 1.97, 95% confidence interval (CI) = 1.27–3.06, P=0.002) and recessive model (AA compared with GA + GG: OR = 1.62, 95% CI = 1.04–2.50, P=0.03). GD susceptibility was significantly detected in European population in all genetic models after ethnicity stratification. In sharp contrast, no significant association could be detected in Asian population. Next, we conducted a meta-analysis for another promoter SNP rs361525. However, SNP rs361525 did not show a significant association with GD in any genetic model before and after ethnicity stratification. Together, our data support that only the promoter single-nucleotide polymorphism (SNP) rs1800629 within the TNF-α gene is associated with increased risk for developing GD, especially in European population. Future large-scale studies are required to validate the associations between TNF-α gene and GD.
Keywords: Graves’ disease, Polymorphism, Tumor Necrosis Factor-α
Introduction
Graves’ disease (GD) is an autoimmune thyroid disease with a 0.5% rate of prevalence in general population [1]. It is characterized by the presence of thyroid-stimulating hormone (TSH) receptor antibodies, leading to hyperthyroidism and goiter. The exact etiology of GD has still remained unknown; however, it is believed that genetic polymorphisms and environmental factors are both involved in the pathogenesis of GD. Since GD is an autoimmune disorder, it is affected by genes, cytokines, and enzymes [2]. Genome-wide scans have identified the human leukocyte antigen (HLA) genomic region of the MHC on chromosome 6p21 linked to GD [3,4]. Tumor necrosis factor-α (TNF-α), residing in the short arm of human chromosome 6 (6p21.3), contains genes encoding HLA molecules. Owing to the biological effect of TNF-α on the thyroid gland and its gene location, TNF-α should be able to affect an individual’s susceptibility to GD [5]. Therefore, TNF-α gene is a functional candidate for studying GD.
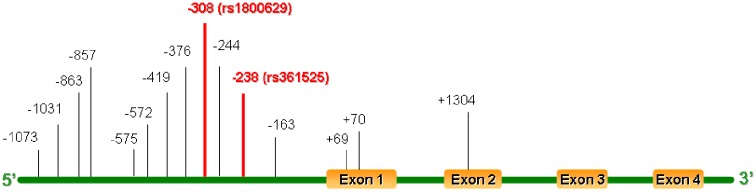
Full-length human TNF-α gene spans 2.76-kb DNA, with four exons and three introns. Single-nucleotide polymorphisms (SNPs) within TNF-α have a potential to cause structural changes within regulatory sites that could affect the function or regulation of TNF-α production. These factors could contribute to the autoimmune process making it an ideal candidate for the development of GD [6]. The TNF-α gene has been noted to be very polymorphic as manifested by the enrichment of many exonic, intronic as well as promoter SNPs (Figure 1) [7]. Although the mechanisms underlying TNF-α modulation of the risks for GD are yet to be fully addressed, elucidation of its genetic predisposition for GD, however, may offer some important clues. Indeed, several variations in the promoter region of the TNF-α gene have been suggested to be associated with increased risks to the development of GD by several genome-wide association studies (GWAS) [8,9]. Particularly, the most widely investigated SNPs of the TNF-α are G-238A (rs361525) and G-308A (rs1800629) in the promoter region, both of them are G to A substitutions. Although similar meta-analyses for the same SNP have already been conducted by Li et al. [10] ~10 years ago, these studies never were comprehensive and the outcomes were found to be conflicting results as well. We, therefore, in the current report, conducted an updated meta-analysis of SNPs rs361525 and rs1800629 in TNF-α gene from all eligible case–control studies to assess the associations amongst reported TNF-α gene with GD.
Figure 1. SNPs in the human TNF-α gene.
Methods
Eligible studies
PubMed, Embase, and ISI Web of Science were searched (the last search was conducted on 25 December, 2017) using the following search terms: ‘TNF-α OR Tumor necrosis factor-alpha’, ‘polymorphism OR variant OR mutation’, and ‘Graves’ disease’. References, which were listed in each identified article, were also searched manually to identify additional eligible studies.
Validity assessment
To be eligible, the following inclusion criteria were established: (i) a human case–control study of a polymorphism associated with GD; (ii) studies that included sufficient genotype data for extraction. Main exclusion criteria for studies were as follows: (i) case reports, letters, reviews, and editorial articles; (ii) literature not containing information regarding diabetes research; (iii) study involving only a case population; and (iv) study not written in English. In the case of multiple studies by the same researchers involving the same or overlapping datasets, we selected the most recent study with the largest number of participants.
Data extraction and quality assessment
Two curators (Y.T. and G.F.) independently extracted information from included studies. Disagreement was resolved by discussion between the two authors. The following data were extracted: first author’s name, year of publication, ethnicities of the individuals involved, the genotyping method, number of cases and controls for each genotype, and the Hardy–Weinberg equilibrium (HWE) amongst the controls. Ethnicity was categorized as Asian and European. A double-check procedure was performed to ensure accuracy of data entry. To evaluate the study quality, we adopted the Newcastle–Ottawa Scale (NOS) with a nine-star system; this scale assesses the quality of cohort and case–control studies. NOS focusses on three separate sections of stars representing the assessment score. The maximal score of NOS is 9 stars: 4 stars for the selection process, 2 stars for comparability, and 3 stars for exposure/outcome. A score of 7 and above was considered to be high-quality study.
Statistical analysis
The strength of associations between SNPs rs1800629 and rs361525 within the TNF-α gene and the risks for GD was assessed by odds ratios (ORs) with 95% confidence intervals (CIs). We explored the association between rs1800629 and GD in homozygote model (AA compared with GG), heterozygote model (GA compared with GG), dominant model (AA + GA compared with GG), recessive model (AA compared with GA + GG), and additive model (A compared with G), respectively. The same genetic models were applied for SNP rs361525 as well. Chi-squared-based Q-statistic test was employed to assess the between-study heterogeneity, and in any case P<0.10 was considered with significant heterogeneity between datasets. Once the effects were assumed to be homogeneous, fixed-effects model was then applied (the Mantel–Haenszel method); otherwise, the random-effects model (DerSimonian and Laird method) was employed appropriately. Sensitivity analysis was performed to assess the influence of each individual study by omitting one study at a time and calculating a pooled estimate for the remainder of the studies. The inverted funnel plots and Egger’s regression test were used to investigate publication bias. Potential publication bias was assessed with funnel plots of the effect sizes compared with the S.E.M.; Begg’s test was used to identify significant asymmetry. If there is evidence of publication bias, funnel plot is noticeably asymmetric. Concerning the significance level of the Begg’s and Egger’s tests was set at 0.05. All statistical tests carried out in the present report were two-tailed. All analyses were conducted using the STATA 11.0 software (STATA Corporation, College Station, TX, U.S.A.).
Results
Workflow for the identification of eligible datasets
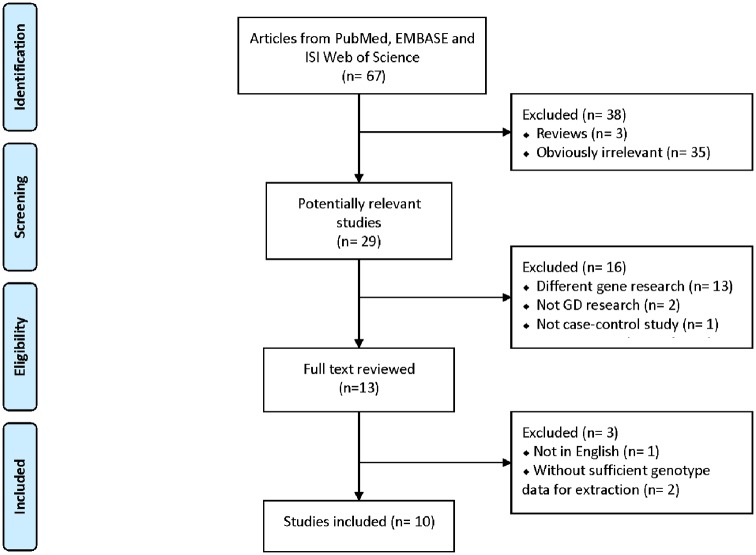
A total of 67 publications were characterized based on our keyword search. After screening the titles and abstracts, 35 studies were identified as irrelevant, and 3 articles were characterized as reviews. Additionally, 16 studies were excluded because 13 of the articles focussed on different genes. Another three articles were excluded because they were not on GD research (two studies) or were not case–control studies (one study). Amongst the remaining thirteen publications, three studies were also rejected as they either failed to provide detailed genotyping information (two articles) or were published in non-English journals (one study) (Figure 2).
Figure 2. PRISMA flow diagram showing the search strategy.
Characteristics of the selected datasets
A total of ten case–control datasets were identified based on our selection criteria. Of these, nine studies were conducted for the rs1800629 polymorphism which included 1980 GD patients and 2636 controls, while six studies were carried out for the rs361525 polymorphism which involved 1869 patients and 2300 controls. The principal characteristics and genotype distributions of the identified studies are shown in Table 1. For SNP rs1800629, six studies were found from Asian [11–16], and three studies were from European population [8,9,17]. For the rs361525 polymorphism, there were four studies originating from Asian [12–14,16], while the rest two studies were from European population [6,8]. Genotypic distribution for both rs1800629 and rs361525 in controls was in consistent with HWE (P>0.05) except for the four datasets highlighted in bold (Table 1). Each study was scored based on the NOS, as shown in Table 2. These nine case–control studies scored 7–8, indicating sufficient quality for inclusion in the meta-analysis.
Table 1. Summary of datasets included for meta-analysis.
| Study ID | Author | Year | Ethnicity | Genotyping method | Study design | Case/control | SNP loci | GD patient | Healthy control | pHWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | |||||||||
| 1 | Duraes et al. [9] | 2014 | European | Taqman | CC | 111/735 | rs1800629 | 72 | 34 | 5 | 562 | 156 | 17 | 0.122 |
| 2 | Kutluturk et al. [11] | 2013 | Asian | PCR-SSP | CC | 100/124 | rs1800629 | 73 | 24 | 3 | 103 | 15 | 6 | 0.000 |
| 3 | Jurecka-Lubieniecka et al. [17] | 2013 | European | PCR-RFLP | CC | 555/341 | rs1800629 | 299 | 231 | 25 | 259 | 71 | 11 | 0.032 |
| 4 | Anvari et al. [12] | 2010 | Asian | PCR-SSP | CC | 105/137 | rs1800629 | 56 | 44 | 5 | 98 | 39 | 0 | 0.052 |
| rs361525 | 74 | 33 | 0 | 79 | 57 | 1 | 0.007 | |||||||
| 5 | Gu et al. [13] | 2010 | Asian | MassArray™ | CC | 426/315 | rs1800629 | 368 | 56 | 2 | 263 | 51 | 1 | 0.369 |
| rs361525 | 408 | 20 | 0 | 281 | 34 | 0 | 0.311 | |||||||
| 6 | Shiau et al. [14] | 2007 | Asian | PCR-RFLP | CC | 187/101 | rs1800629 | 168 | 16 | 3 | 77 | 24 | 0 | 0.175 |
| rs361525 | 50 | 70 | 3 | 186 | 3 | 0 | 0.912 | |||||||
| 7 | Chen et al. [15] | 2005 | Asian | PCR-RFLP | CC | 95/60 | rs1800629 | 85 | 10 | 0 | 49 | 9 | 2 | 0.083 |
| 8 | Bednarczuk et al. [8] | 2004 | European | PCR-SSP | CC | 228/248 | rs1800629 | 122 | 96 | 10 | 172 | 72 | 4 | 0.25 |
| rs361525 | 220 | 8 | 0 | 225 | 22 | 1 | 0.563 | |||||||
| 9 | Simmonds et al. [6] | 2004 | European | PCR-RFLP | CC | 810/836 | rs361525 | 660 | 145 | 5 | 727 | 105 | 4 | 0.92 |
| 10 | Kamizono et al. [16] | 2000 | Asian | PCR-SSOP | CC | 173/575 | rs1800629 | 169 | 4 | 0 | 556 | 18 | 1 | 0.04 |
| rs361525 | 166 | 7 | 0 | 552 | 23 | 0 | 0.62 | |||||||
Abbreviations: CC, case/control; PCR-SSOP, PCR-sequence specific oligonucleotide polymorphism; PCR-RFLP, PCR-restriction fragment length polymorphism; PCR-SSP, PCR-sequence specific primer.
Table 2. Quality assessments of case–control studies according to the NOS.
| Study ID | Authors | Year | Selection | Comparability | Exposure | Total score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | ||||
| 1 | Duraes et al. [9] | 2014 | * | * | / | * | * | / | * | * | * | 7 |
| 2 | Kutluturk et al. [11] | 2013 | * | * | / | * | * | / | * | * | * | 7 |
| 3 | Jurecka-Lubieniecka et al. [17] | 2013 | * | * | / | * | * | / | * | * | * | 7 |
| 4 | Anvari et al. [12] | 2010 | * | * | / | * | * | / | * | * | * | 7 |
| 5 | Gu et al. [13] | 2010 | * | * | / | * | * | * | * | * | * | 8 |
| 6 | Shiau et al. [14] | 2007 | * | * | / | * | * | / | * | * | * | 7 |
| 7 | Chen et al. [15] | 2005 | * | * | / | * | * | * | * | * | * | 8 |
| 8 | Bednarczuk et al. [8] | 2004 | * | * | / | / | * | / | * | * | * | 6 |
| 9 | Simmonds et al. [6] | 2004 | * | * | * | * | * | / | * | * | * | 8 |
| 10 | Kamizono et al. [16] | 2000 | * | * | / | * | * | / | * | * | * | 7 |
Publication quality check list
Selection: a: Is the case definition adequate? b: Representativeness of the cases. c: Selection of controls; d: Definition of controls.
Comparability: e: Study controls for ethnicity. f: Study controls for any additional factor.
Exposure: g: Ascertainment of exposure. h: Same method of ascertainment for cases and controls. i: Non-response rate.
The asterisks (*) represent the stars in the NOS assessment.
Association between TNF-α gene polymorphism and GD
Meta-analysis for the promoter SNP rs1800629 was carried out by including 1980 GD patients and 2636 controls. A significant association was characterized between the rs1800629 polymorphism and GD in the homozygous model (AA compared with GG: OR = 1.97, 95% CI = 1.27–3.06, P=0.002) and recessive model (AA compared with GA + GG: OR = 1.62, 95% CI = 1.04–2.50, P=0.03) (Table 3). For analysis of ethnic stratification, we divided the datasets into two subgroups, Asian and European. GD susceptibility was significantly detected in European population in all genetic models. In sharp contrast, no significant association could be detected in Asian population (Table 4). Next, we conducted a meta-analysis for another promoter SNP rs361525, in which we have included the above identified five datasets (1869 patients and 2300 controls in total). However, SNP rs361525 did not show a significant association with GD in any genetic model before and after ethnicity stratification (Tables 3 and 4). Of note, our meta-analysis for SNP rs1800629 and rs361525 was hampered by the presence of genetic heterogeneity, which could be due to the differences of ethnicities and gene–environmental interactions.
Table 3. Results for meta-analysis of TNF-α polymorphisms with GD risk.
| SNPs | OR (95% CI) | P-value | Test of heterogeneity | p for publication bias1 | |
|---|---|---|---|---|---|
| I2 | P-value | ||||
| rs1800629 (G > A) | |||||
| AA compared with GG | 1.97 [1.27, 3.06] | 0.002 | 10.9% | 0.34 | 0.71 |
| GA compared with GG | 1.26 [0.80, 1.98] | 0.33 | 85.2% | 0.00 | 0.13 |
| AA + GA compared with GG | 1.25 [0.81, 1.94] | 0.32 | 85.0% | 0.00 | 0.08 |
| AA compared with GA + GG | 1.62 [1.04, 2.50] | 0.03 | 4.4% | 0.40 | 0.99 |
| A compared with G allele | 1.20 [0.84, 1.71] | 0.31 | 81.9% | 0.00 | 0.04 |
| rs361525 (G > A) | |||||
| AA compared with GG | 1.67 [0.67, 4.24] | 0.266 | 42.0% | 0.16 | 0.99 |
| GA compared with GG | 1.38 [0.51, 3.74] | 0.522 | 94.0% | 0.00 | 0.91 |
| AA + GA compared with GG | 1.38 [0.51, 3.76] | 0.530 | 94.2% | 0.00 | 0.92 |
| AA compared with GA + GG | 1.47 [0.57, 3.80] | 0.427 | 3.7% | 0.37 | 0.94 |
| A compared with G allele | 1.28 [0.52, 3.16] | 0.587 | 93.5% | 0.00 | 0.96 |
Egger’s test was performed to assess publication bias.
P < 0.05 was considered statistically significant.
Table 4. Subgroup analysis of rs1800629 and rs361525 in TNF-α.
| Polymorphism | Genetic model | Ethnicity | Number of datasets | OR (95% CI) | P-value | Test of heterogeneity | |
|---|---|---|---|---|---|---|---|
| I2 | P-value | ||||||
| rs1800629 | AA compared with GG | Asian | 6 | 1.40 [0.64, 3.06] | 0.396 | 27.3% | 0.230 |
| European | 3 | 2.31 [1.35, 3.95] | 0.002 | 0.0% | 0.713 | ||
| GA compared with GG | Asian | 6 | 0.91 [0.50, 1.69] | 0.772 | 79.9% | 0.000 | |
| European | 3 | 2.14 [1.55, 2.95] | 0.000 | 53.8% | 0.115 | ||
| AA + GA compared with GG | Asian | 6 | 0.90 [0.50, 1.61] | 0.720 | 79.3% | 0.000 | |
| European | 3 | 2.18 [1.67, 2.84] | 0.000 | 38.0% | 0.199 | ||
| AA compared with GA + GG | Asian | 6 | 1.32 [0.60, 2.87] | 0.489 | 25.3% | 0.245 | |
| European | 3 | 1.78 [1.04, 3.02] | 0.000 | 0.0% | 0.605 | ||
| A compared with G | Asian | 6 | 0.89 [0.53, 1.48] | 0.647 | 77.7% | 0.000 | |
| European | 3 | 1.9 [1.60, 2.28] | 0.000 | 0.0% | 0.46 | ||
| rs361525 | AA compared with GG | Asian | 4 | 3.61 [0.75, 17.3] | 0.109 | 72.8% | 0.055 |
| European | 2 | 1.09 [0.33, 3.55] | 0.891 | 0.0% | 0.429 | ||
| GA compared with GG | Asian | 4 | 2.02 [0.32, 12.69] | 0.452 | 96.1% | 0.000 | |
| European | 2 | 0.80 [0.20, 3.16] | 0.746 | 90.0% | 0.002 | ||
| AA + GA compared with GG | Asian | 4 | 2.04 [0.32, 13.07] | 0.453 | 96.2% | 0.000 | |
| European | 2 | 0.78 [0.19, 3.20] | 0.725 | 90.7% | 0.001 | ||
| AA compared with GA + GG | Asian | 4 | 2.82 [0.53, 15.1] | 0.225 | 53.4% | 0.143 | |
| European | 2 | 1.04 [0.32, 3.41] | 0.946 | 0.0% | 0.470 | ||
| A compared with G | Asian | 4 | 1.85 [0.35, 9.87] | 0.469 | 95.8% | 0.000 | |
| European | 2 | 0.76 [0.19, 3.05] | 0.694 | 90.8% | 0.001 | ||
P < 0.05 was considered statistically significant.
Publication bias
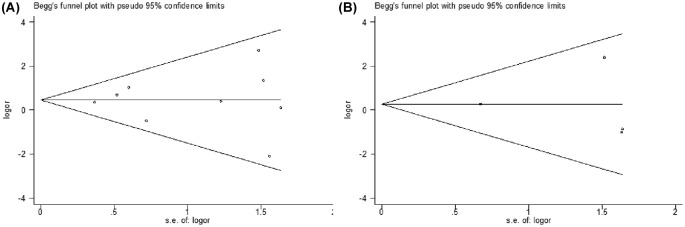
Begg’s funnel plot and Egger’s test were next conducted to assess publication bias. The shape of the funnel plots appeared to be symmetrical [SNP rs1800629: AA compared with (GA + GG); SNP rs361525: AA compared with (GA + GG)] and the Egger’s test did not show any evidence of publication bias (Figure 3). Analysis of sensitivity also revealed that results derived from our study are stable and reliable (data not shown).
Figure 3. Funnel plot analysis to detect publication bias.
Each point represents a separate study for the indicated association. (A) SNP rs1800629: AA compared with (GA + GG), (B) SNP rs361525: AA compared with (GA + GG).
Discussion
TNF-α is an inflammatory cytokine that is produced by intrathyroidal inflammatory cells and thyroid follicular cells and plays a pivotal role in regulating immunological reactions and the development of autoimmune diseases [18]. Upon the recognition of this functional property, TNF-α has thus been considered to be a candidate gene for GD. Nevertheless, no consistent results have been reached so far in terms of its genetic predisposition in GD pathoetiology. To address this question, we conducted a meta-analysis with the aim of concentration on the two SNPs, G-238A (rs361525) and G-308A (rs1800629) in the promoter region. Our studies demonstrated by clear and convincing evidence that only the promoter SNP rs1800629 within the TNF-α gene is associated with an increased risk for developing GD. The results of our overall meta-analysis supported that only G- > A mutation at −308 in TNF-α was a risk factor for GD, while the other SNP did not show a significant association with GD in any genetic model. To exclude the influence of population stratification, we then divided all datasets into two subgroups, Asian population and European population. Much stronger association was noted in the European populations, while the association was undetectable in the Asian population, representing the existence of genetic heterogeneity between different ethnic groups, which could be caused by the differences of gene–environmental interactions. These results were consistent with the findings of Duraes et al. [9] in a Portuguese population and Jurecka-Lubieniecka et al. [17] in a Polish population. TNF-α is produced by monocytes, T cells, natural killer cells, and mast cells, which is an essential contributing factor for the autoimmune thyroid dysfunctions. TNF-α −308 A allele is associated with a higher level of TNF-α transcript, due to the great potency of the promoter region to activate the transcription [19,20]. Therefore, individuals carrying higher TNF-α secreting genotypes may be susceptible to GD development. TNF-α gene polymorphisms at position −238 is another SNP which is commonly studied. Although our meta-analysis did not detect an association between −238 and GD, it was reported that the region between −254 and −230 contains a regulatory sequence that acts as a TNF-α repressor site, and thus a mutation at −238 might be disrupting regulation [19,21].
Our meta-analysis has some key advantages compared with individual studies. First, to guarantee the quality of the present study, we included the most updated literature and used explicit criteria for study inclusion and a strict procedure for data extraction. Additionally, a substantial number of subjects were pooled from individual studies, which significantly increased the statistical power. However, there are several limitations in our study. First, the controls were hospital-based study in our included literatures. Compared with hospital-based study, a population-based case–control study can reduce more selection bias and have higher confidence. Second, our search was limited to published English language studies. Some potential studies which were published in other languages or unpublished have been systematically excluded. This may explain some publication bias in our meta-analysis, which may have affected the results of this meta-analysis in as far as those studies that had produced negative results might not have been published. Third, the study population is limited for meta-analysis. Considering this would lead to low statistical power, future studies with a large dataset would be necessary for fully establishing the impact on susceptibility to GD.
In summary, the results of our meta-analysis identified that only the promoter SNP rs1800629 within the TNF-α gene is associated with increased risk for developing GD, especially in European population. However, future studies with a large dataset focussing on addressing their functional relevance would be necessary for fully establishing their effect on GD susceptibility.
Abbreviations
- CI
confidence interval
- GD
Graves’ disease
- HLA
human leukocyte antigen
- HWE
Hardy–Weinberg equilibrium
- NOS
Newcastle–Ottawa Scale
- OR
odds ratio
- SNP
single-nucleotide polymorphism
- TNF-α
tumor necrosis factor-α
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81500796, 81500620]; and the Research Fund of Wuhan Union Hospital [grant number 02.03.2017-321].
Author contribution
W.K. and X.C. conceived and designed the study strategy. Y.T. and G.F. were responsible for acquisition of data: statistical analysis and interpretation of data; and drafting or revision of the manuscript. T.Z. was responsible for reference collection and data management. Y.T. wrote the manuscript. G.F. prepared the tables and figures. W.K. and X.C. were responsible for study supervision. All authors reviewed the manuscript.
References
- 1.Vanderpump M.P. (2011) The epidemiology of thyroid disease. Br. Med. Bull. 99, 39–51 10.1093/bmb/ldr030 [DOI] [PubMed] [Google Scholar]
- 2.Ganesh B.B., Bhattacharya P., Gopisetty A. and Prabhakar B.S. (2011) Role of cytokines in the pathogenesis and suppression of thyroid autoimmunity. J. Interferon Cytokine Res. 31, 721–731 10.1089/jir.2011.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heward J.M., Allahabadia A., Daykin J., Carr-Smith J., Daly A. et al. (1998) Linkage disequilibrium between the human leukocyte antigen class II region of the major histocompatibility complex and Graves’ disease: replication using a population case control and family-based study. J. Clin. Endocrinol. Metab. 83, 3394–3397 [DOI] [PubMed] [Google Scholar]
- 4.Stenszky V., Kozma L., Balazs C., Rochlitz S., Bear J.C. et al. (1985) The genetics of Graves’ disease: HLA and disease susceptibility. J. Clin. Endocrinol. Metab. 61, 735–740 10.1210/jcem-61-4-735 [DOI] [PubMed] [Google Scholar]
- 5.Matsuno H., Yudoh K., Katayama R., Nakazawa F., Uzuki M. et al. (2002) The role of TNF-alpha in the pathogenesis of inflammation and joint destruction in rheumatoid arthritis (RA): a study using a human RA/SCID mouse chimera. Rheumatology (Oxford) 41, 329–337 10.1093/rheumatology/41.3.329 [DOI] [PubMed] [Google Scholar]
- 6.Simmonds M.J., Heward J.M., Howson J.M., Foxall H., Nithiyananthan R. et al. (2004) A systematic approach to the assessment of known TNF-alpha polymorphisms in Graves’ disease. Genes Immun. 5, 267–273 10.1038/sj.gene.6364066 [DOI] [PubMed] [Google Scholar]
- 7.Hajeer A.H. and Hutchinson I.V. (2000) TNF-alpha gene polymorphism: clinical and biological implications. Microsc. Res. Tech. 50, 216–228 10.1002/1097-0029(20000801)50:3%3c216::AID-JEMT5%3e3.0.CO;2-Q [DOI] [PubMed] [Google Scholar]
- 8.Bednarczuk T., Hiromatsu Y., Seki N., Ploski R., Fukutani T. et al. (2004) Association of tumor necrosis factor and human leukocyte antigen DRB1 alleles with Graves’ ophthalmopathy. Hum. Immunol. 65, 632–639 10.1016/j.humimm.2004.02.033 [DOI] [PubMed] [Google Scholar]
- 9.Duraes C., Moreira C.S., Alvelos I., Mendes A., Santos L.R. et al. (2014) Polymorphisms in the TNFA and IL6 genes represent risk factors for autoimmune thyroid disease. PLoS ONE 9, e105492 10.1371/journal.pone.0105492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., Zhou Z., Liu X., Liu Y., Zhang J. et al. (2008) Association of tumour necrosis factor alpha (TNF-alpha) polymorphisms with Graves’ disease: a meta-analysis. Clin. Biochem. 41, 881–886 10.1016/j.clinbiochem.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 11.Kutluturk F., Yarman S., Sarvan F.O. and Kekik C. (2013) Association of cytokine gene polymorphisms (IL6, IL10, TNF-alpha, TGF-beta and IFN-gamma) and Graves’ disease in Turkish population. Endocr. Metab. Immune Disord. Drug Targets 13, 163–167 10.2174/18715303113139990001 [DOI] [PubMed] [Google Scholar]
- 12.Anvari M., Khalilzadeh O., Esteghamati A., Momen-Heravi F., Mahmoudi M. et al. (2010) Graves’ disease and gene polymorphism of TNF-alpha, IL-2, IL-6, IL-12, and IFN-gamma. Endocrine 37, 344–348 10.1007/s12020-010-9311-y [DOI] [PubMed] [Google Scholar]
- 13.Gu L.Q., Zhu W., Pan C.M., Zhao L., Zhang M.J. et al. (2010) Tumor necrosis factor alpha (TNF-alpha) polymorphisms in Chinese patients with Graves’ disease. Clin. Biochem. 43, 223–227 10.1016/j.clinbiochem.2009.08.012 [DOI] [PubMed] [Google Scholar]
- 14.Shiau M.Y., Huang C.N., Yang T.P., Hwang Y.C., Tsai K.J. et al. (2007) Cytokine promoter polymorphisms in Taiwanese patients with Graves’ disease. Clin. Biochem. 40, 213–217 10.1016/j.clinbiochem.2006.11.009 [DOI] [PubMed] [Google Scholar]
- 15.Chen R.H., Chen W.C., Wang T.Y., Tsai C.H. and Tsai F.J. (2005) Lack of association between pro-inflammatory cytokine (IL-6, IL-8 and TNF-alpha) gene polymorphisms and Graves’ disease. Int. J. Immunogenet. 32, 343–347 10.1111/j.1744-313X.2005.00536.x [DOI] [PubMed] [Google Scholar]
- 16.Kamizono S., Hiromatsu Y., Seki N., Bednarczuk T., Matsumoto H. et al. (2000) A polymorphism of the 5′ flanking region of tumour necrosis factor alpha gene is associated with thyroid-associated ophthalmopathy in Japanese. Clin. Endocrinol. (Oxf.) 52, 759–764 10.1046/j.1365-2265.2000.01011.x [DOI] [PubMed] [Google Scholar]
- 17.Jurecka-Lubieniecka B., Ploski R., Kula D., Krol A., Bednarczuk T. et al. (2013) Association between age at diagnosis of Graves’ disease and variants in genes involved in immune response. PLoS ONE 8, e59349 10.1371/journal.pone.0059349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coperchini F., Pignatti P., Leporati P., Carbone A., Croce L. et al. (2016) Normal human thyroid cells, BCPAP, and TPC-1 thyroid tumor cell lines display different profile in both basal and TNF-alpha-induced CXCL8 secretion. Endocrine 54, 123–128 10.1007/s12020-015-0764-x [DOI] [PubMed] [Google Scholar]
- 19.Fong C.L., Siddiqui A.H. and Mark D.F. (1994) Identification and characterization of a novel repressor site in the human tumor necrosis factor alpha gene. Nucleic Acids Res. 22, 1108–1114 10.1093/nar/22.6.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis E., Franchimont D., Piron A., Gevaert Y., Schaaf-Lafontaine N. et al. (1998) Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin. Exp. Immunol. 113, 401–406 10.1046/j.1365-2249.1998.00662.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayley J.P., de Rooij H., van den Elsen P.J., Huizinga T.W. and Verweij C.L. (2001) Functional analysis of linker-scan mutants spanning the -376, -308, -244, and -238 polymorphic sites of the TNF-alpha promoter. Cytokine 14, 316–323 10.1006/cyto.2001.0902 [DOI] [PubMed] [Google Scholar]