Abstract
We reported previously that a 2′ fluoro-modified (2′ F) phosphorothioate (PS) antisense oligonucleotides (ASOs) with 5–10–5 gapmer configuration interacted with proteins from Drosophila behavior/human splicing (DBHS) family with higher affinity than PS-ASOs modified with 2′-O-(2-methoxyethyl) (2′ MOE) or 2′,4′-constrained 2′-O-ethyl (cEt) did. Rapid degradation of these proteins and cytotoxicity were observed in cells treated with 2′ F PS-ASO. Here, we report that 2′ F gapmer PS-ASOs of different sequences caused reduction in levels of DBHS proteins and hepatotoxicity in mice. 2′ F PS-ASOs induced activation of the P53 pathway and downregulation of metabolic pathways. Altered levels of RNA and protein markers for hepatotoxicity, liver necrosis, and apoptosis were observed as early as 24 to 48 hours after a single administration of the 2′ F PS-ASO. The observed effects were not likely due to the hybridization-dependent RNase H1 cleavage of on- or potential off-target RNAs, or due to potential toxicity of 2′ F nucleoside metabolites. Instead, we found that 2′ F PS-ASO associated with more intra-cellular proteins including proteins from DBHS family. Our results suggest that protein-binding correlates positively with the 2′ F modification-dependent loss of DBHS proteins and the toxicity of gapmer 2′ F PS-ASO in vivo.
INTRODUCTION
Over the past three decades, advanced chemical modifications have resulted in substantial improvements in pharmacological and therapeutic properties of antisense oligonucleotides (1–3). Therapeutic oligonucleotides are modified at backbone, sugar, and heterocyclic base to achieve nuclease stability and favorable pharmacokinetics (1–3). However, certain modifications can detrimentally affect the therapeutic performance of oligonucleotides, and some are associated with oligonucleotide toxicities through hybridization-dependent or independent mechanisms (4,5). For example, concerns over the potential for hybridization-dependent off-target cleavages have been increased for siRNAs and antisense oligonucleotides with sugar modifications, especially the ones that confer ultra-high affinity to nucleic acids (6,7). A possible consequence of this increased affinity is that hybridization can now occur over shorter regions of homology and 1–2 mismatches are tolerated. In terms of hybridization-independent effects, chemical modifications of oligonucleotides increase binding to many plasma and cellular proteins, which facilitates delivery and pharmacological performance of oligonucleotides (4,8–15). However, undesirable oligonucleotide-protein interactions can result in adverse effects. Importantly, adverse effects such as mild pro-inflammatory responses observed in mice due to the undesirable binding to proteins by first generation PS-DNA drugs were significantly mitigated by incorporating 2′ MOE modifications (16), which is at least partially due to the fact that 2′ MOE-modified PS-ASOs associate with proteins less avidly than PS-DNA (14).
In addition to toxicity caused by the intact modified oligonucleotides, toxicities have been attributed to metabolites (17,18). Oligonucleotides are degraded into nucleosides, and some modified nucleosides that are substrates for the nucleotide salvage pathway are potentially toxic, as has been demonstrated for analogues including those that are used as anticancer or antiviral agents (19). For example, the nucleoside analogue Fialuridine (FIAU), developed as a treatment for chronic hepatitis B, causes severe mitochondrial dysfunction, liver toxicities, metabolic acidosis, and death in humans (17,18).
We have recently studied the protein binding of PS-ASOs containing different nucleoside modifications (4,8–12,20,21). To our surprise, we found that the 5–10–5 gapmer PS-ASO containing 2′ F nucleoside modifications (2′ F PS-ASO) bound cellular proteins more promiscuously and with greater affinity than PS-ASOs containing 2′ MOE or cEt modifications (4,9). We also observed that when transfected into cultured HeLa cells, 2′ F PS-ASO caused DNA damage and cell death (4). Such cytotoxic effects were not observed in cells transfected with the same concentration of 2′ MOE PS oligonucleotides with the same sequence (4). In addition, levels of oligonucleotide-binding proteins including P54nrb (Non-POU domain-containing octamer-binding protein), PSF (splicing factor, proline- and glutamine-rich) and PSPC1 (paraspeckle component 1) were rapidly reduced in vitro in mouse or human cells treated with 2′ F PS-ASO either by transfection or by free uptake (4). Four to six 2′ F-modified nucleotides within the PS-oligonucleotide seemed to be sufficient to cause the protein reduction (4). P54nrb, PSF, and PSPC1 belong to the Drosophila behavior/human splicing (DBHS) family and are enriched in nuclear paraspeckle structures (22,23). We found previously that PS oligonucleotides interacted with DBHS proteins and were rapidly recruited to the nuclear paraspeckle structures upon entry into a cell (11). DBHS proteins are involved in many important cellular processes including transcription, splicing, polyadenylation, DNA repair, translation and apoptosis (24–29). Cellular levels of DBHS proteins are highly correlated with cell survival (28,30). As a result, degradation of DBHS proteins upon binding to the 2′ F PS-ASO may at least partially contribute to the observed cytotoxic effects.
To determine whether 2′ F-modified oligonucleotides result in similar effects in vivo, we treated mice with 5–10–5 gapmer PS-ASOs modified with 2′ MOE, cEt, and 2′ F designed to target the Pten transcript. Consistent with our observations in vitro, the 2′ F PS-ASO caused more significant hepatotoxicity in vivo than did PS-ASOs with 2′ MOE or cEt modifications. Potential molecular mechanisms of the adverse effects of 2′ F PS oligonucleotide suggest that the toxicity of these modified ASOs is not dependent on hybridization to RNA transcripts but likely results from binding to proteins.
MATERIALS AND METHODS
Toxicity studies in mice
Studies of up to 96 h in duration were used to evaluate the acute hepatotoxicity of PS-ASOs. Male BALB/c mice aged 6–8 weeks were obtained from Charles River Laboratories. ASOs or saline were administered subcutaneously on study day 1. On study days 2, 3, 4 and 5, animals were anesthetized using 2–4% isoflurane, and blood was collected by cardiac puncture. For long-term low-dose exposure, ASOs or saline were administered subcutaneously at 20 mg/kg per dose, three doses per week, and blood samples were collected weekly by tail-bleeding. Blood samples were processed to plasma and evaluated for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) using a Beckman Coulter AU480 Bioanalyzer. H&E and immunohistochemical staining of relevant tissues was performed as described previously (31). Animal experiments were conducted according to American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the institutions Animal Welfare Committee (Cold Spring Harbor Laboratory's Institutional Animal Care and Use Committee guidelines).
Hepatocyte isolation, cell culture and treatment
Liver perfusion and hepatocyte isolation were performed as described previously (31). Isolated primary hepatocytes were grown at 37°C, 8% CO2 in Williams Medium E supplemented with 10% fetal bovine serum, 1 × Antibiotic/Antimycotic, 10 mM HEPES and 2 mM L-Glutamine. HeLa cells were grown at 37°C, 7.5% CO2 in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. For siRNA treatment, cells at 70% confluency were transfected with 3 nM siRNA using Lipofectamine RNAiMax (Thermo Fisher Scientific) at a 6 μg/ml final concentration. For ASO treatment, cells at 70% confluency were transfected with oligonucleotides at specified concentration using Lipofectamine 2000 (Thermo Fisher Scientific) at a final concentration of 4 μg/ml, and harvested at specified times after transfection for subsequent analyses.
Microarray experiments
Mouse liver punches were homogenized using Bio-Gen PRO200 Homogenizer (PRO Scientific) in TRIzol (Thermo Fisher Scientific). Total RNA was isolated according to protocols supplied by the manufacturer. Three animals were included in each group. Microarray experiments and analyses were performed by Phalanx Biotech. Genes with significantly altered expression was identified as those with absolute log2(ratio) ≥ 1 and P-value < 0.05.
RNA isolation and qRT-PCR
Mouse liver punches were homogenized using Bio-Gen PRO200 Homogenizer (PRO Scientific) in TRIzol (Thermo Fisher Scientific) or RLT buffer from RNeasy Kit (Qiagen). Total RNA was isolated according to protocols supplied by the manufacturers. TaqMan One-step qRT-PCR was performed using AgPath-ID™ One-Step RT-PCR Reagents (Thermo Fisher Scientific). Expression levels of target RNA were normalized to total RNA quantified using Quant-iT RiboGreen RNA Reagent (Thermo Fisher Scientific). Primer-probe sets for mouse Cdkn1a (Mm01303209_m1), mouse Gadd45a (Mm00432802_m1), mouse Mcl1 (Mm01257351_g1), mouse Bcl-xl (Mm00437783_m1), mouse Map3k6 (Mm00522235_m1), mouse Cd68 (Mm03047343_m1), mouse Lgr5 (Mm00438890_m1), mouse Rhbg (Mm00491234_m1), mouse P54nrb (Mm00834875_g1) and mouse Sfpq (Mm01179807_m1), mouse Pten (Mm00477208_m1 and Mm01212532_m1) were purchased from Thermo Fisher Scientific.
Western analysis
Liver samples were homogenized using Bio-Gen PRO200 Homogenizer (PRO Scientific) in RIPA buffer supplemented with 1 × Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), quantitated using a BCA protein assay (Pierce), and were separated on a 4–12% gradient SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane using iBlot Gel Transfer Device (Thermo Fisher Scientific). The membranes were blocked at room temperature for 30 min with blocking buffer containing 5% (w/v) nonfat dry milk in 1 × PBS and incubated with primary antibodies in blocking buffer at room temperature for 2 h or 4°C overnight. After washing three times with washing buffer (0.1% Tween-20 in 1 × PBS) for 5 min each wash, membranes were incubated with secondary antibodies in blocking buffer at room temperature for 1 h. After washing three times with washing buffer for 5 min each wash, proteins were detected based on Enhanced chemiluminescence (Abcam). Antibodies to cleaved PARP (ab32064), P21 (ab7960), HSP90 (ab74248), P54nrb (ab133574) and BIP (ab21685) were purchased from Abcam. Anti-PSF antibody was purchased from Sigma (p2860).
Isolation of ASO-binding proteins
Streptavidin magnetic beads (Millipore; 25 μl per reaction) were washed in binding buffer (1× PBS with 0.2% tween-20) three times and incubated with 25 μM biotinylated cEt-modified PS-ASO (ION-586183) or biotinylated 2′ F-modified PS-ASO (ION- 623496) in 200 μl binding buffer at room temperature for 30 min. The beads were washed three times with binding buffer and incubated at 4°C for 2 h with 500 μg HeLa cell extract prepared in IP lysis buffer (Pierce). The beads were washed with the washing buffer (1× PBS with 0.2% tween-20, with additional 100 mM NaCl) ten times and incubated with competitor PS-ASOs at specified concentrations (0.5×, 1× and 2× of binding PS-ASO) for 5 min at room temperature with constant rotation. Supernatants were collected as eluted proteins. Beads were boiled to release the proteins that were not eluted by competition.
Immunofluorescent staining
Immunofluorescent staining experiments were performed as described previously (32). GFP-PSF was constructed as described previously (33). PS-ASOs were electroporated by Neon Transfection System (Thermo Fisher Scientific). Electroporation parameters are pulse voltage 1005 V, pulse width 35 ms and pulse number 2.
Bioluminescence resonance energy transfer (BRET) binding affinity assay
BRET assays were performed as described previously (9).
RESULTS
2′ F modified PS-ASOs cause hepatotoxicity in mice
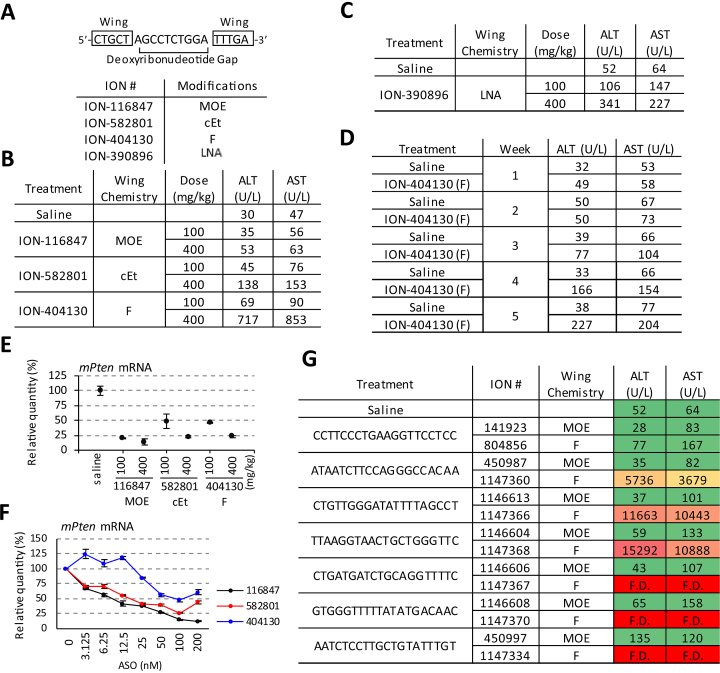
We observed previously that transfection of the PS-ASO containing 2′ F modifications (ION-404130) caused cytotoxicity of HeLa cells, whereas PS-ASOs with the same sequence but modified with 2′ MOE (ION-116847) or cEt (ION-582801) did not (4). Here, we tested the same ASOs in mice to further evaluate the effects of different 2′ modifications on liver functions in vivo. These ASOs are fully PS-modified, and have a 5–10–5 gapmer design (Figure 1A). In this design, the 10 deoxyribonucleotides in the middle allow RNase H1-mediated cleavage of targeted RNA, and the five nucleotides on each wing that are modified with 2′ MOE (ION-116847), cEt (ION-582801) or 2′ F (ION-404130) to impart nuclease stability, reduce proinflammatory responses, and enhance thermodynamic stability of the ASO–RNA duplex (2,16).
Figure 1.
PS-ASOs induce hepatotoxicity in mice in a sugar modification-dependent manner. (A) Schematic representation of the 5–10–5 Pten gapmer ASO. (B) Plasma levels of ALT and AST upon a single administration of Pten PS-ASOs modified with 2′ MOE (ION-116847), cEt (ION-582801) or 2′ F (ION-404130) at 100 or 400 mg/kg for 96 h. N = 3 per group. (C) Plasma levels of ALT and AST upon a single administration of Pten LNA (ION-390896) at 100 or 400 mg/kg for 96 h. N = 3 per group. (D) Plasma levels of ALT and AST upon repeated administration of 2′ F ASO (ION-404130) at 20 mg/kg per dose, three doses per week. S-saline, and F-2′ F PS-ASO (ION-404130). ( E) qRT-PCR of on-target Pten mRNA. Mice were treated as was described in B. The error bars indicate data acquired from three individual animals. (F) qRT-PCR showed dose-dependent reduction of target Pten mRNA in mouse hepatocytes by Pten ASOs modified with 2′ MOE (ION-116847), cEt (ION-582801), or 2′ F (ION-404130). ASOs were delivered by lipid transfection. The error bars represent standard deviation from three separate experiments. (G) Plasma levels of ALT and AST upon a single administration of PS-ASOs modified with either 2′ MOE or 2′ F at 400 mg/kg for 96 h. Seven pairs of 2′ MOE or 2′ F-modified sequences were included. Hepatotoxic potentials of the 2′ F PS-ASOs were rank-ordered and color-coded (red: high hepatotoxic potentials; and green: low hepatotoxic potentials) based on plasma levels of ALT and AST. N = 3 per group. F.D. = found death.
PS-ASOs were administered subcutaneously at 100 or 400 mg/kg on day 1, and BALB/c mice were sacrificed on day 4 (after 96 h). Significant elevations in ALT (717 U/L) and AST (853 U/L) were observed in animals given 2′ F PS-ASO ION-404130 at 400 mg/kg relative to animals treated with saline (ALT 30 U/L and AST 47 U/L) (Figure 1B). In several repeats of the experiments, animal deaths were observed for several mice dosed with 2′ F PS-ASOs at 400 mg/kg before the 96-h time point, whereas no deaths were observed in mice treated with the other ASOs. Moderately elevated levels of ALT (138 U/L) and AST (153 U/L) were observed in animals given the cEt-containing PS-ASO (ION-582801) at 400 mg/kg. Similar to cEt, ASOs containing other bicyclic nucleic acid modifications such as locked nucleic acid (LNA: ION-390896) also caused moderate elevation in levels of ALT (341 U/L) and AST (227 U/L) (Figure 1C). The 2′ MOE PS-ASO (ION-116847) did not cause any significant elevation in ALT or AST at 400 mg/kg dose (Figure 1B). We also reported previously in vitro that at least four to six 2′ F-modifications within a 20-mer PS-oligonucleotide were required to cause cytotoxicity (4). In mice, moderately elevated transaminases were observed with six 2′ F modifications within a 20-mer PS-oligonucleotide (Supplementary Figure S1A). Thus, the hepatotoxic potentials of these ASOs correlated well with their cytotoxicity in cultured cells (4).
Elevations in levels of ALT and AST were also observed in a long-term lower dose tolerability experiment in which BALB/c mice were given three doses of 2′ F PS-ASO ION-404130 at 20 mg/kg every week (Figure 1D). Blood samples were collected weekly by tail-bleeding and animals were sacrificed after week five. Elevated transaminases were observed after week four. This result suggests that 2′ F PS-ASO ION-404130 caused hepatotoxicity upon repeated lower dose exposure.
The observed hepatotoxicity with 2′ F-modified PS-ASO ION-404130 was not due to the reduction of the intended target, Pten mRNA. The 2′ F PS-ASO (ION-404130) was the least potent among the tested Pten ASOs in the reduction of Pten mRNA (Figure 1E), whereas the 2′ MOE PS-ASO (ION-116847) reduced Pten mRNA levels the most of the ASOs tested. This potency rank order correlated well with observations in vitro in cultured mouse hepatocytes after lipid transfection of ASOs (Figure 1F), suggesting that the 2′ F PS-ASO (ION-404130) may be less effective in supporting RNase H1-mediated target cleavage.
We observed previously in cultured cells that some PS-ASOs of different sequences caused reduction in levels of DBHS proteins and cell death (4). To confirm that the observed 2′ F modification-dependent hepatotoxicity was not limited to a single sequence, we tested a total of seven additional matched sequences that are designed as either 5–10–5 MOE or 5–10–5 F gapmers (Figure 1G and Supplementary Figure S1B). These additional sequences targeting genes other than PTEN were included to control for the potential effects of on-target reduction on observed toxicity. Hepatotoxicity of the tested 2′ F PS-ASOs were rank-ordered based on plasma levels of ALT and AST. Severe hepatotoxicity and animal death were observed for six out of the seven tested 2′ F-modified PS-ASOs but not the 2′ MOE PS-ASOs of the same sequences and gapmer design at the same dose and duration. These results suggest that the 5–10–5 gapmer PS-ASOs with 2′ F modifications are in general more toxic than PS-ASOs containing 2′ MOE. In addition, we found that not all 2′ F-containing PS-ASOs were toxic since ION-141923 (2′ MOE-modified) and 804856 (2′ F-modified) did not significantly elevate the levels of transaminases at 400 mg/kg.
2′ F PS -ASO (ION-404130) activates P53 pathway and induces apoptosis
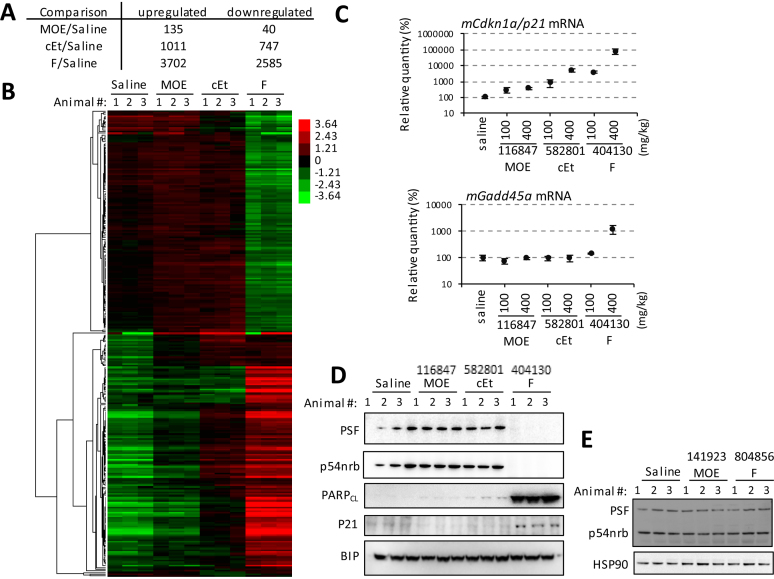
Microarray analyses were performed on the total RNAs isolated from the livers of animals at 96 h after a single administration of 400 mg/kg of 2′ MOE (ION-116847), cEt (ION-582801), or 2′ F (ION-404130)-modified ASOs. Gene expression differed significantly in mice treated with 2′ F PS-ASO compared to mice given saline or 2′ MOE PS-ASO (Figure 2A, B and Supplementary Figure S2). Treatment with 2′ F PS-ASO caused significant upregulation (log2(ratio) ≥1, P-value < 0.05) of 3702 transcripts and significant downregulation (log2(ratio) ≤ –1, P-value < 0.05) of 2585 transcripts compared with saline-treated animals. In mice treated with cEt PS-ASO compared to controls, 1011 genes were upregulated and 747 were downregulated. Only 135 upregulated and 40 downregulated transcripts were identified in mice given the 2′ MOE PS-ASO.
Figure 2.
2′ F PS-ASO (ION-404130) activated multiple stress pathways and induced expression of genes involved in apoptosis. (A) Numbers of genes altered in liver transcriptomes of mice 96 h after the administration of saline or 400 mg/kg 2′ MOE ASO (ION-116847), cEt ASO (ION-582801) or 2′ F ASO (ION-404130), as detected by microarray analyses. (B) Unsupervised hierarchical clustering analysis of 273 genes differentially expressed in ASO-treated mice compared to saline-treated mice. Logarithmic values of treated to untreated expression ratios are shown in the heat map using red and green color codes for up- and down-regulation, respectively. Experiments were done with three animals per group. (C) qRT-PCR of transcripts regulated downstream of P53 pathway, including Cdkn1a/p21 and Gadd45a. The error bars indicate data acquired from three individual animals. (D) Western analysis of liver proteins isolated from animals administrated with saline, 2′ MOE (ION-116847), cEt (ION-582801), or 2′ F (ION-404130) PS-ASOs. ASOs were administered at 400 mg/kg and animals were sacrificed at 96 h after the administration of ASOs. Experiments were done with three animals per individual group. Bip serves as a loading control. PARPCL-cleaved PARP. ( E) Western analysis of liver proteins isolated from animals administrated with saline, 2′ MOE (ION-141923), or F (ION-804856) PS-ASOs. ASOs were administered at 400 mg/kg and animals were sacrificed at 96 h after the administration of ASOs. Experiments were done with three animals per individual group. HSP90 serves as a loading control. PARPCL-cleaved PARP.
Gene Ontology analyses were performed to identify biological processes enriched in differentially expressed genes. Top regulated biological processes for each ASO treatment were listed in Supplementary Tables S1-S3. Significantly altered biological processes for animals receiving 2′ F PS-ASO (ION-404130) included oxidation-reduction pathways, transcription, apoptosis, and cell proliferation (Supplementary Table S3). Genes involved in apoptosis (GO:0006915) were moderately affected by the cEt PS-ASO (k/K = 0.1208) (Supplementary Table S2), but to a significantly lesser extent than the 2′ F PS-ASO (k/K = 0.3713). The 2′ MOE PS-ASO did not significantly alter these biological processes (Supplementary Table S1). KEGG pathway analysis suggests that the most downregulated pathways in 2′ F PS-ASO-treated animals were metabolic pathways, and the most upregulated pathways included ribosomal biogenesis, TNF signaling, and P53 signaling (Supplementary Tables S4–S6). Additional cellular processes and signaling pathways, including DNA replication, RNA transport, RNA splicing, cell cycle, apoptosis, HIF-1 signaling, NF-kappa-B signaling, MAPK signaling, and FOXO signaling, were also significantly upregulated in mice given the 2′ F PS-ASO.
Consistent with the observations for blood chemistry, RNA markers for hepatotoxicity and liver necrosis were significantly differentially expressed in mice treated with the 2′ F PS-ASO (ION-404130) compared to control mice (Supplementary Figures S3 and S4). Moderately increased levels of RNA markers for hepatotoxicity and liver necrosis were observed in cEt PS-ASO (ION-582801)-treated mice. Mice given the 2′ MOE PS-ASO (ION-116847) showed minimal changes in the levels of RNA markers for hepatotoxicity and liver necrosis.
Activation of the P53 signaling pathway in liver was reported in CD-1 mice treated with a hepatotoxic LNA-modified gapmer ASO (34). Liver apoptosis was also observed to be induced by toxic LNA ASOs (35). To analyze and confirm the activation of the P53 pathway by the 2′ F PS-ASO, qRT-PCR was performed to quantify RNA levels of Cdkn1a/P21 and Gadd45a, which are known to be transcriptionally upregulated by P53. Liver mRNA levels of Cdkn1a/P21 were increased to approximately 800-, 48- and 3.6-fold in mice treated with 400 mg/kg PS-ASOs modified with 2′ F (ION-404130), cEt (ION-582801), or 2′ MOE (ION-116847), respectively (Figure 2C), correlating with the elevation in ALT and AST. Activation of the P53 pathway by the 2′ F PS-ASO in vivo was further validated by western blot analysis of P21 protein from liver homogenates (Figure 2D). A significant increase in levels of P21 protein was observed in 2′ F ASO (ION-404130), but not in 2′ MOE (ION-116847) or cEt (ION-582801) ASO treated animals. Induction of caspase-dependent apoptotic cell death by the 2′ F PS-ASO (ION-404130) was also supported by western analysis using an antibody specific for cleaved PARP (25-kD fragment) (Figure 2D). Moderate accumulation of cleaved PARP was detected in mice treated with the cEt ASO (ION-582801), but no significant PARP cleavage was observed in mice treated with the 2′ MOE PS-ASO (ION-116847) (Figure 2D). In addition, consistent with the observation in vitro, 2′ F-modified ION-404130, but not ION-116847 (MOE) or ION-582801 (cEt), caused reduction in levels of P54nrb and PSF proteins in mouse livers (Figure 2D). Reduction in levels of P54nrb and PSF as well as the accumulation of cleaved PARP was also observed from liver homogenates from mice treated with other toxic 2′ F modified PS-ASOs (ION-1147368, ION-1147366, and ION-1147360) but not their MOE counterparts (ION-1146604, ION-1146613 and ION-450987) (Supplementary Figure S5). Importantly, PS-ASOs 2′ modified with MOE (ION-141923) or F (ION-804856), which did not cause significant elevation in levels of ALT and AST, did not reduce the levels of P54nrb and PSF proteins (Figure 2E).
DBHS proteins have been linked to the repair of double-stranded DNA breaks (DSBs) through non-homologous end joining pathways (26,29). As a result, loss of DBHS proteins in cultured cells resulted in the increased sensitivity of cells to DNA damaging agents. In our study, activation of pathways related to DNA damage or DNA replication was observed by microarray analyses in the samples from mice treated with the 2′ F PS-ASO (ION-404130). For example, 144 out of 421 genes (34.20%) annotated as related to cellular responses to DNA damage stimulus (GO: 0006974) were significantly differentially expressed in animals receiving 2′ F PS-ASO (ION-404130) compared to saline-treated mice (Supplementary Table S3). Of these 144 genes, 118 were upregulated and 26 were downregulated. In mice treated with the 2′ MOE PS-ASO (ION-116847), only six genes in the cellular responses to DNA damage stimulus category were upregulated and only two were downregulated (Supplementary Table S1). In mice treated with cEt PS-ASO (ION-582801), no genes involved in this category were significantly differentially regulated (Supplementary Table S2). Thus, these DNA damage responses observed in mice were 2′ modification-dependent. Although all three ASOs tested contain full PS modifications, only 2′ F-modified ION-404130 caused significant activation of DNA damage responses.
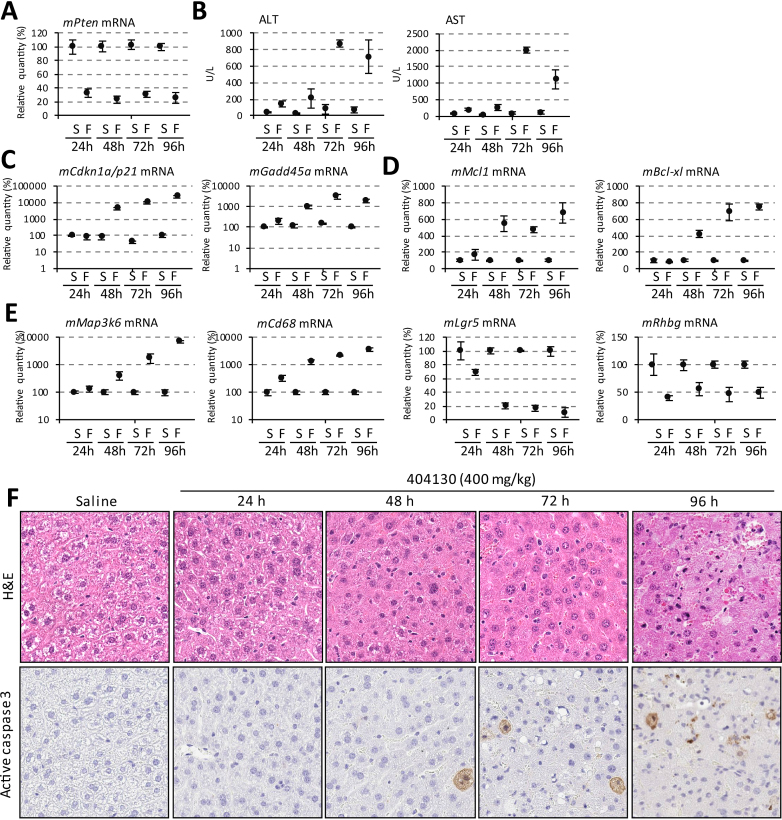
Biomarkers for P53 signaling, necrosis, and apoptosis are observed to be affected at early time points after subcutaneous dosing of mice with 2′ F ASO
We performed a detailed time course in vivo to further evaluate phenotypes related to treatment with 2′ F PS-ASO. Mice were given 400 mg/kg 2′ F ASO (ION-404130) subcutaneously on day 1 and were sacrificed at 24, 48, 72 and 96 h after dosing. Maximal reduction of the mRNA target of this ASO, Pten, was achieved at 24 h (Figure 3A). Time-dependent elevations in transaminases relative to saline-treated controls were observed, and significant elevations in ALT and AST were detected at 72 and 96 h (Figure 3B). Time-dependent increases in levels of the P53-regulated Cdkn1a/P21 and Gadd45a mRNAs, as well as the anti-apoptosis RNA markers Mcl-1 and Bcl-xl were observed 48 h after the administration of the ASO (Figure 3C and D). As a result of liver necrosis, upregulation in levels of Map3k6 and Cd68 mRNAs and downregulation in levels of Lgr5 and Rhbg mRNAs were observed from 24 h after administration of the 2′ F PS-ASO (Figure 3E).
Figure 3.
Time course of mRNA alterations and stress phenotypes induced by 2′ F PS-ASO (ION-404130) in vivo. ( A–F) Mice were treated with a single 400 mg/kg 2′ F PS-ASO (ION-404130) or with saline and indicated parameters were quantified at 24, 48, 72 and 96 h after dosing. N = 3 per group. The error bars indicate data acquired from three individual animals. S-saline, and F-2′ F PS-ASO (ION-404130). (A) qRT-PCR quantification of on-target Pten mRNA levels in liver. (B) Plasma levels of ALT and AST. (C) qRT-PCR of mRNA transcripts Cdkn1a/p21 and Gadd45a. (D) qRT-PCR of liver mRNA levels of anti-apoptosis markers Mcl1 and Bcl-xl. (E) qRT-PCR of liver mRNA levels of necrosis markers that are up-regulated (Map3k6 and Cd68) or down-regulated (Lgr5 and Rhbg) by 2′ F ASO. (F) Representative images of H&E staining and active caspase 3 staining of liver sections.
In agreement with the changes in levels of RNA markers for apoptosis and necrosis, histological analyses performed on the liver tissues from animals dosed with 400 mg/kg of ION-404130 demonstrate that severe hepatocyte swelling, necrosis, and apoptosis was observed as early as 48 h (Figure 3F). These results suggest the acute toxicity in livers of mice treated with 2′ F PS-ASO (ION-404130).
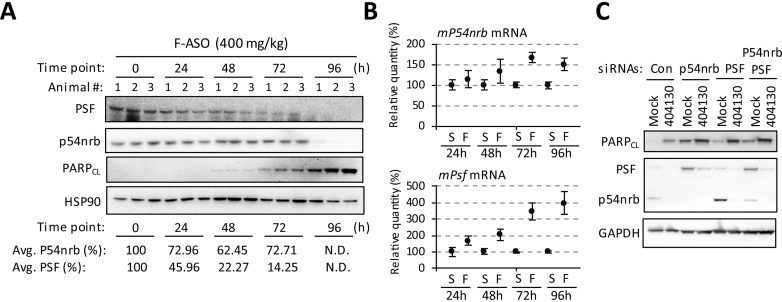
2′ F-ASO (ION-404130) time-dependently reduces levels of P54nrb and PSF proteins in mice
Consistent with the liver histology and the RNA markers for P53 activation and apoptosis, accumulation of cleaved PARP in mouse liver was observed as early as 48–72 h after receiving 400 mg/kg 2′ F PS-ASO ION-404130 (Figure 4A). We reported previously in cultured cells that the loss of DBHS proteins correlated positively with the cytotoxic potentials of PS-ASOs with different 2′ modifications (4). In mice, modest reduction in levels of PSF was observed at 24 and 48 h, and more substantial reduction in levels of PSF protein was observed at 72 and 96 h, whereas P54nrb was reduced at 96 h after dosing. We do not exclude the possibility that the reduction in those proteins is due to apoptotic cleavage. P54nrb has been reported to be the caspase substrate. However, PSF was reported to remain intact during apoptosis (36).
Figure 4.
2′ F PS-ASO causes a reduction of DBHS proteins in vivo. (A and B) Mice were treated with a single dose of 400 mg/kg 2′ F PS-ASO (ION-404130) or with saline and indicated parameters were quantified at 24, 48, 72 and 96 h after dosing. N = 3 per group. (A) Western analyses of isolated liver proteins. HSP90 serves as a loading control. PARPCL-cleaved PARP. Levels of p54nrb and PSF proteins were quantitate by ImageJ. N.D.-band not detected. (B) qRT-PCR of mRNA transcripts of P54nrb and PSF. The error bars indicate data acquired from three individual animals. S-saline, and F-2′ F PS-ASO (ION-404130). (C) HeLa cells were treated with control luciferase siRNA (Con), P54nrb siRNA, PSF siRNA or both P54nrb and PSF siRNAs for 96 h. Treated cells were then either mock-transfected or transfected with 200 nM 2′ F ASO (ION-404130) for another 12 h. Western analysis was performed. GAPDH serves as a loading control.
PS-ASOs localize to paraspeckles within minutes after being delivered into HeLa cells by electroporation (Supplementary Figure S6), suggesting that interactions between PS-ASOs and paraspeckle proteins, such as P54nrb and PSF, occur rapidly when ASOs are released into cells. Although reduction in levels of both P54nrb and PSF proteins were observed at later time points compared with the caspase activation, it is possible that interactions between ASOs and P54nrb/PSF occurred earlier. In this case, western analyses may not be sensitive enough to detect small changes or altered protein functions. To gain insight into early events, we evaluated the levels of mRNAs for P54nrb and PSF. Indeed, an upregulation in liver mRNA levels of P54nrb and PSF was observed as early as 24–48 h in animals receiving 400 mg/kg 2′ F PS-ASO ION-404130 (Figure 4B). Importantly, no significant increase in the levels of PSF mRNA were observed in mice given the 2′ MOE PS-ASO (ION-116847), according to microarray results (Supplementary Figure S7). Levels of mRNAs encoding several other paraspeckle proteins, including RBM14, EWSR1, TAF15, RUNX3, ZC3H8 and KLF4, were also increased in livers of mice treated with the 2′ F PS-ASO (ION-404130) but not with the 2′ MOE PS-ASO (ION-116847) as shown in our microarray analyses (Supplementary Figure S7). It is possible that the mRNA levels of both P54nrb and PSF were increased to compensate for the decreased availability of functional P54nrb and PSF proteins. These results suggest that, consistent with observations in vitro, 2′ F ASO caused a reduction in availability and levels of functional P54nrb and PSF at protein level. The reduction in availability of functional DBHS proteins was an early event and was less likely mediated by the increase hepatotoxicity.
Levels of DBHS proteins have been linked to cell survival (24,25,28,30,37). Depletion/loss of any single DBHS protein resulted in the compensatory upregulation in protein levels of other DBHS proteins in cultured cells or in mice (Figure 4C) (4,26,28,30). Upregulation in levels of PSF and PSPC1 proteins were observed in intellectual disability patients carrying a nonsense mutation in P54nrb (30). In HeLa cells, depletion of either P54nrb or PSF by siRNAs for 72 h resulted in the accumulation of cleaved PARP (Figure 4C). In addition, treatment of P54nrb or PSF-depleted cells with 2′ F PS-ASO (ION-404130) exacerbated 2′ F ASO-related cytotoxicity (Figure 4C). DBHS proteins may not account for all the toxic outcomes observed in cells and in mice since 2′ F PS-ASOs associate with many cellular proteins, and many of these ASO-binding proteins play important roles in maintaining cellular homeostasis. However, the loss of DBHS proteins is toxic to cells and can contribute to the 2′ F PS-ASO-related cyto- and hepatotoxicity.
Hybridization-dependent toxicity is not likely to account for the 2′ F PS-ASO-induced hepatotoxicity
The 2′ F-modified ASO-induced hepatotoxicity might result from hybridization-dependent toxicities due to on- or off-target RNA cleavages or from hybridization-independent toxicities such as increased protein binding. Both hypotheses were tested. In the case of the on-target antisense activity, which is mediated by the hybridization-dependent mechanism, the 2′ F PS-ASO (ION-404130) less effectively reduced levels of the targeted Pten transcript than did the 2′ MOE PS-ASO (ION-116847) or the cEt PS-ASO (ION-582801) in vivo and in cells (Figure 1E and F).
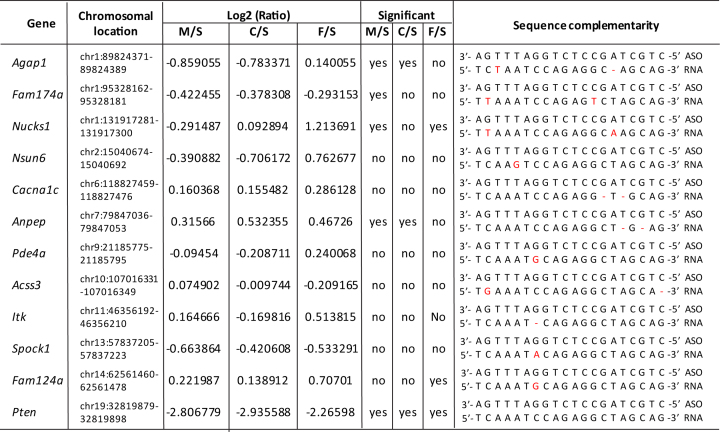
We also attempted to identify potential off-target cleavage sites by searching mouse genomic sequence for regions with partial complementarity to the sequence of the 20-mer ASOs tested here. Potential off-target hybridization sites were selected for those with no more than two mismatches or indels to the ASO, as previous studies have shown that antisense activity reduced dramatically with two or more mismatches (16). A total of 38 sites were identified that met these criteria using GGGenome against the mouse genome (GRCm38/mm10). Of these, 25 sites were mapped to intergenic regions, and 13 were mapped to regions that are transcribed, including the on-target Pten mRNA. Expression of 12 of 13 of these possible transcripts was detected by microarray analysis, and the reduction of the intended target, the Pten mRNA, was confirmed (Figure 5). Interestingly, based on analysis of the microarray data, none of the potential off-target transcripts were reduced by >50% by the 2′ MOE, cEt or 2′ F PS-ASOs targeting Pten. The most affected off-target transcript was Agap1 mRNA, which compared with saline-treated animals, was reduced to 55.13% (log2(ratio) = –0.8591) and 58.10% (log2(ratio) = –0.7834) by the 2′ MOE and by the cEt ASOs, respectively. Agap1 levels were not reduced significantly by the 2′ F PS-ASO (log2(ratio) = 0.1401). These results indicate that no substantial correlation was observed between the 2′ F PS-ASO-induced liver toxicity and hybridization-mediated target reduction.
Figure 5.
Adverse effects observed in vivo with 2′ F PS-ASO are not likely due to the hybridization-dependent off-target cleavage events. Potential off-target liver transcripts identified based on sequence homology. Mismatches and indels on these RNAs relative to the Pten ASO sequence are indicated in red. mRNA levels of these transcripts are based on microarray results (N = 3). Transcripts were significantly altered if P-value <0.05. M-2′ MOE PS-ASO (ION-116847), C-cEt PS-ASO (ION-582801), F-2′ F PS-ASO (ION-404130) and S-saline.
PS-ASOs containing 2′ F modifications associate with significantly more cellular proteins than do ASOs modified with 2′ MOE or cEt
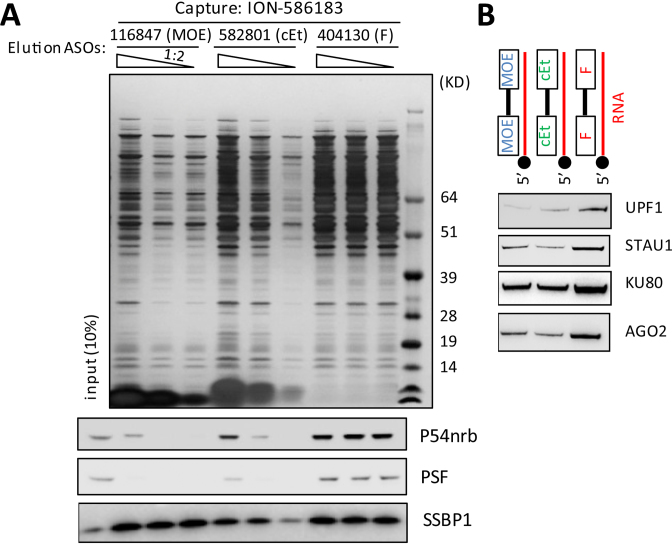
ASOs with PS-backbone modifications have increased binding affinity for proteins relative to ASOs with phosphodiester (PO) backbones (13). For example, using a recently developed bioluminescence resonance energy transfer (BRET)-based ASO–protein interaction assay (9), we found that a PS-DNA oligonucleotide associated with P54nrb 100-fold more tightly than its PO counterpart, and a PS-RNA oligonucleotide associated with P54nrb ∼60-fold more tightly than the PO-RNA (Supplementary Figure S8). To test interactions between global intracellular proteins and PS-ASOs with various 2′ modifications, we captured ASO-binding proteins with a biotinylated Pten-targeted cEt ASO (ION-586183) and eluted proteins by competition using PS-ASOs modified with 2′ MOE (ION-116847), cEt (ION-582801) or 2′ F (ION-404130) at three different concentrations. The eluted proteins were separated on a PAGE gel and visualized by sliver staining (Figure 6A). The 2′ F PS-ASO eluted significantly more proteins even at the lowest concentration than did cEt or 2′ MOE ASOs. Similar results were obtained with a different capture ASO modified with 2′ F (ION- 623496) (Supplementary Figure S9A).
Figure 6.
2′ F PS-ASO associates with more intracellular proteins than do 2′ MOE or cEt PS-ASOs. (A) ASO-binding proteins were pulled down from HeLa lysate with a 5′ biotinylated Pten ASO modified with cEt (ION-586183: 5′-biotinylated-582801) and were eluted by completion using 2′ MOE (ION-116847), cEt (ION-582801) or 2′ F (ION-404130). Sliver staining was performed to show the eluted proteins. Western blot was performed for P54nrb, PSF and SSBP1. (B) ASO-binding proteins were pulled down from HeLa lysate with ION-116847, ION-582801 and ION-404130 duplexed with a 5′ biotinylated complementary RNA. Western blot was performed for UPF1, STAU1, KU80 and AGO2.
2′ F PS-ASO caused rapid degradation of P54nrb and PSF in cells and in mice (4). Indeed, extent of association of PS-ASOs with P54nrb and PSF correlated positively with the observed cyto- and hepatotoxicity (Figure 6A). A similar preference of P54nrb to 2′-F ASO was also reported previously by a BRET binding assay (9). In addition, results from BRET binding assay also suggest a higher level of association of the 2′ F PS-ASO compared to the 2′ MOE and cEt PS-ASOs with other cellular ASO binding proteins such as La (Sjogren Syndrome Antigen B/La Autoantigen) and XRN2 (5′-3′ Exoribonuclease 2) (Supplementary Figure S9B). Although the 2′ F PS-ASO was associated with more cellular proteins in general than were the PS-ASOs with 2′ MOE and cEt modifications, interactions between ASOs and individual proteins varied. For example, comparable levels of association with SSBP1 (Single stranded DNA binding protein 1) were observed for the 2′ F and 2′ MOE PS-ASOs, but the cEt bound less SSBP1. In addition, when duplexed with its complementary RNA, 2′-F PS-ASO associates with more cellular proteins such as STAU1 (Staufen Double-Stranded RNA Binding Protein 1), UPF1 (Regulator of nonsense transcripts 1), and AGO2 (Argonaute 2, RISC Catalytic Component) than other ASO/RNA hybrids (Figure 6B). It is possible that 2′ F ASO/RNA duplex better mimics the conformation of an RNA-RNA duplex than do the duplexes formed between RNA and 2′ MOE or cEt, resulting in interactions with many dsRNA-binding proteins. Our results suggest that the 2′ F PS-ASO binds more cellular proteins and more avidly. The rank order of protein binding of these tested ASOs correlates positively with their cyto- and hepatotoxic potentials.
DISCUSSION
In this study, we report a 2′ F modification-dependent hepatotoxicity of PS-ASOs in vivo. This is a continuation of our previous study in vitro in which we observed that a 2′ F PS-ASO, but not 2′ MOE or cEt PS-ASOs with the same sequence and gapmer design, caused a rapid degradation of DBHS proteins in cultured cells and resulted in cell death (4). While it has been documented previously that the 2′ F PS-oligonucleotide-related cytotoxicity is cell-line and delivery method-dependent (38,39), our observations in vivo and previously reported work in vitro (4) in both human and mouse cells and by both transfection and free-uptake suggest that 2′ F-modified PS-oligonucleotides can cause adverse effects in different cell types regardless of delivery methods. Indeed, in mice, many 2′ F gapmer PS-ASOs with different sequences caused reduction of DBHS proteins, elevation in transaminases, and apoptotic liver death. 2′ F PS-ASO ION-804856, which did not reduce levels of DBHS proteins, did not caused elevation in levels of liver transaminases. The PS-ASOs modified with cEt (ION-582801) or LNA (ION-390896) was less hepatotoxic compared with the 2′ F PS-ASO (ION-404130), while the 2′ MOE PS-ASO (ION-116847) did not increase levels of ALT/AST at any given dose and time points. The hepatotoxic potentials of the 2′ F ASO correlate well with its increased binding to intracellular proteins, suggesting that undesirable ASO–protein interactions can contribute to the loss of DBHS proteins and hepatotoxicity.
Many previous studies suggest that phosphorothioate backbone modification contribute to ASO–protein interactions (4,8–12). A minimum of 9–10 continuous PS modifications allow the association of PS-ASOs with proteins (11,13). We and others also reported that 2′ modifications influenced the binding of ASOs to cellular proteins (4,8,9,11,33,40). We found that the association of total intracellular proteins with a 2′ MOE PS-ASO (ION-116847) was significantly less than with a cEt PS-ASO (ION-582801) or with a 2′ F PS-ASO (ION-404130), confirming that 2′ modifications can further impact ASO–protein interactions in addition to the PS backbone modification. This result agrees with our previous hypothesis that hydrophobicity of 2′ modification of oligonucleotides is one of the factors that contribute to ASO–protein interactions (10). For many but not all proteins, affinity increased with increasing hydrophobicity (2′ F > cEt > 2′ MOE) of wing modifications of a gapmer PS-ASO. At the individual protein level, we reported recently that the dissociation constants (Kds) for ION-116847 (MOE), ION-582801 (cEt), and ION-404130 (F) to P54nrb were 82.9, 9.3 and 2.1 nM, respectively (9). This ∼40-fold difference in binding affinity to P54nrb of the 2′ MOE PS-ASO versus the 2′ F PS-ASO is evidence of the significant impact of 2′ modification on ASO–protein interactions.
In this study, we found that the extents of ASO–protein interactions were correlated positively with the observed transaminase elevation, suggesting that excessive interactions of ION-404130 (F) with intracellular proteins contribute to hepatotoxicity. A positive correlation between increased intracellular protein binding and more severe hepatotoxicity was also previously reported for LNA ASOs (41). It has also been reported that a 2′ O-Methyl (2′ O-Me) modified PS-oligonucleotide associate with less proteins such as RPA (replication protein A) than the PS-DNA, resulting in less non-specific adverse effects in cells (40). This result also serves as another example that hydrophobicity of 2′ modification of PS-ASOs affect ASO–protein interactions (2′ H in DNA is more hydrophobic than 2′ O-Me). Since hydrophobicity is the primary force that drives the folding of proteins and the formation of protein complexes, high-affinity binding of PS-ASOs containing hydrophobic 2′ modifications such as 2′ F to hydrophobic residues likely alters the protein folding and/or protein–protein interaction, impairing their stability and/or functions. In addition, in our study, only 2′-F PS-ASO induced a significant reduction in levels of P54nrb and PSF proteins in vitro and in vivo, suggesting that 2′ modifications can affect the consequence(s) of ASO–protein interactions. In addition to the increased binding affinity, it is also possible that proteins such as P54nrb and PSF interact with 2′ F PS-ASO at different binding sites or with different capacities, which require further evaluation.
In addition to hydrophobicity, other factors, such as charge, sequence, and design can also contribute to the ASO–protein interactions. For example, despite being highly hydrophobic, neutral morpholino and peptide nucleic acid (PNA) oligomers do not significantly binds extracellular or intracellular proteins (Kds > 1 mM), resulting in rapid plasma clearance in vivo (42). In our recent study, we also found that Kds of cEt PS-ASOs for P54nrb vary from 1 nM to 3 μM depending on sequence (9), suggesting the sequence-dependent protein-binding. Importantly, many studies suggest that the hepatotoxic potentials of therapeutic oligonucleotides are predictable bioinformatically from their sequence and are determined by certain sequence motifs independent of hybridization (34,43), suggesting that sequence-dependent protein binding may contribute to the sequence-dependent hepatoxicity. Indeed, LNA gapmer ASOs containing toxic sequence motifs bind to more hepatocellular proteins than do LNA gapmer ASOs without toxic sequence motifs (34). Moreover, very different protein-binding profiles were observed for a single-stranded ASO versus an ASO–RNA duplex (10,11). Although cytotoxicity, in terms of P54nrb and PSF reduction, was observed with both single-stranded and duplexed oligonucleotides containing 2′ F modification (Supplementary Figure S10) (4), detailed quantitative comparison of toxic potentials in vitro and in vivo of 2′ F-containing duplexed versus single-stranded oligonucleotides requires further evaluation. All these results demonstrate the complexity of ASO–protein interactions. Undesirable interactions between ASOs and proteins can be a general mechanism of oligonucleotide toxicity.
In this study, we tested in vivo a total of eight pairs of 5–10–5 gapmer PS-ASOs comparing 2′ F versus 2′ MOE modifications. Seven out of the eight tested 2′ F PS-ASOs are highly hepatotoxic, causing substantial elevation in levels of liver transaminases or animal deaths after a single administration. In contrast, no significant elevation in levels of liver transaminases were observed for the 2′ MOE counterparts with the same sequences and gapmer design at the same dose and duration. However, we did find one sequence for which neither 2′ F nor 2′ MOE-modified PS-ASOs elevated levels of liver transaminases at 400 mg/kg dose for 96 h. These results suggest that (i) the tested 2′ F-modified PS-ASOs are more toxic than their 2′ MOE-modified counterparts, and (ii) not all 2′ F-modified PS-ASOs are toxic and (iii) ASO sequences contribute to their hepatotoxic potentials of 2′ F PS-ASOs in addition to ASO modifications. However, the sequence rules related to the toxic potentials of 5–10–5 gapmer 2′ F ASOs were not currently understood due to the limited numbers of sequences tested. With the limited sets of ASOs tested, we observed a correlation between 2′ F modification and increased hepatotoxicity. Follow-up studies to include large numbers of 5–10–5 gapmers modified with 2′ MOE versus 2′ F will allow us to further establish the correlation between 2′ F modifications and ASO toxicity. However, with the sequences tested here, we are confident that we are reporting a general trend rather than a specific case. It is clear that in addition to the 2′ modifications, many factors including charge, sequence, and configurations can also contribute to the ASO–protein interactions and consequently, cyto- and hepato- toxic potentials of ASOs. We and others observed that ASO–protein interactions and hepatotoxic potentials of PS-ASOs can be substantially altered by changing even a single sugar modification (5,34,43) (our unpublished data). It is possible that a single modification or a mismatch nucleotide can profoundly affect the structure and molecular electrostatic potentials of oligonucleotides, which may contribute to the ASO–protein interaction and toxicity. Importantly, the FDA-approved Pegaptanib sodium (Macugen), a 2′ F-containing RNA aptamer directed against vascular endothelial growth factor (VEGF) showed an excellent safety profile (44). Thus, it is entirely possible that the increased hepatotoxic potentials observed for 2′ F modifications in a 5–10–5 gapmer configuration may not be applicable to other therapeutic oligonucleotides with different configurations such as double-stranded siRNAs, aptamers, or splicing modulation ASOs.
The lack of off-target cleavages by the tested ASOs was not surprising, since for the sequence-specific hybridization-dependent off-target antisense events to occur, the homologous region of the unintended RNAs must be accessible to ASOs. Many factors, including RNA structures, protein binding, and RNA processing rates (e.g. splicing rate or degradation rate), and RNase H1 cleavage efficiency, influence ASO activity (45). Furthermore, for hybridization-dependent off-target cleavage to cause toxicity, the off-target transcripts must be crucial for cellular functions. All these requirements render the toxicity due to off-target cleavage improbable for the ASOs tested here. In this case, all three tested ASOs have the same sequence, and the 2′ MOE PS-ASO most effectively reduces levels of targeted Pten, therefore toxicity due to the hybridization-based on- or off-target cleavages are not likely to be the primary cause of toxicity by the 2′ F PS-ASO in vivo. In our experience, even at the doses used in this study, meaningful reduction in RNAs is difficult to be observed for sites with two or more mismatched, particularly if these mismatches are in the DNA gap. However, we do not completely rule out the contribution of the hybridization-based off-target RNA reduction to the observed hepatotoxicity since it is still possible that RNase H1-mediated cleavage events may occur at sites with more than two mismatches or indels to the ASO.
One additional possible mechanism of the hepatotoxicity of 2′ nucleoside modified oligonucleotides is through 2′ nucleoside metabolites that may be processed through the salvage pathway. This is an important consideration given the hepatotoxicity observed with FIAU (17,18). A gapmer PS-ASO containing 2′ F nucleosides on the wings is less nuclease-resistant than a 2′ MOE or cEt modified PS-ASO (our unpublished data) and the incorporation of 2′ F-modified nucleosides into nascent DNA/RNA has been observed (46–48). However, in this study, metabolites are unlikely to contribute to the observed toxicities. The metabolism of a 2′ F PS oligonucleotide to 2′ F nucleoside is slow, whereas hepatotoxic effects of a 2′ F PS-ASO were observed as early as 24 h after subcutaneous administration, and some mice died of hepatotoxicity between 72 and 96 h after a single dose. Moreover, many studies suggest that humans are more sensitive than mice to the FIAU-induced mitochondrial dysfunction and hepatotoxicity due to the presence of a mitochondria nucleoside transporter specific to humans (49). This protein is believed to be responsible for the transport of FIAU into the mitochondria in human cells (50). However, in longer term studies or in human trials in which 2′ F nucleoside could significantly accumulate in mitochondria, it is entirely possible that 2′ F nucleoside metabolites could contribute to toxicities.
Therapeutic ASOs must interact with proteins to have systemic distribution and ultimate uptake of the drugs into desired tissues and cells. However, high affinity binding of ASOs to either plasma or intracellular proteins clearly contributes to toxic effects in vitro and in vivo. 2′ modifications play an important role in determining these interactions and the degrees of hydrophobicity of these 2′ modifications seem to correlate positively with both protein binding and toxic responses. As an example, we found here that a 2′ F gapmer PS-ASO associated with more cellular proteins at a global level than did 2′ MOE or cEt ASOs, which may impair the localization and functions of intracellular proteins such as DBHS proteins (and perhaps other proteins as well) and lead to the hepatotoxicity. Our results emphasize that caution should be taken when incorporating 2′ modifications into therapeutic oligonucleotides and detailed structure activity relationship studies need to be performed to better evaluate characteristics of modified oligonucleotides to yield safe, potent and efficacious drugs.
Supplementary Material
ACKNOWLEDGEMENTS
For their excellent technical assistance, we would like to thank Beatrice DeBrosse-Serra and Gene Hung for histopathology evaluations, Mark Andrade for ASO synthesis, Donna Sipe for handling of animal facilities. In addition, we are grateful to Jeffery Engelhardt, Hans Gaus, Eric Swayze, Sebastien Burel, Thazha P. Prakash, Shiyu Wang, and Jeffrey Bailey for valuable discussions and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Ionis Pharmaceuticals. Funding for open access charge: Ionis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1. Crooke S.T.V., T. A., Lima W.F., Wu H.-J.. Crooke ST. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 3–46. [Google Scholar]
- 2. Swayze E.E., Bhat B.. Crooke ST. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 143–182. [Google Scholar]
- 3. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017; 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shen W., Liang X.H., Sun H., Crooke S.T.. 2′-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 2015; 43:4569–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., Kreeger J., DiMattia D., Chen T. et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012; 22:344–359. [DOI] [PubMed] [Google Scholar]
- 6. Burel S.A., Hart C.E., Cauntay P., Hsiao J., Machemer T., Katz M., Watt A., Bui H.H., Younis H., Sabripour M. et al. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2016; 44:2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kasuya T., Hori S., Watanabe A., Nakajima M., Gahara Y., Rokushima M., Yanagimoto T., Kugimiya A.. Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci. Rep. 2016; 6:30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liang X.H., Shen W., Sun H., Kinberger G.A., Prakash T.P., Nichols J.G., Crooke S.T.. Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2′-modifications and enhances antisense activity. Nucleic Acids Res. 2016; 44:3892–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vickers T.A., Crooke S.T.. Development of a quantitative BRET affinity assay for nucleic acid-protein interactions. PLoS One. 2016; 11:e0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang X.H., Sun H., Shen W., Crooke S.T.. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015; 43:2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen W., Liang X.H., Crooke S.T.. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014; 42:8648–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T.. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014; 42:7819–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown D.A., Kang S.H., Gryaznov S.M., DeDionisio L., Heidenreich O., Sullivan S., Xu X., Nerenberg M.I.. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994; 269:26801–26805. [PubMed] [Google Scholar]
- 14. Watanabe T.A., Geary R.S., Levin A.A.. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302). Oligonucleotides. 2006; 16:169–180. [DOI] [PubMed] [Google Scholar]
- 15. Weidner D.A., Valdez B.C., Henning D., Greenberg S., Busch H.. Phosphorothioate oligonucleotides bind in a non sequence-specific manner to the nucleolar protein C23/nucleolin. FEBS Lett. 1995; 366:146–150. [DOI] [PubMed] [Google Scholar]
- 16. Henry S.P., Kim T.-W., Kramer-Stickkand K., Zanardi T.A., Fey R.A., Levin A.A.. Crooke ST. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 305–326. [Google Scholar]
- 17. Cui L., Yoon S., Schinazi R.F., Sommadossi J.P.. Cellular and molecular events leading to mitochondrial toxicity of 1-(2-deoxy-2-fluoro-1-beta-D-arabinofuranosyl)-5-iodouracil in human liver cells. J. Clin. Invest. 1995; 95:555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McKenzie R., Fried M.W., Sallie R., Conjeevaram H., Di Bisceglie A.M., Park Y., Savarese B., Kleiner D., Tsokos M., Luciano C. et al. Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N. Engl. J. Med. 1995; 333:1099–1105. [DOI] [PubMed] [Google Scholar]
- 19. Jordheim L.P., Durantel D., Zoulim F., Dumontet C.. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013; 12:447–464. [DOI] [PubMed] [Google Scholar]
- 20. Crooke S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017; 27:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. [DOI] [PubMed] [Google Scholar]
- 22. Fox A.H., Lam Y.W., Leung A.K., Lyon C.E., Andersen J., Mann M., Lamond A.I.. Paraspeckles: a novel nuclear domain. Curr. Biol.: CB. 2002; 12:13–25. [DOI] [PubMed] [Google Scholar]
- 23. Fox A.H., Lamond A.I.. Paraspeckles. Cold Spring Harbor Perspect. Biol. 2010; 2:a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shav-Tal Y., Zipori D.. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002; 531:109–114. [DOI] [PubMed] [Google Scholar]
- 25. Knott G.J., Bond C.S., Fox A.H.. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016; 44:3989–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S., Li Z., Shu F.J., Xiong H., Phillips A.C., Dynan W.S.. Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res. 2014; 42:9771–9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cobbold L.C., Spriggs K.A., Haines S.J., Dobbyn H.C., Hayes C., de Moor C.H., Lilley K.S., Bushell M., Willis A.E.. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2008; 28:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen W., Liang X.H., Sun H., De Hoyos C.L., Crooke S.T.. Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS One. 2017; 12:e0173494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li S., Kuhne W.W., Kulharya A., Hudson F.Z., Ha K., Cao Z., Dynan W.S.. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009; 37:6746–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mircsof D., Langouet M., Rio M., Moutton S., Siquier-Pernet K., Bole-Feysot C., Cagnard N., Nitschke P., Gaspar L., Znidaric M. et al. Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat. Neurosci. 2015; 18:1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lima W.F., Murray H.M., Damle S.S., Hart C.E., Hung G., De Hoyos C.L., Liang X.H., Crooke S.T.. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016; 44:5299–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen W., Sun H., De Hoyos C.L., Bailey J.K., Liang X.H., Crooke S.T.. Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Res. 2017; 45:10672–10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey J.K., Shen W., Liang X.H., Crooke S.T.. Nucleic acid binding proteins affect the subcellular distribution of phosphorothioate antisense oligonucleotides. Nucleic Acids Res. 2017; 45:10649–10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burdick A.D., Sciabola S., Mantena S.R., Hollingshead B.D., Stanton R., Warneke J.A., Zeng M., Martsen E., Medvedev A., Makarov S.S. et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid–modified antisense oligonucleotides. Nucleic Acids Res. 2014; 42:4882–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F.. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007; 35:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shav-Tal Y., Lee B., Bar-Haim S., Vandekerckhove J., Zipori D.. Enhanced proteolysis of pre-mRNA splicing factors in myeloid cells. Exp. Hematol. 2000; 28:1029–1038. [DOI] [PubMed] [Google Scholar]
- 37. Yarosh C.A., Iacona J.R., Lutz C.S., Lynch K.W.. PSF: nuclear busy-body or nuclear facilitator. Wiley Interdiscipl. Rev. RNA. 2015; 6:351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Janas M.M., Jiang Y., Schlegel M.K., Waldron S., Kuchimanchi S., Barros S.A.. Impact of oligonucleotide structure, chemistry, and delivery method on in vitro cytotoxicity. Nucleic Acid Ther. 2017; 27:11–22. [DOI] [PubMed] [Google Scholar]
- 39. Garber K. Worth the RISC. Nat. Biotechnol. 2017; 35:198–202. [DOI] [PubMed] [Google Scholar]
- 40. Yoo B.H., Bochkareva E., Bochkarev A., Mou T.C., Gray D.M.. 2′-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004; 32:2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kakiuchi-Kiyota S., Whiteley L.O., Ryan A.M., Mathialagan N.. Development of a method for profiling protein interactions with LNA-modified antisense oligonucleotides using protein microarrays. Nucleic Acid Ther. 2016; 26:93–101. [DOI] [PubMed] [Google Scholar]
- 42. Wan W.B., Seth P.P.. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016; 59:9645–9667. [DOI] [PubMed] [Google Scholar]
- 43. Hagedorn P.H., Yakimov V., Ottosen S., Kammler S., Nielsen N.F., Hog A.M., Hedtjarn M., Meldgaard M., Moller M.R., Orum H. et al. Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther. 2013; 23:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ng E.W., Shima D.T., Calias P., Cunningham E.T. Jr., Guyer D.R., Adamis A.P.. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006; 5:123–132. [DOI] [PubMed] [Google Scholar]
- 45. Lima W.F., Vickers T.A., Nichols J., Li C., Crooke S.T.. Defining the factors that contribute to on-target specificity of antisense oligonucleotides. PLoS One. 2014; 9:e101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richardson F.C., Zhang C., Lehrman S.R., Koc H., Swenberg J.A., Richardson K.A., Bendele R.A.. Quantification of 2′-fluoro-2′-deoxyuridine and 2′-fluoro-2′-deoxycytidine in DNA and RNA isolated from rats and woodchucks using LC/MS/MS. Chem. Res. Toxicol. 2002; 15:922–926. [DOI] [PubMed] [Google Scholar]
- 47. Aurup H., Williams D.M., Eckstein F.. 2′-Fluoro- and 2′-amino-2′-deoxynucleoside 5′-triphosphates as substrates for T7 RNA polymerase. Biochemistry. 1992; 31:9636–9641. [DOI] [PubMed] [Google Scholar]
- 48. Aoyama H., Sarih-Cottin L., Tarrago-Litvak L., Litvak S., Guschlbauer W.. 2′-Fluoro-2′-deoxycytidine triphosphate as a substrate for RNA- and DNA-dependent DNA polymerases. Biochim. Biophys. Acta. 1985; 824:218–224. [DOI] [PubMed] [Google Scholar]
- 49. Xu D., Nishimura T., Nishimura S., Zhang H., Zheng M., Guo Y.Y., Masek M., Michie S.A., Glenn J., Peltz G.. Fialuridine induces acute liver failure in chimeric TK-NOG mice: a model for detecting hepatic drug toxicity prior to human testing. PLoS Med. 2014; 11:e1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee E.W., Lai Y., Zhang H., Unadkat J.D.. Identification of the mitochondrial targeting signal of the human equilibrative nucleoside transporter 1 (hENT1): implications for interspecies differences in mitochondrial toxicity of fialuridine. J. Biol. Chem. 2006; 281:16700–16706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.