Abstract
The piggyBac transposase (PB) is distinguished by its activity and utility in genome engineering, especially in humans where it has highly promising therapeutic potential. Little is known, however, about the structure–function relationships of the different domains of PB. Here, we demonstrate in vitro and in vivo that its C-terminal Cysteine-Rich Domain (CRD) is essential for DNA breakage, joining and transposition and that it binds to specific DNA sequences in the left and right transposon ends, and to an additional unexpectedly internal site at the left end. Using NMR, we show that the CRD adopts the specific fold of the cross-brace zinc finger protein family. We determine the interaction interfaces between the CRD and its target, the 5′-TGCGT-3′/3′-ACGCA-5′ motifs found in the left, left internal and right transposon ends, and use NMR results to propose docking models for the complex, which are consistent with our site-directed mutagenesis data. Our results provide support for a model of the PB/DNA interactions in the context of the transpososome, which will be useful for the rational design of PB mutants with increased activity.
INTRODUCTION
Transposable elements (TEs) are DNA segments that use TE-encoded proteins to move or copy themselves from donor to target sites within their host genomes. TE insertion into a functional gene may result in gene inactivation, and incorrect rejoining of the newly exposed ends of the flanking donor site following TE excision can result in chromosomal aberrations. TEs thus have profound effect on host gene expression and are intimately involved in genome evolution (1).
TEs can be grouped into two major classes: class I (retrotransposons) and class II (DNA transposons). The class II piggyBac transposon was originally isolated from a cell line of the cabbage looper moth Trichoplusia ni (T.ni) (2). It encodes the piggyBac transposase (PB), which catalyses cut-and-paste transposition. PB excises the transposon from its donor site without leaving a DNA footprint (3), using a mechanism that involves the formation of DNA transposon-end hairpins (4), and inserts the transposon into its specific TTAA target site. TEs closely related to the T. ni piggyBac transposon are called piggyBac-like elements (PLEs) and have been found in numerous organisms, such as fungi, plants, insects, fishes and mammals (5–21). Some PLEs were shown to be active (7,9,12,21) and others are likely active (5,8,10,11,16).
Because of their high transposition efficiency and large cargo capacity (>100 kb) (22), piggyBac-based systems (23,24) are very useful tools for efficient integration of transgenes into the genomes of a wide range of invertebrate and vertebrate species (25–33). Engineered transgenes integrated into host genomes via piggyBac vectors may be potential therapeutic agents.
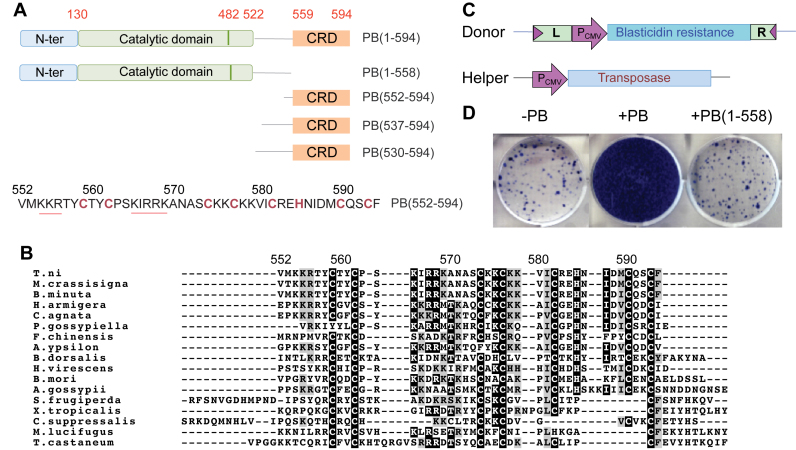
PB contains 594 amino acids organized into several distinct domains (Figure 1A) (23,34). Its conserved RNase H-like catalytic core, PB(130–482), like that of many transposases and retroviral integrases (35,36), contains a conserved acidic amino acid triad DD(D/E) (D268, D346, D447) that is required for all steps of transposition (4,37). The C-terminus of PB contains a highly conserved (5,8,10,11,20,37) Cysteine-Rich Domain (CRD) extending from PB(559) to the very C-terminus of PB (Figure 1A), which has been proposed to form a Really Interesting New Gene (RING)-finger motif (37) or a Plant Homeo Domain (PHD) finger (4). It overlaps with a nuclear localization signal (NLS), which was mapped within PB(551–571) (38) (underlined in red in Figure 1A).
Figure 1.
Role of the C-terminal domain of PB. (A) Schematic representation of the piggyBac transposase PB(1–594) and various constructs used in this study. The catalytic domain is in green, the C-terminal Cysteine-Rich Domain (CRD) in orange and the N-terminal domain in blue. The sequence of the CRD corresponding to PB(552–594) is represented below. The cysteine and histidine residues implicated in Zn2+ binding are in red and the two 554-KKR-556 and 565-KIRRK-569 stretches of residues belonging to the bipartite NLS are underlined in red. (B) Multiple sequence alignments of the piggyBac transposase T.ni and 16 other piggyBac transposase-like sequences from various species: S. frugiderpa (Spodoptera frugiperda), T. castaneum (Tribolium castaneum), X. tropicalis (Xenopus tropicalis), M. lucifugus (Myotis lucifugus), C. suppressalis (Chilo suppressalis), F. chinensis (Fenneropenaeus chinensis), A. gossypii (aphis gossypii), H. virescens (heliothis virescens), B. dorsalis (Bactrocera dorsalis), B. mori (Bombyx mori), P. gossypiella (Pectinophora gossypiella), B. minuta (Bactrocera minuta), M. crassisigna (Macdunnoughia crassisigna), A. ypsilon (Agrotis ypsilon), H. armigera (Helicoverpa armigera), C. agnata (Ctenoplusia agnata). Identical amino acids are shown in black boxes and similar amino acids are in grey boxes. (C) Schematic representation of plasmids used in the mammalian cell integration assay. Pcmv is the cytomegalovirus promoter. ‘L’ corresponds to the 328-bp piggyBac Left-End (LE), and ‘R’ to the 361-bp Right-End (RE). (D) Integration assays of a transposon expressing Blasticidin resistance, in absence of PB (left), in presence of PB (middle) and in presence of PB deleted of its C-terminal domain, PB(1–558) (right). The frequency of integration is indicated by blue colonies.
In this study, we show that the PB CRD is required in vivo for transposition. We demonstrate that it is required in vitro for DNA breakage and joining and that it binds in vitro to specific 19-bp DNA regions (LE35 and RE63) (Figure 2A) located within the transposon ends that are required for transposition. DNase I footprinting studies allowed us to identify conserved palindromic sequence motifs within these regions and an additional internal protected region at the left end. Using nuclear magnetic resonance (NMR) spectroscopy, we determined the 3D structure of the PB CRD, revealing that this domain adopts a compact fold and binds two Zn2+ ions with a C3H (ZF1) and C4 (ZF2) coordination mode in a cross-brace zinc finger (ZF) motif. NMR interaction studies of PB(559–594) with short DNA oligonucleotides and NMR-driven molecular-docking simulations allow us to propose specific structural models of PB(559–594)/DNA interactions. We performed PB(559–594) site-directed mutation experimental studies to validate the proposed PB(559–594)/DNA interaction surface.
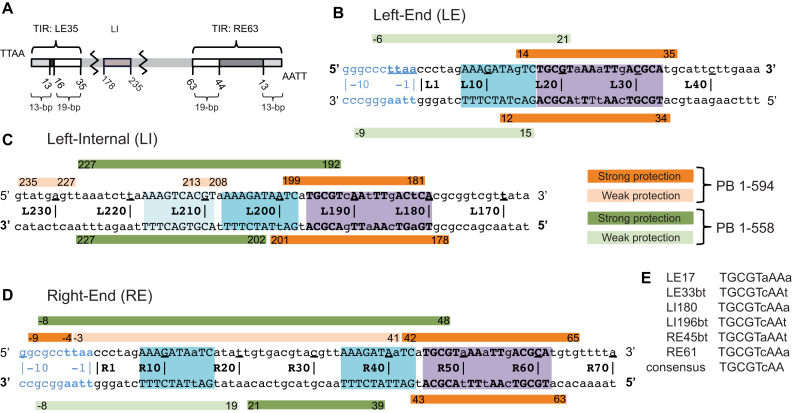
Figure 2.
Summary of DNase I footprinting results. (A) Schematic representation of the piggyBac transposon. The piggyBac left (LE1–35) and right (RE1–63) ends consist of a 13-bp terminal inverted repeat (light gray) and a 19-bp internal inverted repeat (white) separated by a 3-bp spacer and a 31-bp spacer respectively. The left internal domain (LI178–235) is highlighted in light brown. (B) Left-End (LE), (C) Left-Internal (LI) and (D) Right-End (RE) protection from DNase I cleavage in presence of full-length protein PB(1–594) (in orange) or truncated PB(1–558) which lacks the CRD (in green). Strong and weak protections are indicated by dark and light bars, respectively. Purple boxes highlight the 19-bp internal inverted repeat targeted by the PB CRD, and the blue boxes the DNA sequence that interact with the truncated PB(1–558). The DNA sequences are numbered from the TTAA target sequence that flanks each transposon end. (E) Comparison of the conserved palindromic DNA sequences belonging to the 19-bp repeat of the LE, LI and RE fragments. In (B–E), the conserved nucleotides between the three LE, LI and RE DNA segments are in uppercase and the others in lowercase. The 5′ to 3′ top strand direction is noted with 5′ and 3′ in bold in (B–D).
MATERIALS AND METHODS
Expression and purification of PB and its derivatives
Full length PB(1–594) and PB(1–558) were produced in E. coli and purified as described in (4). MBP-PB(530–594) was expressed in E. coli Top10 cells. Cultures at 19°C were induced with 0.2% arabinose once the OD600 reached 0.8, and grown for 5 h prior to harvesting. Cells were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris pH 7.5, 1 M NaCl, 0.5 mM TCEP, one cOmplete™ EDTA-free protease inhibitor tablet) supplemented with 0.2 mM AEBSF and lysed by sonication. The clarified lysate was passed over amylose resin (NEB) and washing with binding buffer containing 0.5 M NaCl and 80 mM Tris pH 8.0. Bound proteins were eluted with binding buffer containing 20 mM maltose, peak fractions were combined, concentrated, and dialyzed overnight against TSK buffer (20 mM HEPES pH 7.5, 0.5 M NaCl, 0.5 mM TCEP, 10% (w/v) glycerol). Gel filtration was carried out as described above, followed by cleavage of the fusion protein by PreScission protease at 4°C overnight. The remaining fusion protein and protease were separated from PB(530–594) by heparin affinity chromatography: proteins were bound in 0.25 M NaCl, 20 mM HEPES pH 7.5, 10% glycerol and PB(530–594) eluted in the same buffer containing 1 M NaCl. Final peaks fractions were concentrated and dialyzed into DNA binding buffer (20 mM HEPES pH 7.5, 0.25 M NaCl, 10% (w/v) glycerol).
A DNA fragment encoding the CRD region (residues 537–594) of the T. ni PB was PCR amplified and cloned between EcoRI and XhoI restriction sites of plasmid pGEX6p1. Protein expression was performed in GOLD(DE3) E. coli cells in LB medium supplemented with 0.1 mM ZnSO4. Exponentially growing cells were induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 16°C. Cells were suspended in buffer A (20 mM Tris–HCl, pH8.0), 0.15 M NaCl, 10% glycerol, 0.5 mM EDTA, 1 mM DTT) supplemented with protease inhibitor (cOmplete™, Roche). Cells were lysed with French press and the clear supernatant was filtered through a 0.45 μm syringe filter, then loaded onto GST-Trap FF (1 ml column) with a syringe. The column was washed with 15 ml of buffer A. GST-tagged PB(537–594) was eluted with the same buffer containing also 10 mM reduced glutathione and tested by SDS-PAGE and Coomassie Blue staining, which showed that the GST-CRD fusion is >95% pure. Protein concentration was established by 280 nm absorbance measurement using the extinction coefficient of GST. GST expression and purification was performed using the same protocol but with the pGEX6p1 plasmid.
Peptide synthesis and DNA substrates
Due to the purification difficulties encountered during the production of the CRD 15N and 13C-enriched peptide in E. coli, non-isotopically labelled chemically synthetized PB(552–594) peptide was purchased from Proteogenix (Oberhausbergen, France). For NMR studies, oligonucleotides corresponding to both strands of LE14–25, LE22–36 and LE14–36 were purchased from Eurofins Genomics. For EMSA experiments, oligonucleotides corresponding to both strands of LE1–35, its randomized version (5′-ACAATAGTATAGAGCGCCACAACTTGGTATGGGCA-3′) and the 3 MUT mutant (5′-CCCTAGAAAGATAGTCTGGCAAAAATTGTGCCATG-3′) were ordered either from IDT (Coralville, IO) or Eurofins Genomics. For EMSA experiments complementary oligonucleotides were annealed in 10 mM Tris pH 8.0 by heating to 95°C for 10 min followed by slow cooling to room temperature.
Integration and excision assays
The piggyBac excision assay and the in vitro cleavage assay have been previously described (39). Briefly, for the latter, either purified PB (0.59 μM) or PB(1–558) (1×: 0.78 μM or 2×: 1.57 μM) was incubated with 5 nM 32P-radiolabeled linearized pXL-PB-D-GFP/Bsd in 25 mM MOPS, pH 7.6, 4% (v/v) glycerol, 150 mM NaCl, 10 mM MgCl2, 2 mM DTT, 0.01% BSA in a final volume of 10 μl at 30°C for the indicated times. Reactions were stopped by adding EDTA to 40 mM, then 10% SDS was added to the reaction mixture to a final concentration of 1.2% and incubated at 65°C for 20 min prior to the addition of 4 μl of 6× loading dye. Samples were run on 1% agarose gels in 1× TBE. Gels were dried and exposed to a PhosphorImage screen, and analyzed by Imagequant software.
In vitro strand transfer assay
Either purified PB (0.59 μM) or PB(1–558) at 1.25 μM (‘low’) or 6.3 μM (‘high’) was incubated with 0.4 pmol of 32P-radiolabeled double stranded oligonucleotides in 25 mM MOPS, pH 7.6, 4% (v/v) glycerol, 150 mM NaCl, 10 mM MgCl2, 2 mM DTT, 0.01% BSA and 300 ng pUC19 plasmid as target DNA in a final volume of 10 μl at 30°C for 120 min. Reactions were stopped by adding EDTA to 40 mM, then 1.4 μl of 10% SDS was added to the reaction mixture and incubated at 45°C for 20 min prior to the addition of 4 μl of 6× loading dye. The samples were run on 1% agarose gels in 0.5× TBE. Gels were dried and exposed to a PhosphorImage screen.
Electrophoretic mobility shift assay (EMSA)
Annealed Cy3-labeled oligonucleotides were mixed with purified proteins in binding buffer consisting of 20 mM HEPES pH 7.5, 125 mM NaCl and 10% (w/v) glycerol and incubated at room temperature for 30 min in a total volume of 10 μl (see figure legends for details). Unlabeled competitor DNA, when present, was included at a final concentration of 10 μM. Samples were loaded onto a 10% acrylamide 1× TBE gel (Invitrogen), and run at 100 V at 4°C for 2 h. Gels were visualized using a GE Typhoon Trio variable mode imager.
DNase I footprinting
Purified PB at 40, 20, 10 and 5 ng/μl was combined with 50 fmol/μl PCR-amplified piggyBac ends (end primers were labelled with 5′-FAM for one and 5′-VIC for the other) in 20 μl final volume buffer containing 25 mM MOPS pH 7.6, 0.2 μg/μl herring sperm DNA (Invitrogen), 3% glycerol, 2 mM DTT, 0.1 μg/μl BSA. The reactions were incubated for 30 min at 25°C, then 1 μl of diluted DNase I was added (1 μl in 250 μl 200 mM MgCl2), and incubated for one additional minute. EDTA was added to a final concentration of 100 mM, the samples were cleaned using a QIAquick nucleotide removal kit, and eluted in 20 μl elution buffer. Control reactions were performed using bovine serum albumin (BSA). The resulting fragment sizes were obtained by automated capillary electrophoresis by the Promoter Characterization Service of the Plant-Microbe Genomics Facility at Ohio State University as described by Zianni et al. (40).
Sequence analysis
Sequence alignment was carried out using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and the alignment was shaded with BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Determination of the zinc equivalent using atomic emission spectroscopy
We used flame atomic absorption spectrophotometry to determine the concentrations of Zn2+ in two GST-tagged PB(537–594) samples (6.8 μM). A ZnCl2 solution was used as calibration standard and purified GST alone as a negative control. GST-tagged PB(537–594) and GST samples were dialysed with a ZnCl2-free buffer. The concentration of GST-tagged PB(537–594) was determined by absorbance at 280 nm using a theoretical extinction coefficient (45000 L⋅mol−1⋅cm−1).
Sample preparation and NMR experiments
The peptide PB(552–594) was first dissolved at pH 3.5 at a final concentration of 500 μM in the presence of 5 mM DTT (to avoid oxidation of the cysteine residues) and in the presence of 0, 50, 100 or 200 mM NaCl. Three equivalents of ZnCl2 with respect to peptide concentration were added and the pH was then adjusted to 6.5.
Complementary oligonucleotides corresponding to the three DNAs (LE14–25, LE22–36 and LE14–36) were annealed and prepared in the same conditions as those used for PB(552–594). The LE14–25/PB(552–594), LE22–36/PB(552–594) and LE14–36/PB(552–594) complexes were prepared by titrating 250 μM of each DNA with 0.25–2 equivalents of a concentrated solution (1 mM) of PB(552–594).
Two-dimensional phase-sensitive 1H Clean-TOCSY (41) with 70 ms spin lock, and NOESY experiments with 100 and 200 ms mixing times (42) were recorded at 20°C, 30°C and 40°C on an AVANCE Bruker 800.13 and 950.13 MHz spectrometers, with a spectral width of 9615 and 12 335 Hz respectively, without sample spinning, with 2k real points in t2 and 512 t1-increments. Pulsed-field gradient-based WATERGATE (43) was used for water suppression. The data were processed using TopSpin 3.5 software (Bruker). A π/6 phase-shifted sine bell window function was applied prior to Fourier transformation in both dimensions (t1 and t2).
The diffusion-ordered NMR spectroscopy (DOSY) spectra (44,45) were acquired on 100% D2O and 95% H2O/ 5% D2O solutions of PB(552–594) and only on 100% D2O solution of aprotinin (the concentrations of PB(552–594) and aprotinin were 588 and 462 μM respectively). The aprotinin molecular weight is only 20% superior of this of PB(552–594) (MW = 6500 Da for aprotinin and MW = 5235 Da for PB(552–594) plus two Zn2+). The two proteins were prepared in the same conditions as those used for PB(552–594), i.e. 200 mM NaCl, 5 mM DTT, 3 equivalents of ZnCl2 with respect to the protein concentrations and the pH was adjusted to 6.5. The DOSY spectra were recorded at 20°C, using a 5-mm triple resonance z-gradient TCI probe head which delivers a maximum gradient strength of 53.5 G/cm. The strength of the gradient pulses, of 1 ms duration, was incremented from 2–98% in 20 experiments, with a diffusion time of 200 ms. A π/2 phase-shifted squared sine bell window function was applied before the Fourier transformation (FT) and a baseline correction was then conducted after the FT.
The diffusion coefficients were calculated using the Bruker Topspin 3.5 DOSY software and the T1/T2 and Dynamics Center packages. The apparent molecular mass (M) of PB(552–594) and aprotinin were calculated from the translational diffusion coefficient measured on the DOSY spectrum using the Stokes–Einstein equation: D = kT/(6ηπrH); with the Boltzmann constant k, the viscosity η of the solvent, the temperature T and the protein hydrodynamic radius rH. In the case of PB(552–594) we also used the following equation that is adapted for non spherical particles: M = (kT/6πηFD)3[4πNA/[3(ν2 + δ1ν1)]]; where k is the Boltzmann constant, T the temperature, η the viscosity of the solution, F the shape factor, D the diffusion coefficient, NA the Avogadro's number, ν2 the partial specific volume of the molecule, ν1 the partial specific volume of the solvent, and δ1 the fractional amount of water bound to the protein (Supplementary Table S1 and Supplementary Figures S5–S8). The viscosity of the solutions η at T = 20°C and in 100% D2O, 200 mM NaCl, 10 mM DTT (condition 1) and in 95% H2O/5% D2O, 200 mM NaCl, 10 mM DTT (condition 2) were set to 1.272 × 10−3 Pa⋅s and 1.024 × 10−3 Pa⋅s respectively to account for distinct contributions of D2O and H2O to viscosity. We used as partial specific volumes, ν2 = 0.71063 × 10−3 m3/kg with two zinc atoms for PB(552–594), ν1 = 0.9054034 × 10−3 m3/kg for the D2O solvent and δ1 = 0.379 as calculated value for the fractional amount of water bound to the peptide. A value of 1.147944 was calculated for the shape factor F (46) using the equation: F = (1 – p2)1/2/(p2/3 ln{[1 + (1 – p2)1/2]/p}) since PB(552–594) is approximately a prolate spheroid (with P = b/a, b is the equatorial radius of the molecule and a the semi-axis revolution measured on the PB(552–594) structures).
NMR structure of PB(552-594)
NOE cross-peak volumes measured on a NOESY spectrum (with a 100 ms mixing time and at 20°C) using CcpNmr 2.1.3 (47) were converted into distances to generate PB(552–594) structures with ARIA version 2.3 (48). For the ensemble calculation, two copies of the protein were calculated per run, and all the restraints involving atoms belonging to residues K576, I587 or F594 were treated as ensemble restraints. For the ensemble restraints, the NOE distances were computed as r−6 averages over the two copies, which corresponds to the treatment of ambiguous restraints in ARIA. All other restraints were applied on each protein copy separately, as in standard ARIA calculations. No force-field terms (vdW energy) were evaluated between atoms of the two protein copies of one ensemble.
A log-harmonic potential was used for the second Cartesian cooling phase of the simulated annealing. We increased the number of torsion angle dynamics steps in the high-temperature phase to 20 000 K, in the torsion angle cooling step to 10 000 K and in the first and second Cartesian dynamics cooling phase to 50 000 K and 40 000 K, respectively. The starting temperature of the torsion angle dynamics was set to 20 000. Otherwise, standard parameters of ARIA version 2.3 were employed (49). Two restraint classes, whose weights were iteratively updated independently of each other, were used for the ensemble and the non-ensemble restraints. All runs started from elongated protein structures, and 2000 ensembles were calculated per iteration. Equivalent protons (methyl groups etc.) were treated as ambiguous restraints instead of using floating assignments. The geometry of the Zn2+-coordination was imposed with fixed distances and angles between the Zn2+ ion and the coordinating atoms (cysteine Sγ or histidine Nδ1).
As a fully assigned cross-peak list was provided as input data already in the first iteration, we used an iteration scheme that differs from the standard ARIA treatment. In iteration 0, the elongated template structure was used for deriving the NOE restraint target distances from the NOE cross-peak intensities. The best 20 single structures from this first iteration were used to calibrate the target distances for the next iteration (iteration 1). The best 1000 ensembles of iteration 1 were then used to identify highly violated restraints, which were not used in the final iteration (iteration 2). For this violation analysis, we used a violation tolerance of 0.5 Å, as no spin diffusion correction was employed. Thirteen restraints, for which the distance between the restrained atoms was larger than the restraint upper-bound in >85% of the structures, were deleted. Just as in the structure calculation itself, the violations of single-copy restraints were analysed for each copy separately, whereas the ensemble restraint distances were taken as ensemble-averages over the two copies of one ensemble.
From the 50 ensembles with lowest total ARIA energy, 23 two-copy ensembles were chosen for water refinement according to the following criteria that had to be met by each copy separately: a percentage of residues in the Ramachandran plot core region ≥70% (according to PROCHECK), presence of the characteristic features of the two structural families, that is with the side chain of N586 turned towards Q591 in PB(552–594)KF (family in which the F594 and K576 NOEs are satisfied) and away from Q591 in PB(552–594)IF (family in which the F594 and I587 NOEs are satisfied), and a backbone RMSD < 0.6Å of residues C559-E584 to the lowest energy structure fulfilling those criteria. For hydrogen bonds that were systematically found in all generated structures, hydrogen bond restraints were added in the subsequent water refinement run (Supplementary Table S2). Two independent water refinements were performed for each ensemble and the solution with lower percentage of residues in the Ramachandran plot core region or with higher backbone RMSD was discarded. Here and in the following analysis, alignment was performed separately for both structure families i.e. either PB(552–594)KF or PB(552–594)IF, using residues T557 to Q591, and backbone RMSD values to the respective average structure were calculated for residues C559 to E584. Out of the 23 water-refined ensembles, 12 two-copy ensembles were chosen as final ensembles, based on the following selection criteria: both copies had a backbone RMSD to the average structure ≤0.5 Å, a percentage of residues in the Ramachandran plot core region ≥70% and at least one of the two copies had a percentage of residues in the Ramachandran plot core region ≥80% (Supplementary Table S3).
Quality of structures was evaluated on the Biological Magnetic Resonance Data Bank site using the Validation Tools http://www.bmrb.wisc.edu/. The ARIA force field energy terms can be broken down to the single copies and are given separately in Table 1, whereas the data energy terms are only evaluated for the two copies simultaneously. To quantify violations of the restraints, a standard RMS criterion was applied, with a restraint potential that is harmonic in the differences between target and effective distances (50). For ensemble restraints, the effective distances were averaged over the two copies.
Table 1. NMR and refinement statistics for PB(552–594) structures.
| Restr. Chain A | Restr. Chain B | Restr. Ens. | |
|---|---|---|---|
| NMR distance and dihedral constraints | |||
| Distance constraints | |||
| Total NOE | 648 | 648 | 96 |
| Intra-residue | 258 | 258 | 33 |
| Sequential (|i – j| = 1) | |||
| 146 | 146 | 21 | |
| Medium-range (|i – j| < 4) | 53 | 53 | 20 |
| Long-range (|i – j| > 5) | 191 | 191 | 22 |
| Intermolecular | 0 | 0 | 0 |
| Violations (RMS) | |||
| Distance constraints (Å) | 0.67 | 0.67 | 1.20 |
| Max. distance restraint violation (Å) | 3.62 | 3.27 | 5.29 |
| PB(552–594) | PB(552–594)KF | PB(552–594)IF | |
| ARIA energy terms (kcal/mol) | |||
| E bond | 14.0±1.0 | 6.7±0.6 | 7.3±0.9 |
| E angle | 105.7±6.9 | 48.0±4.9 | 57.6±5.8 |
| E improper | 59.3±5.4 | 24.7±3.0 | 34.6±3.6 |
| E dihedral | 427.3±3.4 | 216.0±3.1 | 211.3±2.0 |
| E vdW | -297.8±12.8 | -150.1±8.3 | -147.7±12.2 |
| E electr | -2780.7±78.6 | -1399.5±76.1 | -1381.2±73.7 |
| E data | -257.7±4.5 | ||
| E total | -2730.0±76.5 | ||
| Structure statistics | |||
| Deviations from idealized geometry | |||
| Bond lengths (Å) | 0.004 | 0.004 | 0.004 |
| Bond angles (°) | 0.7 | 0.7 | 0.8 |
| Impropers (°) | 1.5 | 1.4 | 1.7 |
| Average pairwise r.m.s. deviation** (Å) | |||
| Heavy | 1.9 (1.1)e,f(1.2)g | 1.7(1.0)e,f,g | 1.6(1.1)e(1.0)f,g |
| Backbone | 1.1 (0.4)e,f(0.5)g | 1.1(0.4)e,g(0.3)f | 1.0(0.3)e,f,g |
| Ramachandran analysis of residuese | |||
| favored region (%) | 91.2 | 91.4 | 91.1 |
| additional allowed regions (%) | 8.6 | 8.3 | 8.9 |
| generously allowed regions (%) | 0.1 | 0.3 | 0.0 |
| disallowed regions (%) | 0.0 | 0.0 | 0.0 |
** ‘Pairwise r.m.s. deviation was calculated among 24 refined structures.’
e: Ordered residue ranges: 556A–565A, 568A–584A, 590A–592A.
f: residue range: 559A–585A.
g: residue range: 559A–590A.
PB(552-594)/LE15-24 and PB(552-594)/LE21-36 docking using HADDOCK
PB(552–594)IF/LE15–24, PB(552–594)IF/LE21–36, PB(552–594)KF/LE15–24, PB(552–594)KF/LE21–36 structure calculations were performed at the HADDOCK webserver (http://haddock.science.uu.nl/services/HADDOCK/) (51).
Ambiguous distance restraints based on chemical shift perturbations and the disappearance of some of the PB(552–594) and oligonucleotide NMR signals were used to drive the docking. Two structures (one structure among the twelve belonging either to PB(552–594)KF or to the PB(552–594)IF families) were used as input structures. The two LE14–25 and LE22–36 DNA sequences were constructed as B-DNA according to the NMR results using the http://haddock.chem.uu.nl/services/3DDART/ webserver (52). During the rigid-body docking, 1000 structures were calculated, and 200 during both simulated annealing and water refinement. All 200 water-refined structures were analysed, and cut-off for clustering was 7.5 Å (interface RMSD), with four structures per cluster. The ranking of the clusters is based on the average score of the top 4 members of each cluster. The HADDOCK score is calculated as function of the intermolecular van der Waals energy, the intermolecular electrostatic energy, the empirical desolvation energy term and the Ambiguous Interaction Restraints (AIRs) energy. The cluster numbering reflects the size of the cluster, with cluster 1 being the most populated cluster. After successful docking, the best complex models (clusters 1) were selected on the basis of the HADDOCK score (Supplementary Tables S4 and S5).
RESULTS
Sequence comparison of the C-terminal CRD of PB-related transposases
Active or potentially active PB-related transposases have been found in a variety of insect species and other organisms (5–17,19–21,53). The C-terminal CRD is well conserved among PB family members (Figure 1B). Sequence comparisons revealed that PB-related transposases possess mostly a CX2CX11CX2CX4CX2HX4CX2C motif with seven cysteines and one histidine that are potential ligands for two Zn2+ ions (4,37). In addition to the potential zinc-coordinating residues, several other basic amino acids are highly conserved.
The C-terminal CRD of PB is required for piggyBac transposition
To determine if the C-terminal domain of PB is required for transposition in mammalian cells, we compared the ability of full-length PB(1–594) and truncated PB(1–558) lacking the C-terminal CRD (Figure 1A) to promote transposition of a piggyBac element containing several hundred bp (L-End = 328 bp; R-End = 361 bp) from the Left and Right transposon ends flanked by piggyBac’s specific TTAA target site duplications. As shown in Figure 1C and D, full-length PB(1–594) was highly active for transposition whereas transposition promoted by PB(1–558) was no higher than background levels.
DNase I footprinting suggests that the C-terminal domain is involved in specific DNA binding
Previous work by others showed that piggyBac elements containing short ends, for example LE1–35 (LE standing for Left-End) and RE1–63 (RE standing for Right-End) can transpose in mammalian cells (54,55), and we have shown that short piggyBac ends can be efficient substrates for transposition in vitro (4). It has also been observed however, that transposition frequency can be increased in some in vivo assays by using longer LE and RE ends (56), leaving it undefined whether other transposon end sequences might be important. The LE1–35 and RE1–63 termini of piggyBac contain two inverted repeats (23): each end contains a 13-bp terminal inverted repeat and an internal 19-bp inverted repeat, but with different spacing between them. In LE1–35, the repeats are separated by a 3-bp spacer, whereas in RE1–63, they are separated by a 31-bp spacer (Figure 2A).
To directly determine the positions of specific interactions of PB with transposon ends, we carried out DNase I footprinting on either LE or RE DNA fragments using both full-length PB(1–594) and truncated PB(1–558) lacking the C-terminal CRD (Supplementary Figures S1 to S3). The results, summarized in Figure 2B to D, revealed strong protection in LE (Figure 2B) around the 19-bp repeat (LE14–35) in the presence of full-length PB as well as at a ‘Left-Internal’ (LI) site, LI181–199 (Figure 2C). This LI site contains a previously unidentified 19-bp repeat-like sequence (highlighted in purple). In RE, full-length PB strongly protects the region containing the 19-bp repeat (RE43–63; Figure 2D). Weak protection (Figure 2D) on one strand is also found to extend slightly beyond the TTAA target sequence flanking the transposon end.
With truncated PB(1–558), the protection on the three DNA fragments is very different: there is little protection of the regions containing the 19-bp repeat-like sequences, strongly suggesting that the C-terminal domain interacts with these repeats (Figure 2 and Supplementary Figures S1–S3). As shown in Figure 2B to D, close inspection of the 19-bp repeat regions reveals that each contains a 17-bp sequence (LE17–33, LI180–196 and RE45–61) composed of 5′-TGCGT(c/a)AA-3′ inverted motifs (Figure 2E).
There are other notable differences in the protection patterns obtained with truncated PB(1–558) compared to full-length PB(1–594). With truncated PB(1–558), weak LE protection extends from outside the TTAA target sequence flanking the 5′ transposon tip to the interior of the 19-bp repeat. In RE, PB(1–558) protection extends from near the external edge of the 19-bp repeat (RE48), into the flanking DNA beyond the TTAA target site duplication. Similarly, although protection at the LI 19-bp repeat is not observed, strong protection is observed extending 35-bp towards the interior of the transposon.
Further inspection of the protected sequences at piggyBac ends reveals that, concealed within the standard description of the organization of piggyBac LE and RE (Figure 2A), there is an additional 10-bp repeat sequence (AAAGATAaTC; highlighted in blue in Figure 2). This sequence is present not only near the transposon tips (LE7–16 and RE7–16) but also within the ‘spacer’ region on RE (RE34–43) as well as adjacent to the LI 17-bp 5′-TGaGTcAAaTTgACGAC-3′ inverted repeat (LI198–207 and LI209–218). The region protected by PB(1–558) also includes the transposon tips flanked by the specific piggyBac target site 5′-TTAA-3′ (Figure 2B to D and Supplementary Figures S1–S3).
PB(530–594) containing the C-terminal CRD binds specifically to piggyBac ends
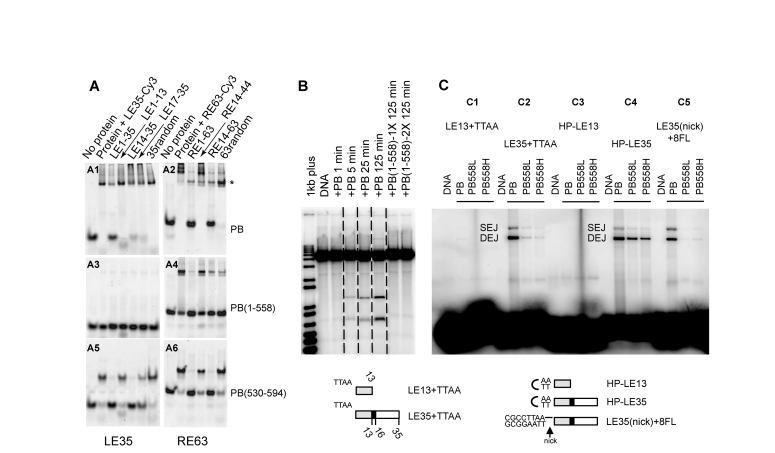
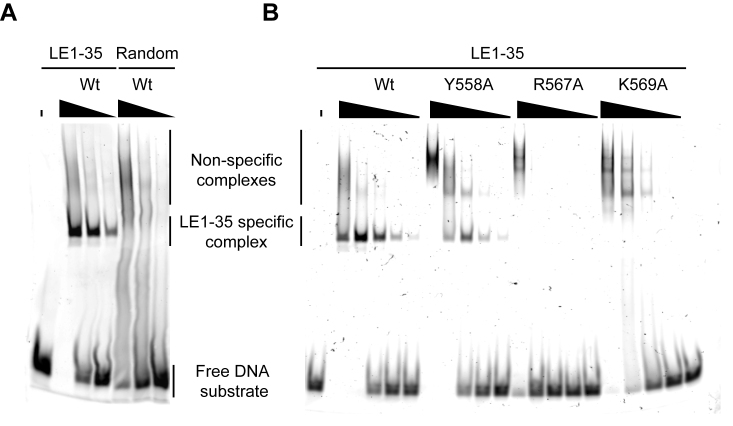
The above DNA protection experiments suggest that the PB CRD binds specifically to sequences within piggyBac ends. To test this hypothesis, we performed competition gel shift assays using Cy3-labeled LE1–35 and RE1–63 probes and various unlabelled double-stranded competitor oligonucleotides. We compared the DNA binding properties of the full-length PB(1–594) (Figure 3A1 and A2), truncated PB(1–558) which lacks the CRD (Figure 3A3 and A4), and PB(530–594), which contains the CRD (Figure 3A5 and A6). We confirmed that the PB C-terminal domain binds specifically to DNA. Indeed, PB(530–594) binds to both LE1–35 and RE1–63, forming a shifted complex in the presence of random competitor DNA but not in the presence of unlabelled cognate oligonucleotides (Figure 3A5 and A6). Notably, the presence of oligonucleotide competitors containing the 19-bp repeat significantly decreases PB(530–594) end binding: LE14–35 and LE17–35 decrease binding to LE1–35, and RE14–63 abolishes binding to RE1–63 whereas RE14–44, which lacks the 19-bp repeat, does not. This suggests that the 19-bp sequence protected in the DNase I footprinting experiments is critical for the specific binding of PB(530–594) to the left and right ends. The observation that LE1–13 (identical to RE1–13) has little effect on complex formation with either Cy3-labeled LE1–35 or RE1–63 confirms our conclusion.
Figure 3.
Role of the C-terminal domain of PB. (A) PB specifically binds its LE-TIR and RE-TIR. For each panel, the first lane is labelled DNA in the absence of protein, and the second lane is upon incubation with either LE35-Cy3 (LE35 for LE1–35) (A1, A3, A5) or RE63-Cy3 (R63 for RE1–63) (A2, A4, A6) (50 nM final concentration). The remaining lanes in each panel indicate the effect of adding various unlabelled competitor DNA oligonucleotides indicated at the top of the figure. The protein is either the full-length PB(1–594) (A1, A2), PB(1–558) (A3, A4) or PB(530–594) (A5, A6) (1 μM final concentration). (B) In vitro cleavage assay. All samples were run on the same gel, and dashed lines indicate where different lanes were combined to prepare the figure. PB was at a final concentration of 0.59 μM, and PB(1–558) 1X and 2X correspond to 0.78 μM and 1.57 μM, respectively. (C) Target joining of LE-TIR oligonucleotide substrates: LE13+TTAA (C1), LE35+TTAA (C2), HP-LE13 (C3), HP-LE35 (C4), LE35(nick)+8FL (C5), into a target plasmid. The Single End Join (SEJ) and Double End Join (DEJ) products were visualized on a native agarose gel. Either PB (0.59 μM) or PB(1–558) at 1.25 μM (’L’ for low concentration) or 6.3 μM (’H’ for high concentration) was used. Schematic representation of LE13+TTAA, TE35+TTAA, HP-LE13, HP-LE35, LE35(nick)+8FL (a phosphodiester bond is present between the 5′-CGCCTTAA-3′ sequence and the LE35 top strand but not between the complementary sequence of 5′-CGCCTTAA-3′ and the LE35 bottom strand; the black horizontal line indicates the intact phosphodiester bond).
The PB C-terminal domain is required for DNA breakage and joining at piggyBac ends
We used in vitro DNA breakage and joining assays to determine whether the PB C-terminal domain is indeed required for these activities at piggyBac ends. To evaluate DNA double strand cleavage in vitro, we used an end-labelled linear DNA fragment containing several hundred bp piggyBac LE and RE ends (LE1–673 and RE1–400 as described in Li et al. (39)). While PB(1–594) promoted breaks at both transposon ends, no DNA double strand breaks were observed at either end with PB(1–558) (Figure 3B). Thus, the PB C-terminal domain is crucial for transposon excision in vitro, exactly as in the above-described transposition assays performed in mammalian cells (Figure 1D).
We also compared the ability of PB(1–594) and PB(1–558) to promote DNA breakage and target joining by measuring the joining of a 5′ end-labelled double-stranded LE oligonucleotide to an unlabelled target plasmid DNA. The joining of one LE oligonucleotide to the target plasmid should yield a nicked circular plasmid (Single End Join – SEJ) and the concerted joining of two LE oligonucleotides should yield a linearized plasmid with one LE attached to each end (Double End Join – DEJ). When PB(1–594) was incubated with an oligonucleotide consisting of LE1–35 and the flanking TTAA target site duplication both SEJ and DEJ products were observed (‘LE35+TTAA’, Figure 3C2). In contrast, only a very low level of target joining was observed with PB(1–558). Similarly, target joining was seen with PB(1–594), but not with PB(1–558), with a ‘nicked’ TTAA-LE1–35 oligonucleotide (‘LE35(nick)+8FL’; Figure 3C5) in which the 3′OH transposon end is exposed as in the first strand cleavage step in piggyBac transposition. In the next step of transposition, the free 3′OH end attacks the 5′ end of the TTAA sequence, forming a TTAA hairpin on the transposon end and releasing the transposon from the flanking DNA. Notably PB(1–594) and PB(1–558), albeit less efficiently than PB(1–594), can both nick the TTAA hairpin to re-expose the 3′OH transposon end, which can then join to the target plasmid (‘HP-LE35′, Figure 3C4), suggesting that the C-terminal domain is most critical for the initial strand cleavage and hairpin formation step. No product was observed if the substrate oligonucleotides contained only the first 13-bp of LE (‘LE13+TTAA’ and ‘HP-LE13′, Figure 3C1 and C3).
NMR study reveals that the C-terminal CRD, PB(552-594), forms a monomeric folded Zn2+-complexed structure
It has been proposed that the C-terminal CRD of PB can adopt a PHD/RING-finger fold (4,37) or at least that this domain is able to interact with one or two Zn2+ ions (38). We verified using flame atomic absorption spectrophotometry that GST-tagged PB(552–594) is able to complex 1.89 ± 0.09 eq of Zn2+ per monomer. To gain insight into the structure of this domain, we performed 1D 1H NMR analysis of a synthetic PB(552–594) peptide. The collected spectra in the absence or presence of Zn2+ at acidic pH 3.5, and in absence of Zn2+ at pH 6.5, are characteristic of a peptide in a random coil conformation with a poorly dispersed NMR spectrum (Supplementary Figure S4A–C). On the contrary at pH 6.5 in the presence of Zn2+ and 5 mM DTT, the spectrum exhibits marked dispersion of the signals, reflecting the presence of a folded metal-bound protein. The quality of the spectra was drastically increased by the addition of 200 mM NaCl (Supplementary Figure S4D–G).
Using diffusion-ordered spectroscopy (DOSY) (44,45), we probed the oligomeric state of PB(552–594) in solution. From the diffusion coefficient of PB(552–594) measured in 100% D2O in the presence of Zn2+, 5 mM DTT and 200 mM NaCl at pH 6.5 (≈1.15 × 10–10 m2/s−1 ± 1.17 × 10–12 m2/s−1), we could derive the molecular weight of the diffusion species to 5000–5400 Da depending on the method used to calculate it (Supplementary Table S1). This is in very good agreement with the theoretical molecular weight of a monomeric PB(552–594) bound to two Zn2+ (MW = 5235 Da), indicating that PB(552–594) is predominantly a monomer in solution (Supplementary Figures S5 to S8 and Supplementary Table S1).
3D structure of the C-terminal CRD of the PB transposase
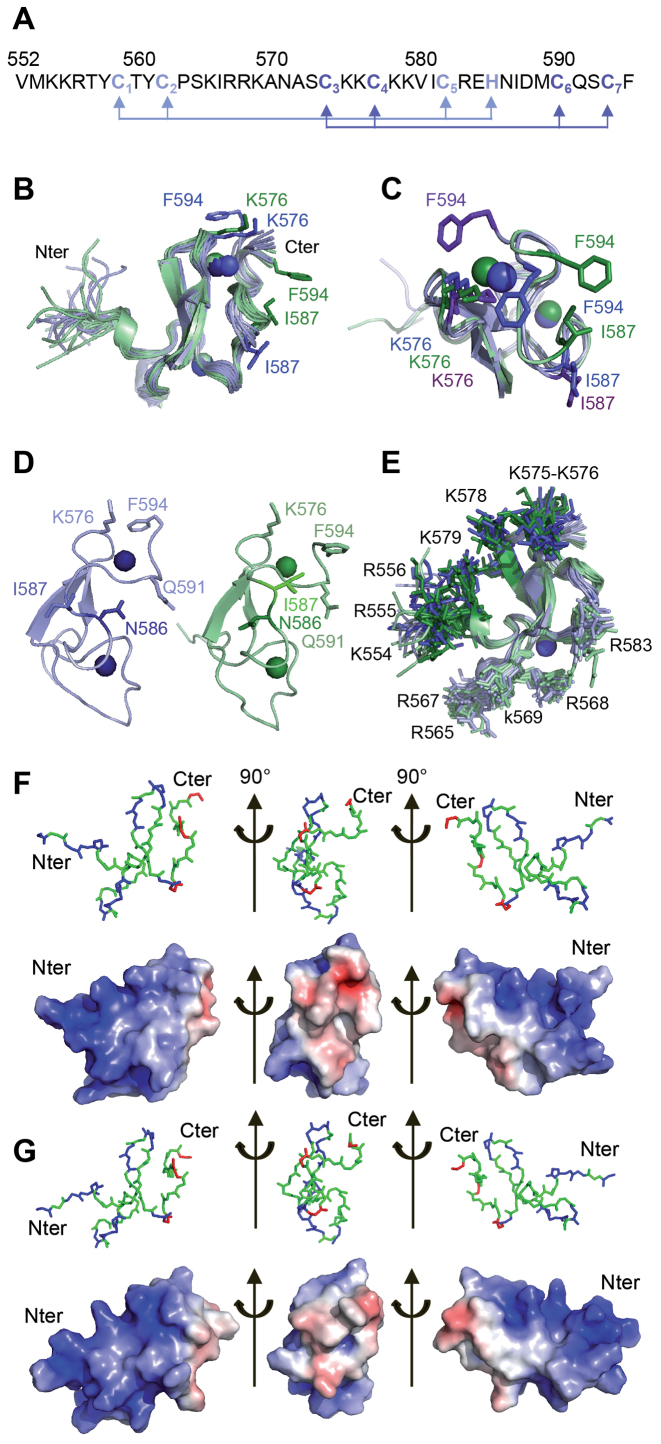
All proton resonances of the PB(552–594) peptide were assigned using the homonuclear 2D NMR spectroscopy strategy (57) (Supplementary Figure S9). The first structures calculated with ARIA 2.3 (Ambiguous Restraints for Iterative Assignment) (48), using only nuclear Overhauser enhancements (NOE) derived distance restraints extracted from the nuclear Overhauser enhancement spectroscopy (NOESY) spectra, are consistent with a cross-brace arrangement of two zinc-binding motifs and identified the candidate residues for zinc coordination (Figure 4A). Analysis of the structures indicated that Nδ1 of H585 could be implicated in the coordination of Zn2+ since it is correctly oriented pointing towards three cysteine sulfur (Cys S) atoms, compatible with a tetrahedral coordination of one Zn2+. In order to confirm this hypothesis, 1H-13C HSQC spectrum was recorded to determine the H585 Cδ2 and Cϵ1 chemical shifts since the identification of the coordination mode of histidine can be based on the 13Cδ2 and 13Cϵ1 aromatic carbon chemical shifts (58). The observed H585 Cδ2 and Cϵ1 chemical shifts (118.7 and 138.76 ppm respectively (Supplementary Figure S10A and B) indicated that H585 Nδ1 is implicated in Zn2+ coordination. Once the topology of the Zn2+-coordinating residues was confirmed, subsequent ARIA structure calculations were performed using distance and angle restraints that imposed tetrahedral Zn2+-coordination to His Nδ1 and Cys S atoms. In parallel, structure calculations were performed using distance and angle restraints that imposed tetrahedral Zn2+-coordination to His Nϵ2 and Cys S atoms. In this latter case, several restraints implicating H585 and several adjacent residues were not satisfied, confirming that H585 Nδ1 is one of the ligands of Zn2+. The final structures show that C559, C562, C582 and H585 on the one hand, and C574, C577, C590 and C593 on the other hand are oriented to coordinate two Zn2+ with a C3H (ZF1) and C4 (ZF2) coordination mode in a cross-brace ZF motif (Figure 4A).
Figure 4.
Structure of the PB C-terminal Cysteine-Rich Domain (A) Sequence of the PB(552–594) peptide: the light blue and dark blue arrows highlight the Cys3-His (ZF1) and Cys4 (ZF2) coordination mode. (B) Superposition of the twenty-four structures generated using ARIA ensemble showing the I587/F594 and K576/F594 proximities in PB(552–594)IF (in green) and in PB(552–594)KF (in blue). (C) Superimposition of one structure of the PB(552–594)IF family (in green) and two of the PB(552–594)KF family (in blue) showing the two possible K576/F594 orientations in PB(552–594)KF (the K576 and F594 side chains are in blue for one orientation and in purple for the other one). (D) Comparison of one structure of each family highlights the relative orientation of N586 and I587: in family PB(552–594)KF (in blue) the side chain of residue N586 is turned towards residue Q591, whereas in family PB(552–594)IF (in green) the side chain of residue N586 points in the other direction. (E) Superposition of the twenty-four structures, highlighting the two positively charged clusters of Arg and Lys residues in dark blue (PB(552–594)KF family) and dark green (PB(552–594)IF family) on one side (554-KRR-556, K575, K576, K578, K579) and in light green and blue on the other side (K565, 567-RRK-569, R583). The two 554-KKR-556 and 565-KIRRK-569 stretches of residues belonging to the bipartite NLS are localised on the same face of PB(552–594). (F) Electrostatic potential calculated on one structure of the PB(552–594)KF family. (G) Electrostatic potential calculated on one structure of the PB(552–594)IF family. The red and blue colors in surface representations denote negative and positive charges, respectively. Graphic representations were performed with PyMOL. Each view corresponds to a 90° rotation of the previous one.
PB(552-594) can adopt two different conformations
Despite several cycles of structure calculation, we observed NOE signals that cannot be satisfied together with a single PB(552–594) conformation. For example, the NOEs involving F594 and I587 on the one hand and F594 and K576 (Supplementary Figure S10C) on the other hand were never simultaneously satisfied. We found, however, that NOEs between F594 and I587 could only be satisfied if those implicating F594 and K576 were removed from the restraint list and vice-versa. Because PB(552–594) is monomeric under our conditions, we propose that these conflicting NOEs are consistent with rapid dynamic behaviour at the NMR chemical shift timescale, of the nine last residues of PB(552–594), which direct F594 either towards K576 or I587. To sort NOEs that are compatible with each other, we used ARIA 2.3 with a modified protocol to adapt the program to ensemble-based calculations (48,49). The restraints are expected to be fulfilled only on average over several structures instead by a single conformer. We found that only two copies are sufficient to account for all observed NOEs. The two copies of the protein chain might be extreme conformations representative of a complex conformational space sampled by PB(552–594) in fast exchange (at the NMR chemical shift timescale). After water refinement, 12 two-copy ensembles, with both copies having a backbone RMSD ≤ 0.5 Å, calculated for residues C559-E584 to the average of the structures of the same family, were chosen as representative structural ensemble of PB(552–594) (Figure 4B).
The structure of PB(552–594) reveals a well-defined globular domain (Table 1) with the two interwoven ZFs knitted together around an antiparallel β-sheet (Figure 4B to E). The flexibility of the last eight residues of PB(552–594) is highlighted in each generated copy, by the F594 orientations (Figure 4B) satisfying either the F594 and K576 (family PB(552–594)KF; shown in blue) or the F594 and I587 (family PB(552–594)IF; shown in green) NOEs. In the PB(552–594)IF structures, the interaction mode between the side chains of I587 and F594 is always very similar, whereas in PB(552–594)KF, the side chain of F594 can be positioned on either side of the side chain of K576 (Figure 4C). Moreover the generated structures show that the proximity of the I587 and F594 residues in PB(552–594)IF goes hand in hand with a different overall conformation of residues 582 to 588, resulting in two alternative orientations of the side chain of residue N586 with respect to residue Q591. In the majority of the PB(552–594)KF structures, N586 is turned towards residue Q591, whereas in PB(552–594)IF, due to the reversal of backbone geometry, the side chain of residue N586 points exactly in the other direction (Figure 4D). In some structures of PB(552–594)IF, residues 583 to 585 form a 3/10 helix, as detected by the program DSSP, whereas for all structures of PB(552–594)KF, DSSP identifies a hydrogen-bonded turn for these residues. The largest consistent difference between dihedral angles of the two structure families can be seen for the psi angle of H585. It adopts values of about -60° for PB(552–594)KF and of about +20° for PB(552–594)IF.
The electrostatic surface potential calculated on one structure of each family, PB(552–594)KF (Figure 4F) and PB(552–594)IF (Figure 4G), reveals two positively charged clusters located exactly on opposite sides on the surface of the peptide (Figure 4E), suggesting that two faces of PB(552–594) are accessible for electrostatic interactions whatever the family. Nevertheless, the flexibility of the last 8 residues of PB(552–594) does generate differences on the electrostatic surfaces implicating the K576, I587 and F594 residues in PB(552–594)KF (middle view of Figure 4F) and PB(552–594)IF (middle view of Figure 4G) with an increasing of hydrophobicity in PB(552–594)IF due to the I587 and F594 proximity.
NMR analysis of the PB CRD in interaction with the 5′-TGCGT-3′/3′-ACGCA-5′ motif of the 19-bp repeats
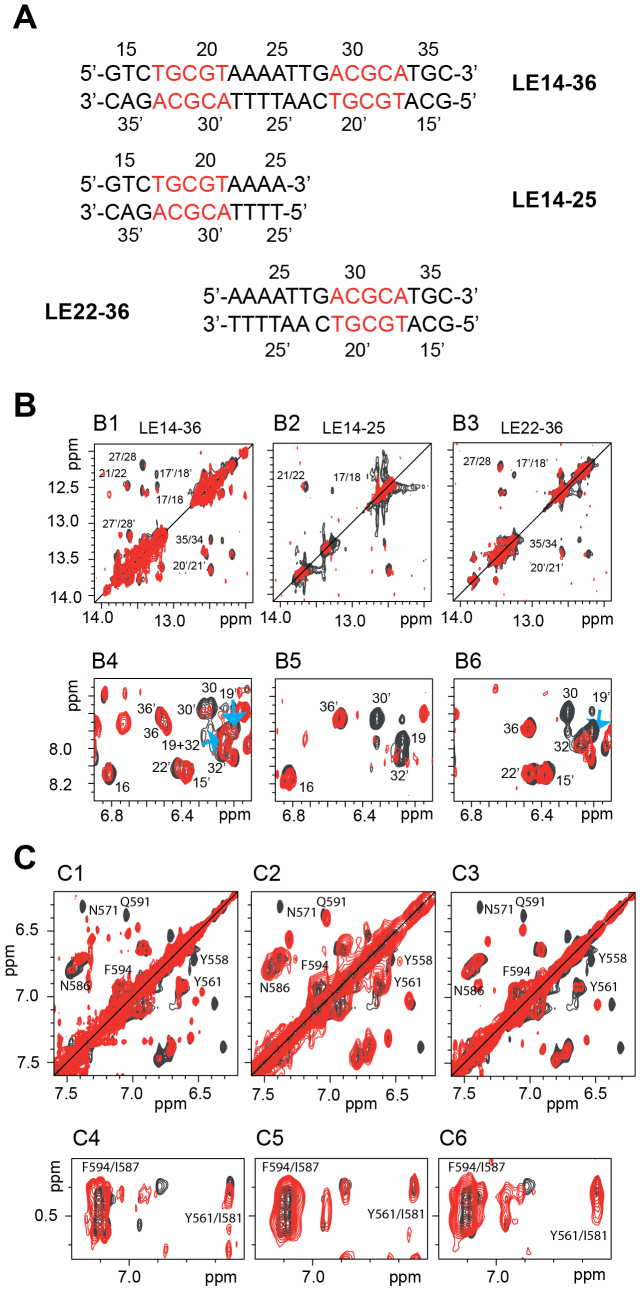
The DNase I protection and DNA binding experiments above suggested that the PB C-terminal CRD binds specifically to the 19-bp repeat within the ends of piggyBac. Inspection of the sequence of the 19-bp repeat (purple boxes in Figure 2B and D) reveals that it contains inverted sequence motifs: 5′-TGCGT-3′/3′-ACGCA-5′ (LE17–21) and 5′-ACGCA-3′/3′-TGCGT-5′ (LE29–33) separated by 7-bp. To determine the specific interaction interfaces between the PB CRD and the 19-bp repeat, we used NMR to analyse complexes formed between PB(552–594) and three DNA sequences from the 19-bp repeat: LE14–36, which contains both the LE17–21 and LE29–33 motifs, and two shorter oligonucleotides each containing either LE17–21 (LE14–25) or LE29–33 (LE22–36) (Figure 5A).
Figure 5.
NMR analysis of DNA binding by the PB C-terminal Cysteine-Rich Domain (A) The three DNA sequences used for the 1H NMR studies. (B) Selected regions of the NOESY spectra showing the imino-imino (B1, B2, B3) and amino-amino (B4, B5, B6) regions of LE14–36 (B1, B4), LE14–25 (B2, B5) and LE22–36 (B3, B6) in absence (black) and in presence of 2 equivalents of PB(552–594). In B1, B2, B3 on one hand and in B4, B5, B6 on the other hand are shown the inter nucleotides imino/imino and intra nucleotides amino/amino NOEs respectively. Some of the cross-peaks disappeared in presence of PB(552–594) whereas others were always visible with approximately the same intensities. (C) Selected regions of the NOESY spectra showing some of the PB(1–558) protons which are either perturbed by the DNA interactions (C1, C2, C3) such as the aromatic protons of Y558, the δ protons of N571 and the ϵ protons of Q591, and those which undergo no significant chemical shift variation in presence of all three DNAs such as the aromatic protons of Y561 and F594, the δ protons of the N586 (C1, C2, C3), the γ and δ protons of I581 and I587 (C4, C5, C6).
We first analysed the three oligonucleotides alone in solution. The presence of almost all imino- and amino-proton resonances in the 1H NMR spectra indicates the presence of base pairing all over the three DNA sequences (Figure 5B and Supplementary Figure S11). Analysis of the standard connectivities in the TOCSY and NOESY spectra led to a nearly complete assignment of the base and deoxyribose protons. We observed sequential 1H–1H NOEs indicative of a B-form helical geometry throughout the three DNAs.
Interpretable DNA NMR spectra were obtained in all cases when PB(552–594) was added to LE14–25, LE22–36 and LE14–36 despite the fact that increasing concentrations of PB(552–594) resulted in partial precipitation, even in the presence of 200 mM NaCl (Supplementary Figure S12A–C). Based on the observed chemical shift variations and the disappearance of some of the LE14–36 (Supplementary Figure S13A), LE14–25 (Supplementary Figure S13B) and LE22–36 DNA sequences (Supplementary Figure S13C) and PB(552–594) 1H resonances (Supplementary Figure S14A–C), we determined the DNA/peptide interfaces i.e. the 5′-TGCGT-3′/3′-ACGCA-5′ motif present in the three DNA oligonucleotides and the T557, Y558, S564, K565, R567, R568, K569, N571 and R583 residues of PB(552–594) (Figure 5B and C and Supplementary Figure S14A–C). On the contrary, we did not observe significant 1H chemical shift variations or disappearance of the Y561, I581, I587, and F594 signals suggesting that these residues are not implicated in the peptide/DNA interfaces. We verified that DNA binding does not strongly affect the folding of the peptide as, despite the limited quality of the spectra, many NOEs resulting from the tertiary folding of PB(552–594) were maintained (Figure 5C4 to C6 and Supplementary Figure S14A–C). We verified also that the DNAs conserved the B-conformation in the complexes since the observed sequential 1H-1H NOEs (medium range H6[i],H1’[i-1]; H8[i],H1’[i-1]) (57) are characteristic of a B-form helical geometry (Supplementary Figure S15).
The interaction sites of PB(552–594) are best delineated on the shortest DNA sequences LE14–25 and LE22–36, which each contains the single 5′-TGCGT-3′/3′-ACGCA-5′ repeat. With the longer LE sequence (LE14–36), the observed chemical shift variations show that the two 5′-TGCGT-3′/3′-ACGCA-5′ inverted motifs can be bound by PB(552–594) (Supplementary Figure S13A to S13C). Additional non-specific interactions cannot be excluded, as suggested by additional interaction signals all over the LE14–36 sequence (Supplementary Figure S13A).
PB(552–594) is therefore able to bind the unique 5′-TGCGT-3′/3′-ACGCA-5′ motif on the two shorter oligonucleotides (LE14–25, LE22–36), and the two 5′-TGCGT-3′/3′-ACGCA-5′ inverted motifs of LE14–36, through one of its highly positively charged surfaces. Due to the disappearance of most of the 1H NMR signals from the protein residues and from the DNA nucleotides at the interface, no intermolecular nuclear Overhauser Effect (NOE) could be detected, thus preventing the structure determination of PB(552–594)/DNA complexes using standard NMR methods.
Reconstruction of the PB/DNA binding interfaces by molecular docking simulations
To understand the molecular basis for DNA recognition by PB(552–594), structural models of the PB(552–594)/LE14–25 and PB(552–594)/LE22–36 complexes were constructed using molecular-docking simulations in the data-driven program HADDOCK (High Ambiguity Driven biomolecular DOCKing) (51,59). The 5′-TGCGT-3′/3′-ACGCA-5′ DNA sequence belonging on LE14–25 and LE22–36 and the T557, Y558, S564, K565, R567, R568, K569, N571 and R583 PB(552–594) residues were used as ambiguous interaction restraints in docking calculations. In the case of the DNA sequences the passive residues were defined automatically around the active residues. For the protein, T560, I566, A571, A572 and E584 were defined as the passive residues surrounding the protein surface interaction. Moreover we defined the N-ter domain of PB(552–594), V552-Y558 as a fully flexible segment.
The best solvent-refined models from the HADDOCK calculations were automatically grouped into clusters based on the interface PB(552–594)/LE14–25 and PB(552–594)/LE22–36 RMSDs which, are automatically defined based on an analysis of all contacts made in all models (Supplementary Tables S4 and S5). A structural comparison between minimum energy structures of all the clusters of each complex show that either PB(552–594) is oriented head to toe relative to the DNA by comparison with that happens in cluster 1, either PB(552–594) is shifted of one base pair toward the 3′ end of the DNA top strand or PB(552–594) is positioned in the minor groove (Supplementary Figures S16 and S17).
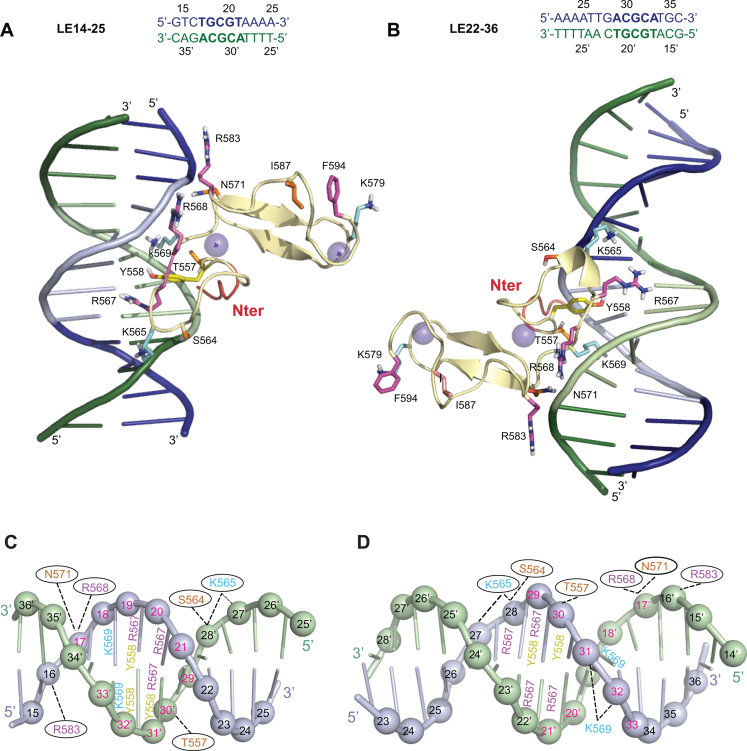
We selected the PB(552–594)/LE14–25 and PB(552–594)/LE22–36 structures of clusters 1 for further analysis since, clusters 1 are the largest clusters (they represent approximately 50% of the clustered structures) and also show the lowest HADDOCK score (Supplementary Table S5). In the two models the strongly positively charged PB(557–571) loop targets the major groove of LE14–25 (Figure 6A and C) and LE22–36 (Figure 6B and D) both containing one of inverted recognition motifs: 5′-TGCGT-3′/3′-ACGCA-5′ or 5′-ACGCA-3′/3′-TGCGT-5′ respectively. The dynamic C-terminal domain of PB(552–594) is oriented toward the opposite side of the DNA interaction surface (Figure 6A and B). Y558, R567, K569 are positioned in the major groove and interact with the edges of the base pairs via hydrogen bond interactions, while T557, S564, N571 on the one hand, and K565, R568, R583 on the other hand contact the sugar phosphate backbone by hydrogen bond or electrostatic interactions, respectively (Figure 6 and Supplementary Tables S6 and S7). As expected from the relative orientations of the recognition motifs, 5′-TGCGT-3′/3′-ACGCA-5′ or 5′-ACGCA-3′/3′-TGCGT-5′, PB(552–594) has reversed orientations on substrates LE14–25 and LE22–36, with its N-terminal extremity oriented towards the 3′ end in LE14–25 and towards the 5′ end in LE22–36 (Figure 6A–D). For the full length substrate, LE15–36, NMR showed that the two inverted recognition motifs interact with PB(552–594). Because the PB(552–594)/DNA complex is dynamic in solution we cannot certify that two PB CRDs bind simultaneously to the same DNA molecule. However, we propose a 2:1 protein/DNA model (Supplementary Figure S18A), in which the two CRDs are oriented in an opposite direction to each other relative to the axis of the DNA without any steric hindrance (Supplementary Figure S18A).
Figure 6.
Comparison of the top cluster 1 models for the PB(552–594)IF/LE14–25 and PB(552–594)IF/LE22–36 complexes. (A) and (C) show the model constructed with LE14–25. (B) and (D) show the model construct with LE22–36. Only the PB(552–594)IF interacting residues (corresponding to those used as ambiguous interaction restraints in docking calculations in HADDOCK) and the residues belonging to the flexible C-terminal domain are represented. K565, K569 and K579 are in cyan, R567, R568, R583 and F594 are represented in pink, T557, S564, N571 and I587 are coloured in orange, Y558 is in yellow. The five N-terminal residues ribbon is coloured in red. (A and B) The top strand is in blue and the complementary strand in green, and the 5′-TGCGT3′-/3′-ACGCA-5′ and 5′-ACGCA3′-/3′-TGCGT-5′ sequences highlighted in light blue and light green respectively. (C and D) LE14–25 and LE22–36 are represented in a cartoon representation and the phosphorus atoms are shown as spheres. The top strand is in light blue and the complementary strand in light green. The 5′-TGCGT-3′/3′-ACGCA-5′ and 5′-ACGCA-3′/3′-TGCGT-5′ inverted sequences are highlighted with the number in red. The PB(552–594)IF interacting residues are highlighted and positioned on the nucleotide with which they interact. When the interactions take place in the major groove with the edges of the base the numbers are localized on the bases (except for K569), and when the contacts are at the level of the sugar phosphate backbone, the numbers are inside of black ovals.
Mutational analysis of the PB CRD/DNA complex
The structural models of Figure 6 predict that the PB CRD interacts with the 5′-TGCGT3′-/3′-ACGCA-5′ motifs of LE1–35. We therefore tested whether this DNA motif is indeed essential for the specific interaction with PB CRD. Using competition gel shift assays (Supplementary Figure S19) we tested the ability of a mutant competitor DNA (LE1–35 3-Mut), in which the two 5′-TGCGT3′-/3′-ACGCA-5′ motifs (see Figure 5A) were replaced by 5′-TGGCA-3′/3′-ACCGT-5′, to compete with a wild-type Cy3-labeled LE1–35 probe for the binding of a GST-PB(537–594) fusion. While an unlabelled wild-type LE1–35 substrate efficiently competed with the labelled probe for complex formation, the 3-Mut substrate, exactly like a random competitor DNA, had no effect on complex formation, indicating that the 5′-TGCGT3′-/3′-ACGCA-5′ motif is a sequence-specific determinant for PB CRD binding (Supplementary Figure S19). Direct EMSA analysis of GST-PB(537–594) binding to Cy3-labeled LE1–35, compared with a Cy3-labeled random substrate, revealed two types of shifted complexes (Figure 7A). A major complex was observed only with the wild-type LE1–35 sequence, indicative of a specific protein/DNA interaction. In addition, multiple higher molecular weight complexes were detected for both DNA substrates at high protein concentrations, suggesting that they result from non-specific interactions. The binding affinities were measured using EMSA. The binding curve (Supplementary Figure S20A) quantifies the binding of GST-PB(537–594) to LE1–35 and the apparent calculated Kd value, ≈ 0.64 μM (Kd=protein concentration when 50% of DNA is free), is compatible with a complex in ‘intermediate exchange’ on the NMR chemical shift timescale (Figure 5B–C and Supplementary Figures S12 and S14).
Figure 7.
DNA binding properties of wild-type and mutants PB Cysteine-Rich Domain. (A) Wild type PB CRD (GST-PB(537–594)) formed different types of complexes in the presence of the LE1–35 and Random substrate. Complexes were assembled in the presence of LE1–35-Cy3 or Random-Cy3 substrates (100 nM) and 0.33 μM, 1 or 3 μM concentration of GST-PB(537–594) before analysis on a 5% acrylamide, 0.5× TBE gel. (B) Wild-type or mutants GST-PB(537–594)/LE1–35 complexes were assembled in the presence of 100 nM of LE1–35-Cy3 substrate and 0.03 μM, 0.11, 0.33, 1 or 3 μM of GST fused PB(537–594). Complexes were further analysed by EMSA on a 5% acrylamide, 0.5× TBE gel.
We performed alanine mutagenesis of the three residues involved in the proposed PB CRD/DNA specific interface (Y558, R567 and K569). The ability of each single mutant to bind a Cy3-labeled LE1–35 substrate was assessed using EMSA (Figure 7B). The R567A mutant exhibited a sharp decrease in its apparent binding affinity (Kd ≈ 2.36 μM), with a complete disappearance of the specific complex and the detection of non-specific shifted complexes only at the highest protein concentration. A similar loss of specific DNA interaction was observed for K569A and, to a lesser extent, for Y558A mutants, as attested by the complete disappearance or strong decrease in the amount of the specific complex (Figure 7B and Supplementary Figure S20A). However, the global apparent affinity for LE1–35 DNA is not significantly decreased for these two mutants (Kd ≈ 0.81 μM or Kd ≈ 0.33 μM, respectively) (Supplementary Figure S20A). All three mutants form similar high molecular shifted complexes with a random DNA substrate (Supplementary Figure S20B). These data confirm that residues Y558, R567 and K569 participate in a sequence-specific mode of interaction with LE1–35. It also supports the notion that loss of specificity reveals an additional non-specific mode of interaction of PB CRD with DNA. Further work is needed to clarify the nature and conformation of the non-specific PB CRD/DNA complexes.
DISCUSSION
Here, we have provided the first structural insight into a functional domain of PB transposase. We have shown by DNase I footprinting and gel shift assays that its C-terminal CRD binds specifically to DNA sequences at the ends of piggyBac, which are required for transposition. We have used NMR to determine the structure of its C-terminal site-specific DNA binding domain in solution and proposed a structural model for the complex formed by the CRD with its target DNA sequence. Our site-directed mutagenesis data further validated the proposed interaction model.
The PB C-terminal CRD is a cross-brace ZF and interacts specifically with DNA sequences
The solution structure of PB CRD revealed that PB(552–594) adopts the specific fold of the cross-brace ZF protein family, which includes various types of PHD, RING and FYVE domains that interact with very diverse targets (60–62). In PB(552–594), the cross-brace ZF is formed by an unusual C3H (ZF1) and C4 (ZF2) coordination mode. Its particular sequential motif of cysteine and histidine Zn2+ ligands, C5HC2, was previously observed only in the RING-type E3 ubiquitin ligase (63) and in the PHD domain of the human JARID1B, a histone specific demethylase (64). Our experiments establish that the PB CRD interacts with a specific sequence on DNA as shown by DNase I footprinting (Figure 2), competition gel shift assays (Figure 3A), and NMR (Figure 5). There are only a few other known examples of DNA-interacting cross-brace ZFs. Among those are the paralogous transcription factors BRPF2 (65) and BRPF1 (66) constituted by two PHD fingers linked by a zinc knuckle: the isolated PHD1 ZF binds to unmodified H3K4me0 and the second PHD2 ZF binds DNA non-specifically. Similarly, the PHD ZF from the PHF6 protein does not interact with histones but rather binds dsDNA or RNA (67). In all these cases, contrary to the results we obtained on the PB CRD, the role and the mechanism of the interaction are not yet well understood (65,66).
DNase I footprinting shows that PB targets unexpectedly three DNA segments on the transposon
Unexpectedly, using DNase I footprinting experiments, we observed that PB not only interacts with the LE and RE Terminal Inverted Repeats (TIRs), but also with an unsuspected internal site at the left end of the transposon (i.e. LI178–227). Interestingly, we noted that the 19-bp repeats of LE and LI are separated by approximately 147-bp, which corresponds to the length of DNA wrapped around one histone octamer within a nucleosome. Thus, the biological relevance of the LI/PB interaction might be explained in the context of chromatin architecture. To explore and help interpret our DNase I footprinting results, we built a model in which a nucleosome is positioned between LE and LI. In such a model, the LE TIR (LE1–35) and LI (beginning at LI180) would be localised in linker DNA and would be brought together as a consequence of chromatin structure (Supplementary Figure S18B). Analysis of several PLEs shows that for each species the same nucleotide sequence is found at the left and right transposon ends but also, as in the case of the piggyBac transposon, in an LI site located around position 180–200 (Supplementary Figure S21), suggesting a conserved role of this LI site.
Piggybac TIRs are schematically made of two distinct repeats: a terminal 13-bp repeat at the very end of the transposon and the internal 19-bp repeat, specifically recognized by PB CRD (Figure 2A). Surprisingly, for full-length PB, strong DNase I protection is mostly observed on the 19-bp repeat on LE, LI and RE, whereas with the truncated PB(1–558) strong protection is shifted towards the terminal 13-bp repeat (Figure 2B–D). We speculate that PB CRD is the driver that targets PB binding to piggyBac TIRs, but that PB harbors other DNA recognition domains. One potential target is a conserved 5′-AAAGATAATC-3′ box found between the 13-bp and 19-bp repeats (shown in blue in Figure 2B–D) and the other includes the tips of the transposon and the flanking TTAA target site duplications, which are specific targets for cleavage by the PB catalytic core. The internal LI site and the right end RE have a more complex organisation than LE, with a longer spacer between the 13-bp and the 19-bp repeats. In both cases, an additional 5′-AAAGATAATC-3′ box is found in the spacer. It remains to be determined whether and how PB binds specifically to the protected sequences located in the spacer between the two 5′-AAAGATAATC-3′ boxes found in LI and RE (Figure 2D).
How can PB transposases interact with DNA in the transpososome context?
Four transpososomes assembled by DD(D/E) transposases have been structurally characterized: the prokaryotic Tn5 transposase in complex with Tn5 transposon end DNAs (68); the Mu transposase in complex with bacteriophage DNA ends and target DNA (69); the eukaryotic mariner Mos1 transposon (70) and Hermes, a member of the hAT transposon family (71). For all of them, the crystal structure reveals that these transposases possess at least a dimerization domain and multiple specific and nonspecific DNA-binding domains. The transposase/DNA complexes all show a trans arrangement, in which each transposon end is bound by the catalytic domain of one of the transposases (at the DNA cleavage site) and by the DNA-binding domain of another transposase. Our DNase I footprinting experiments show that PB contains several DNA-binding domains that recognize different DNA elements. But the strong protection of the 19-bp repeats of LE, LI and RE DNA sequences by the PB CRD is incompatible with a strong protection of 5′ DNA sequences located beside the 19-bp repeat on the same DNA segment. On the contrary, this could be compatible with a transposase/DNA trans arrangement in transpososomes.
It has been shown that truncated versions of piggyBac vectors result in a decrease in transposition efficiency (56). LI is an additional piggyBac transposase binding site, that could increase the transposition efficiency by co-operative binding of the transposase to generate the synaptic complex (72). The three DNA segments targeted by PB (LE, LI and RE) could be held together through a series of PB/DNA interactions, and probably PB/PB contacts, to assemble into a higher-order transpososome structure, in which DNA breakage and joining would occur.
The structure of PB CRD/DNA complexes can be used to design transposase mutants with modified integration and excision capacities
Isolation and characterization of mutant transposases with increased or altered activity is useful for the dissection of protein structure–function relationships and can also be very useful in genome engineering applications. We have previously reported the isolation of a number of hyperactive PB mutants (S509G/N571S, Q591P, Q591R, F594L) (73) and hyperactive Exc+Int−(R372A/K375A) mutants (T560A, S564P, N571S, S573A, M589V, S592G, F594L) that have amino acid changes in their C-terminal tail. Two of the modified residues (S564 and N571) are directly implicated in DNA binding as judged from the structural model presented here, which might contribute to the hyperactive phenotype. Other mutations do not appeared to play a role in DNA binding: T560 and S573 on the one hand, and M589, Q591, S592 and F594 that are located in the C-terminal flexible tail on the other hand (Supplementary Figure S22).
The flexible C-terminus is not implicated in DNA recognition and is oriented on the opposite side of the structure relative to the DNA interacting domain (Figure 6A and B). This hydrophobic and acidic domain (I587, D588, M589, Q591, S592 and F594) harbours relatively well conserved residues (I587 and F594) or residues that have similar chemical properties (as M589, replaced by I or V in other PB-like transposase) (Figure 1B). This suggests that the flexible C-terminus of PB may fulfil other important functions, e.g. promoting protein/protein interactions, that remain to be determined. The structure of PB(552–594) and the structural models of the PB(552–594)/DNA complexes will likely be useful for the rational design of transposase mutants with increased activity and selectivity.
DATA AVAILABILITY
Atomic coordinates of the structures of PB(552–594) have been deposited to the Protein Data Bank under accession code 5LME (http://www.rcsb.org/pdb/home/home.do).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Emeline Dubois for stimulating discussions during the early phase of this work and François Bontems for many stimulating discussions and helpful advice. We thank Dr Yves Mechulam of Ecole polytechnique, Laboratoire de biochimie, Palaiseau, France, for his help on the flame atomic absorption spectrophotometry experiments.
The FP7 WeNMR (project# 261572) and H2020 West-Life (project# 675858) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure and the DIRAC4EGI service with the dedicated support of CESNET-MetaCloud, INFN-PADOVA, NCG-INGRID-PT, RAL-LCG2, TW-NCHC, IFCA-LCG2, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan and the US Open Science Grid
Authors’ contributions: N.M. and E.G. designed and directed the NMR experiments. N.M. and S.M. collected, processed and analyzed the NMR data. N.M. and E.L. generated and analyzed the electrostatic surfaces of the PB CRD and the HADDOCK peptide/DNA complexes. N.A. discussed the results and interpreted the gel shift assay. N.M. generated the protein/DNA complexes and wrote the manuscript with editorial input from all the co-authors.
X.L. designed, performed and interpreted the mammalian cell integration assay and the DNase I footprinting experiments, and the in vitro DNA breakage and joining assays experiments. N.L.C. directed the project and contributed to the writing of the paper.
S.A.W. designed a new ARIA protocol and generated and analysed the PB(552–594) structure. B.B. and M.N. helped to design a new ARIA protocol and interpreted the results.
J.L.T. performed and interpreted the gel shift assay. A.B.H. and F.D. designed, interpreted and directed the gel shift assay and the in vitro DNA breakage and joining experiments, and contributed to the writing of the manuscript.
N. Mathy and J.B. expressed the PB CRD in E. coli and conducted early purification attempts. J.B. obtained the first evidence that PB binds Zn2+ using flame emission spectroscopy and performed the series of point mutations in PB(537–594). J.B. and M.B. discussed the results and implications of the experimental work, and commented on the manuscript at all stages. M.B. contributed to the writing of the main text.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Intramural Research Program of the NIH and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (to J.L.T., A.B.H., F.D.); The Centre National de la Recherche Scientifique (CNRS) and the Agence Nationale de la Recherche (ANR) [ANR-14-CE10-0005-01 (PIGGYPACK) to N.M., E.L., N.A., E.G., N. Mathy, J.B., M.B.]; The Institut de Chimie des Substances Naturelles (ICSN) (to S.M); European Union [FP7-IDEAS-ERC 294809 to M.N.; Howard Hughes Medical Institute, Department of Molecular Biology & Genetics, Johns Hopkins University School of Medicine (to X.L. N.L.C.). Funding for open access charge: Laboratoire de Biologie et Chimie Structurales, Institut de Chimie des Substances Naturelles, CNRS UPR 2301, Université Paris-Saclay, 91198 Gif sur Yvette cedex, France.
Conflict of interest statement. None declared.
REFERENCES
- 1. Feschotte C., Pritham E.J.. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007; 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fraser M.J., Smith G.E., Summers M.D.. Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J. Virol. 1983; 47:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraser M.J., Ciszczon T., Elick T., Bauser C.. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect. Mol. Biol. 1996; 5:141–151. [DOI] [PubMed] [Google Scholar]
- 4. Mitra R., Fain-Thornton J., Craig N.L.. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008; 27:1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun Z.C., Wu M., Miller T.A., Han Z.J.. piggyBac-like elements in cotton bollworm, Helicoverpa armigera (Hubner). Insect. Mol. Biol. 2008; 17:9–18. [DOI] [PubMed] [Google Scholar]
- 6. Luo G.H., Li X.H., Han Z.J., Guo H.F., Yang Q., Wu M., Zhang Z.C., Liu B.S., Qian L., Fang J.C.. Molecular characterization of the piggyBac-like element, a candidate marker for phylogenetic research of Chilo suppressalis (Walker) in China. BMC Mol. Biol. 2014; 15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu C., Wang S.. PLE-wu, a new member of piggyBac transposon family from insect, is active in mammalian cells. J. Biosci. Bioeng. 2014; 118:359–366. [DOI] [PubMed] [Google Scholar]
- 8. Wu M., Sun Z., Luo G., Hu C., Zhang W., Han Z.. Cloning and characterization of piggyBac-like elements in lepidopteran insects. Genetica. 2011; 139:149–154. [DOI] [PubMed] [Google Scholar]
- 9. Luo G.H., Wu M., Wang X.F., Zhang W., Han Z.J.. A new active piggyBac-like element in Aphis gossypii. Insect. Sci. 2011; 18:652–662. [Google Scholar]
- 10. Daimon T., Mitsuhiro M., Katsuma S., Abe H., Mita K., Shimada T.. Recent transposition of yabusame, a novel piggyBac-like transposable element in the genome of the silkworm, Bombyx mori. Genome. 2010; 53:585–593. [DOI] [PubMed] [Google Scholar]
- 11. Wang J., Miller E.D., Simmons G.S., Miller T.A., Tabashnik B.E., Park Y.. piggyBac-like elements in the pink bollworm, Pectinophora gossypiella. Insect. Mol. Biol. 2010; 19:177–184. [DOI] [PubMed] [Google Scholar]
- 12. Wu M., Sun Z.C., Hu C.L., Zhang G.F., Han Z.J.. An active piggyBac-like element in Macdunnoughia crassisigna. Insect. Sci. 2008; 15:521–528. [Google Scholar]
- 13. Ray D.A., Feschotte C., Pagan H.J., Smith J.D., Pritham E.J., Arensburger P., Atkinson P.W., Craig N.L.. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008; 18:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J., Ren X., Miller T.A., Park Y.. piggyBac-like elements in the tobacco budworm, Heliothis virescens (Fabricius). Insect. Mol. Biol. 2006; 15:435–443. [DOI] [PubMed] [Google Scholar]
- 15. Zimowska G.J., Handler A.M.. Highly conserved piggyBac elements in noctuid species of Lepidoptera. Insect. Biochem. Mol. Biol. 2006; 36:421–428. [DOI] [PubMed] [Google Scholar]
- 16. Xu H.F., Xia Q.Y., Liu C., Cheng T.C., Zhao P., Duan J., Zha X.F., Liu S.P.. Identification and characterization of piggyBac-like elements in the genome of domesticated silkworm, Bombyx mori. Mol. Genet. Genomics. 2006; 276:31–40. [DOI] [PubMed] [Google Scholar]
- 17. Sarkar A., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H.. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genomics. 2003; 270:173–180. [DOI] [PubMed] [Google Scholar]
- 18. Handler A.M., McCombs S.D.. The piggyBac transposon mediates germ-line transformation in the Oriental fruit fly and closely related elements exist in its genome. Insect. Mol. Biol. 2000; 9:605–612. [DOI] [PubMed] [Google Scholar]
- 19. Handler A.M., Zimowska G.J., Armstrong K.F.. Highly similar piggyBac elements in Bactrocera that share a common lineage with elements in noctuid moths. Insect. Mol. Biol. 2008; 17:387–393. [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Du Y., Wang S., Brown S.J., Park Y.. Large diversity of the piggyBac-like elements in the genome of Tribolium castaneum. Insect. Biochem. Mol. Biol. 2008; 38:490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitra R., Li X., Kapusta A., Mayhew D., Mitra R.D., Feschotte C., Craig N.L.. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li M.A., Turner D.J., Ning Z., Yusa K., Liang Q., Eckert S., Rad L., Fitzgerald T.W., Craig N.L., Bradley A.. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011; 39:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cary L.C., Goebel M., Corsaro B.G., Wang H.G., Rosen E., Fraser M.J.. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989; 172:156–169. [DOI] [PubMed] [Google Scholar]
- 24. Fraser M.J., Cary L., Boonvisudhi K., Wang H.G.. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995; 211:397–407. [DOI] [PubMed] [Google Scholar]
- 25. Ding S., Wu X., Li G., Han M., Zhuang Y., Xu T.. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005; 122:473–483. [DOI] [PubMed] [Google Scholar]
- 26. Lobo N.F., Fraser T.S., Adams J.A., Fraser M.J. Jr. Interplasmid transposition demonstrates piggyBac mobility in vertebrate species. Genetica. 2006; 128:347–357. [DOI] [PubMed] [Google Scholar]
- 27. Wilson M.H., Coates C.J., George A.L. Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007; 15:139–145. [DOI] [PubMed] [Google Scholar]
- 28. Lu Y., Lin C., Wang X.. PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res. 2009; 37:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yusa K., Rad R., Takeda J., Bradley A.. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009; 6:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saridey S.K., Liu L., Doherty J.E., Kaja A., Galvan D.L., Fletcher B.S., Wilson M.H.. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol. Ther. 2009; 17:2115–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hamalainen R., Cowling R., Wang W., Liu P., Gertsenstein M. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009; 458:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakanishi H., Higuchi Y., Kawakami S., Yamashita F., Hashida M.. piggyBac transposon-mediated long-term gene expression in mice. Mol. Ther. 2010; 18:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Di Matteo M., Matrai J., Belay E., Firdissa T., Vandendriessche T., Chuah M.K.. PiggyBac toolbox. Methods Mol. Biol. 2012; 859:241–254. [DOI] [PubMed] [Google Scholar]
- 34. Elick T.A., Bauser C.A., Fraser M.J.. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996; 98:33–41. [DOI] [PubMed] [Google Scholar]
- 35. Rice P.A., Baker T.A.. Comparative architecture of transposase and integrase complexes. Nat. Struct. Biol. 2001; 8:302–307. [DOI] [PubMed] [Google Scholar]
- 36. Hickman A.B., Chandler M., Dyda F.. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010; 45:50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keith J.H., Schaeper C.A., Fraser T.S., Fraser M.J. Jr. Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol. Biol. 2008; 9:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keith J.H., Fraser T.S., Fraser M.J. Jr. Analysis of the piggyBac transposase reveals a functional nuclear targeting signal in the 94 c-terminal residues. BMC Mol. Biol. 2008; 9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B. Jr et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F.R.. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 2006; 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 41. Griesinger C., Otting G., Wüthrich K., Ernst R.R.. Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Soc. 1988; 110:7870–7872. [Google Scholar]
- 42. Kumar A., Ernst R.R., Wüthrich K.. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980; 95:1–6. [DOI] [PubMed] [Google Scholar]
- 43. Piotto M., Nicholson J.K., Huang H., Sklenar V., Mao X.A.. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992; 2:661–665. [DOI] [PubMed] [Google Scholar]
- 44. Morris K.F., Johnson J.C.S.. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 1992; 114:3139–3141. [Google Scholar]
- 45. Johnson J.C.S. Diffusion ordered nuclear magnetic resonance spectroscopy: Principles and applications. Prog. Nucl. Magn. Reson. Spectrosc. 1999; 34:203–256. [Google Scholar]
- 46. Yao S., Howlett G.J., Norton R.S.. Peptide self-association in aqueous trifluoroethanol monitored by pulsed field gradient NMR diffusion measurements. J. Biomol. NMR. 2000; 16:109–119. [DOI] [PubMed] [Google Scholar]
- 47. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 48. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T.E., Nilges M.. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007; 23:381–382. [DOI] [PubMed] [Google Scholar]
- 49. Bardiaux B., Malliavin T., Nilges M.. ARIA for solution and solid-state NMR. Methods Mol. Biol. 2012; 831:453–483. [DOI] [PubMed] [Google Scholar]
- 50. Bernard A., Vranken W.F., Bardiaux B., Nilges M., Malliavin E.T.. Bayesian estimation of NMR restraint potential and weight: a validation on a representative set of protein structures. Proteins. 2011; 79:1525–1537. [DOI] [PubMed] [Google Scholar]
- 51. de Vries S.J., van Dijk M., Bonvin A.M.J.J.. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010; 5:883–897. [DOI] [PubMed] [Google Scholar]
- 52. van Dijk M., Bonvin A.M.J.J.. a DNA structure modelling server. Nuclear Acids Res. 2009; 37:W235–W239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hikosaka A., Kobayashi T., Saito Y., Kawahara A.. Evolution of the Xenopus piggyBac transposon family TxpB: domesticated and untamed strategies of transposon subfamilies. Mol. Biol. Evol. 2007; 24:2648–2656. [DOI] [PubMed] [Google Scholar]
- 54. Meir Y.J., Weirauch M.T., Yang H.S., Chung P.C., Yu R.K., Wu S.C.. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011; 11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Solodushko V., Bitko V., Fouty B.. Minimal piggyBac vectors for chromatin integration. Gene Ther. 2014; 21:1–9. [DOI] [PubMed] [Google Scholar]
- 56. Li X., Harrell R.A., Handler A.M., Beam T., Hennessy K., Fraser M.J. Jr. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect. Mol. Biol. 2005; 14:17–30. [DOI] [PubMed] [Google Scholar]
- 57. Wüthrich K. NMR of Proteins and Nucleic Acids. 1986; USA: Wiley/Interscince. [Google Scholar]
- 58. Barraud P., Schubert M., Allain F.H.. A strong 13C chemical shift signature provides the coordination mode of histidines in zinc-binding proteins. J. Biomol. NMR. 2012; 53:93–101. [DOI] [PubMed] [Google Scholar]
- 59. van Zundert G.C., Rodrigues J.P., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S., van Dijk M., de Vries S.J., Bonvin A.M.. The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016; 428:720–725. [DOI] [PubMed] [Google Scholar]
- 60. Schindler U., Beckmann H., Cashmore A.R.. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993; 4:137–150. [DOI] [PubMed] [Google Scholar]
- 61. Joazeiro C.A., Weissman A.M.. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000; 102:549–552. [DOI] [PubMed] [Google Scholar]
- 62. Gaullier J.M., Simonsen A., D’Arrigo A., Bremnes B., Stenmark H., Aasland R.. FYVE fingers bind PtdIns(3)P. Nature. 1998; 394:432–433. [DOI] [PubMed] [Google Scholar]
- 63. Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X.. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007; 39:623–630. [DOI] [PubMed] [Google Scholar]
- 64. Guo X., Xu Y., Wang P., Li Z., Xu Y., Yang H.. Crystallization and preliminary crystallographic analysis of a PHD domain of human JARID1B. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011; 67:907–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu L., Qin S., Zhang J., Ji P., Shi Y., Wu J.. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J. Struct. Biol. 2012; 180:165–173. [DOI] [PubMed] [Google Scholar]
- 66. Klein B.J., Muthurajan U.M., Lalonde M.E., Gibson M.D., Andrews F.H., Hepler M., Machida S., Yan K., Kurumizaka H., Poirier M.G. et al. Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res. 2015; 44:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu Z., Li F., Ruan K., Zhang J., Mei Y., Wu J., Shi Y.. Structural and functional insights into the human Borjeson-Forssman-Lehmann syndrome-associated protein PHF6. J. Biol. Chem. 2014; 289:10069–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I.. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000; 289:77–85. [DOI] [PubMed] [Google Scholar]
- 69. Montano S.P., Pigli Y.Z., Rice P.A.. The mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012; 491:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Richardson J.M., Colloms S.D., Finnegan D.J., Walkinshaw M.D.. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009; 138:1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hickman A.B., Ewis H.E., Li X., Knapp J.A., Laver T., Doss A.L., Tolun G., Steven A.C., Grishaev A., Bax A. et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell. 2014; 158:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saedler H., Gierl A.. Transposable Elements. 1996; Berlin Heidelberg: Springer-Verlag. [Google Scholar]
- 73. Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L.. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the structures of PB(552–594) have been deposited to the Protein Data Bank under accession code 5LME (http://www.rcsb.org/pdb/home/home.do).