Abstract
Although subtelomeric regions in humans are heterochromatic, the epigenetic nature of human telomeres remains controversial. This controversy might have been influenced by the confounding effect of subtelomeric regions and interstitial telomeric sequences (ITSs) on telomeric chromatin structure analyses. In addition, different human cell lines might carry diverse epigenetic marks at telomeres. We have developed a reliable procedure to study the chromatin structure of human telomeres independently of subtelomeres and ITSs. This procedure is based on the statistical analysis of multiple ChIP-seq experiments. We have found that human telomeres are not enriched in the heterochromatic H3K9me3 mark in most of the common laboratory cell lines, including embryonic stem cells. Instead, they are labeled with H4K20me1 and H3K27ac, which might be established by p300. These results together with previously published data argue that subtelomeric heterochromatin might control human telomere functions. Interestingly, U2OS cells that exhibit alternative lengthening of telomeres have heterochromatic levels of H3K9me3 in their telomeres.
INTRODUCTION
Telomeres guaranty the replication of chromosome ends and prevent genome instability. In most eukaryotes, telomeric DNA is composed of short double-stranded repeats (1,2). These repeats are also present at interstitial chromosomal loci in a wide variety of organism and have been related to chromosomal aberrations, fragile sites, hot spots for recombination and diseases caused by genomic instability. However, the biological functions of interstitial telomeric sequences (ITSs) remain unknown (3–6).
The epigenetic characteristics of telomeric regions, which include telomeres and subtelomeres, influence telomere functions (2,7–10). The study of the epigenetic modifications present in subtelomeres can be easily achieved by chromatin immunoprecipitation (ChIP) followed by polymerase chain reaction of single-copy sequences. In contrast, the study of telomeres is challenged by subtelomeres and/or ITSs, which are usually present at pericentromeric and subtelomeric regions (5). Whereas telomeres and subtelomeres cannot be differentiated by microscopy techniques, ITSs might interfere with the analysis of telomeric chromatin structure by ChIP followed by hybridization with a telomeric probe (ChIP-hyb). Moreover, ITSs might be identified as telomeres in massively parallel DNA sequencing studies (ChIP-seq) (11,12). Hence, the analysis of the epigenetic modifications present at telomeres should be carefully designed.
The study of telomeres independently of ITSs may be facilitated by the fact that they usually have different sequence organizations. Whereas telomeres are essentially composed of tandem arrays of perfect telomeric repeats, ITSs usually contain perfect telomeric repeats interspersed with degenerate repeats. In fact, it is uncommon for ITSs to contain long stretches of perfect tandem telomeric repeats (6,13).
We have previously studied the epigenetic modifications of Arabidopsis thaliana telomeres independently of ITSs by analyzing genome-wide ChIP-seq data. Since Arabidopsis ITSs contain few arrays of perfect telomeric repeats, we could identify as telomeric reads those that contained four perfect tandem telomeric repeats. The analyses of the ChIP-seq experiments and other results led us to establish that Arabidopsis telomeres are not heterochromatic. In turn, subtelomeric regions and ITSs exhibit heterochromatic features in Arabidopsis (11,12,14,15).
The epigenetic nature of human telomeres remains controversial (5,8,9,16). While some groups propose that telomeres are heterochromatic, other groups have reported that telomeres have low levels of heterochromatic marks and exhibit euchromatic modifications. We consider that these differences might have arisen, at least in part, from experimental limitations like those mentioned above. In addition, different human cell lines might have distinct epigenetic features at telomeres.
Here, we have refined our previous analyses of ChIP-seq data to study the chromatin structure of telomeres in human cells. We have performed statistical analyses of multiple ChIP-seq experiments. Our results reveal that human telomeres have lower levels of H3K9me3 than the heterochromatic Satellites II and III in most of the commonly studied laboratory cell lines. Besides, we have found that human telomeres are enriched in the euchromatic H4K20me1 and H3K27ac marks. In contrast, U2OS cells that maintain their telomeres through ALT exhibit heterochromatic levels of H3K9me3.
MATERIALS AND METHODS
Determination of the relative amounts of perfect telomeric sequences at telomeres and ITSs
To identify telomeric reads in human ChIP-seq studies, we first determined the number of times that sequences containing 4, 5 or 6 perfect tandem telomeric repeats are contained in human ITSs and telomeres. We analyzed ITSs running a Mega Blast search at the NCBI Mapviewer home (http://www.ncbi.nlm.nih.gov/mapview/), using the Build reference database. The search parameters were the following: automatically adjust parameters for short input sequences; expect threshold = 10; max matches in a query range = 0; match/mismatch scores = 1,-2; gap cost = linear; word size = 256 and without filters or repeat masking.
To determine the number of times that telomeric sequences containing 4, 5 or 6 perfect tandem telomeric repeats were present at telomeres, we multiplied the number of human haploid chromosomes by 2 and by the average telomere length previously reported for humans expressed in bp (17,18). Then, we divided the resulting values by the number of base pair in each perfect telomeric sequence.
The relative abundance of telomeric sequences containing 4, 5 or 6 tandem repeats at ITSs versus telomeres is indicated in Supplementary Table S1. We considered that sequences rendering ratios equal or lower than 2% could represent human telomeres in genome-wide ChIP-seq studies. Following a similar procedure we determined the number of telomeric repeats that could represent telomeres in ChIP-seq experiments performed with other model systems including Mus musculus, Danio rerio, Chlamydomonas reinhardtii and Caenorhabditis elegans (Supplementary Table S1).
Determination of enrichment values
The enrichment levels of the different epigenetic marks were first calculated using multiple ChIP-seq experiments released by one study of the ENCODE consortium (SRP006944; Supplementary Table S2) (19). This study reported the analysis of 10 different epigenetic marks in 9 cell lines. Additionally, enrichment values were also calculated using experiments released by other studies (Supplementary Tables S3 and 4). For each study, the Fastq files of all the experiments, including the input and the immunoprecipitated samples, were downloaded from the ENA (European Nucleotide Archive) repository and aligned with the human hg19 reference genome using Bowtie2 (20). Experiments with more than 25% of unmapped reads were not considered for further analyses. We decided to discard low mappability experiments because they often arise from defective DNA sequencing, which most probably affects differentially to specific DNA sequences like those found at telomeres.
The reads containing the sequences (CCCTAA)5 or (TTAGGG)5 were identified as telomeric since these sequences were essentially found at telomeres (Supplementary Table S1). Both kinds of telomeric reads were counted and used to calculate enrichment values. In addition, the reads containing the sequences ATTCCATTCGATTCCATTCG or ATTCCATTCCATTCCATTCCATTCCATTCC were used to calculate the enrichment levels of Satellites II or III, respectively (21). Thus, four different kinds of reads were analyzed. The frequency of each kind of read was calculated for every ChIP-seq experiment by dividing the corresponding number of reads by the total number of mapped reads. However, when the number of reads was below 40 the corresponding frequencies were not used for further calculations to avoid low coverage bias.
Enrichment values for each kind of read and cell type were calculated by dividing the immunoprecipitation frequencies between the frequencies of the corresponding input samples and expressed as log2. Then, for every specific mark, enrichment values were pooled together and statistical levels of significance were determined using the Student’s t-test or the test of Wilcoxon, depending on whether the distributions of enrichments were normal or not according to the Shapiro–Wilk test.
RESULTS
Identification of telomeric reads in human ChIP-seq studies
To identify the reads that represent telomeres in ChIP-seq experiments, we first determined the number of times that an array containing n perfect telomeric repeats [(CCCTAA)n] is present in human telomeres or ITSs. We found that telomeric arrays containing more than four repeats are essentially located in telomeres (Supplementary Table S1). Thus, reads containing four or more tandem telomeric repeats should represent telomeres, and not ITSs, in genome-wide ChIP-seq experiments. Considering that the reads in the experiments analyzed here contained 36 bp or more, we decided to identify as telomeric reads those that contained five perfect tandem telomeric repeats. In addition, and for comparison, we determined the number of reads arising from the heterochromatic Satellites II and III using their consensus sequences (21).
Epigenetic features of human telomeres
To decipher the epigenetic modifications that label human telomeres, we first decided to analyze multiple ChIP-seq experiments released by one study of the ENCODE consortium (19). Different epigenetic states were defined in this study based on the genome-wide distribution of 10 epigenetic marks in different cell lines. The marks were H3K9me3, H3K27me3, H3K36me3, H4K20me1, H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K9ac and H3K72me2. The combination of these marks in all the cell lines analyzed rendered 15 epigenomic states. One of these states corresponds to heterochromatin and is characterized by the presence of H3K9me3. The rest of the states correspond to euchromatin and associate with specific combinations of the epigenetic marks (19).
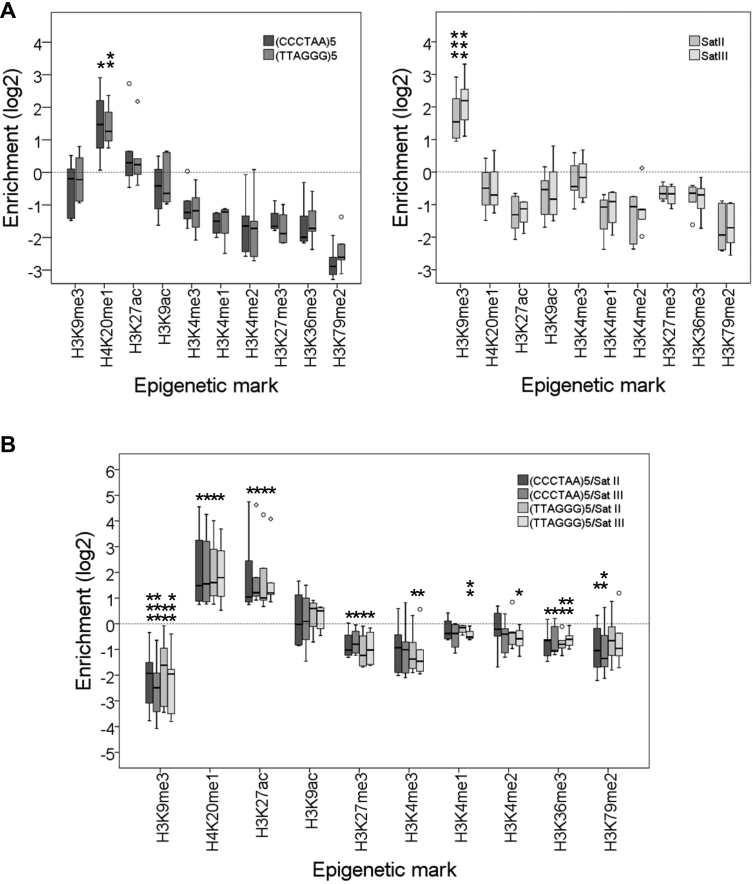
We have analyzed the telomeric enrichment of all the epigenetic marks mentioned above pooling together the data obtained for different cell lines (Supplementary Table S2). Thus, our analyses also reveal the consensus epigenetic characteristics of different human cell lines. These cell lines included embryonic stem cells (H1 ES), cancer cells like K562 or HepG2 and primary cells like HSMM, NHLF and HMEC. In agreement with the aforementioned classification of epigenetic states, we found that the heterochromatic Satellites II and III are enriched in H3K9me3 (Figure 1A). However, we did not find enriched levels of H3K9me3 at telomeres. Instead, telomeres exhibit increased levels of H4K20me1 (Figure 1A). In addition, we found that telomeres are enriched in H4K20me1 and H3K27ac with regard to Satellites II and III in all the cellular lines analyzed. Conversely, Satellites II and III are enriched in H3K9me3 with regard to telomeres (Figure 1B). Thus, our results argue that human telomeres are not heterochromatic but associate with the euchromatic H4K20me1 and H3K27ac marks.
Figure 1.
Human telomeres are not heterochromatic in commonly studied human cell lines. (A) Box plot representation of the enrichment levels of different histone modification at telomeres [(CCCTAA)5 and (TTAGGG)5] and at Satellites II and III. Asterisks label enriched marks. (B) Box plot representation of the enrichment levels of different histone modifications at telomeres versus Satellites II and III. Asterisks label enriched and depleted marks. Enrichment levels are expressed as the log2 of enrichment values. *P< 0.1, **P< 0.05, ***P< 0.01. These representations have been performed using data released by one study of the ENCODE consortium (Supplementary Table S2). Although nine different cell lines were analyzed in this study, the representations shown above only include data from six cell lines because the inputs of the remaining lines did not fulfill our mappability requirements (see ‘Materials and Methods section’). The cell lines analyzed include H1 embryonic stem cells, cancer cells like K562 or HepG2 and primary cells like HSMM, NHLF and HMEC.
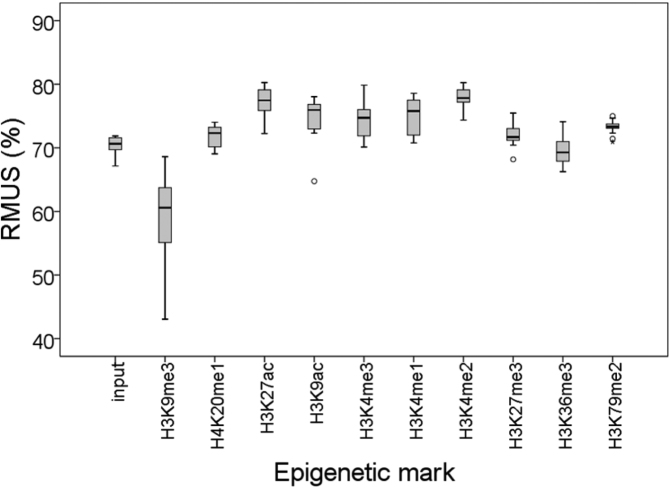
Since heterochromatin is known to be enriched in different kinds of repetitive elements, we decided to analyze whether H3K9me3 preferentially associates with repetitive sequences. Not surprisingly, we found that H3K9me3 had a higher tendency to associate with repetitive sequences than the rest of the epigenetic marks analyzed here, which corroborates that H3K9me3 is the hallmark that label heterochromatin (Figure 2).
Figure 2.
H3K9me3 preferentially associates with repetitive sequences. A box plot representation of the percentage of Reads Mapping to Unique Sequences (RMUS) for each epigenetic modification analyzed in Figure 1 is shown (see Supplementary Table S2).
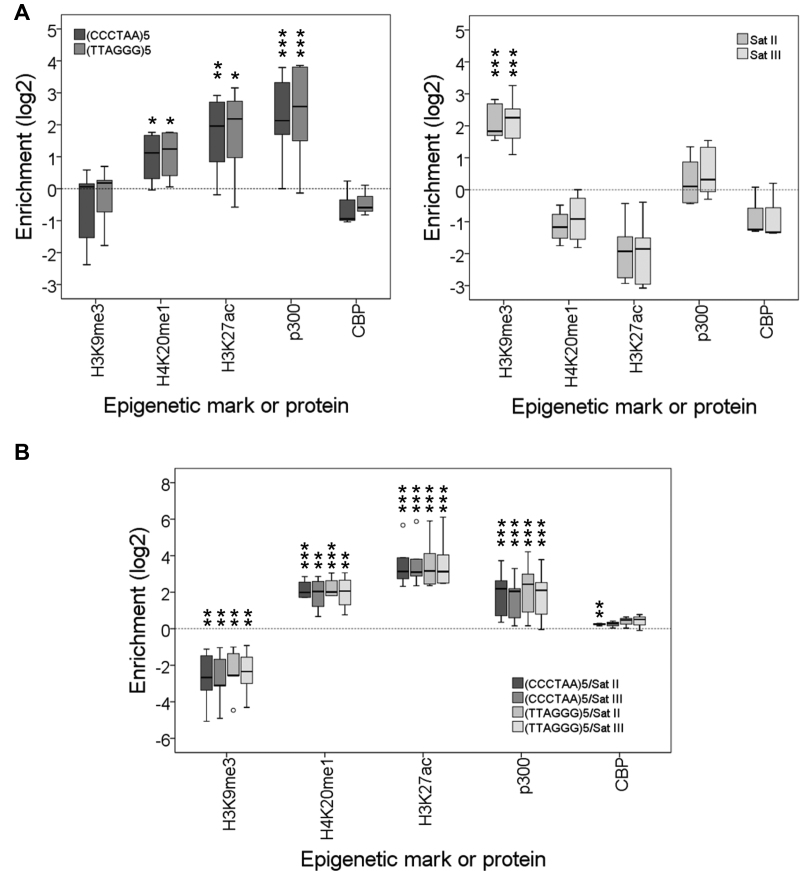
To further verify that telomeres do not have heterochromatic levels of H3K9me3 in commonly studied laboratory cell lines, we decided to study additional ChIP-seq experiments (Supplementary Table S3). For that purpose, we pooled together and analyzed data obtained from different cell lines including HELA, SEM, A549, Dnd41, Osteoblasts and CD14+ (Figure 3). The results obtained confirmed our previous findings corroborating that telomeres in most of the commonly studied human cell lines are not heterochromatic according to their epigenetic features. Instead, they are labeled with H4K20me1 and H3K27ac. Interestingly, a recent report also describes the presence of H3K27ac in HepG2, K562 and CD14+ cell lines (see Supplementary comments) (22).
Figure 3.
Human telomeres are labeled with H4K20me1 and H3K27ac and associate with p300. (A) Box plot representation of the enrichment levels of different histone modifications or proteins at telomeres [(CCCTAA)5 and (TTAGGG)5] and at Satellites II and III. Asterisks label enriched marks. (B) Box plot representation of the enrichment levels at telomeres versus Satellites II and III. Asterisks label enriched and depleted marks. Enrichment levels are expressed as the log2 of enrichment values. *P<0.1, **P<0.05, ***P<0.01. These representations have been performed using data released by multiple labs using different human cell lines (Supplementary Table S3). The cell lines analyzed include HELA, SEM, A549, Dnd41, CD14+, LP-1, H1, NCC, HEK293, MCF-7, T47-D, MADS brite, MADS white and Osteoblasts.
Since H3K27ac is known to be established by p300 and CBP acetyltransferases (23–25), we studied different genome-wide studies that addressed the presence of these proteins in human cells. The cell lines analyzed were H1, NCC, HEK293, MCF-7, T47-D, LP-1, MADS brite, MADS white and Osteoblasts (Supplementary Table S3). Our results show that p300, but not CBP, associates with telomeres and support that this enzyme catalyzes the acetylation of lysine 27 in histone H3 (Figure 3).
Telomeres in U2OS cells exhibit heterochromatic levels of H3K9me3
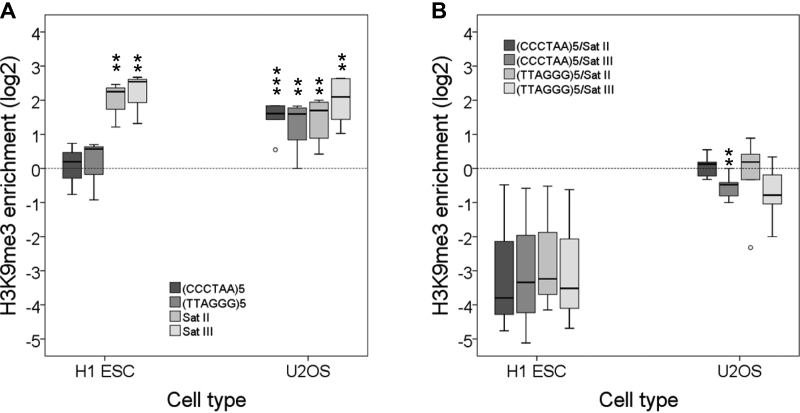
Certain cancer cell lines do not maintain their telomeres through the action of telomerase. Instead, these cells elongate their telomeres by a recombinational mechanism that involves changes in DNA sequence and protein composition. Hence, this alternative lengthening of telomeres (ALT) alters telomeric chromatin structure (26). We took advantage of several genome-wide experiments that addressed the presence of H3K9me3 in U2OS cells, which elongate their telomeres through ALT, to study the presence of this heterochromatic mark at telomeres (Supplementary Table S4). Interestingly, we found that the telomeres of U2OS cells have heterochromatic levels of H3K9me3, similar to those associated with Satellites II and III. As a control we verified that the telomeres of H1 stem cells are not enriched in H3K9me3 (Figure 4).
Figure 4.
Heterochromatic levels of H3K9me3 are found in the telomeres of U2OS cells. (A) H3K9me3 enrichment levels at telomeres [(CCCTAA)5 and (TTAGGG)5] and at Satellites II and III in U2OS and H1 embryonic stem cells. (B) H3K9me3 enrichment levels at telomeres versus Satellites II and III. Enrichment levels are expressed as the log2 of enrichment values. **P< 0.05, ***P< 0.01. These representations have been performed using data released by different laboratories (Supplementary Table S4).
DISCUSSION
Telomeres in most commonly studied human cell lines exhibit euchromatic features
As mentioned above, the analysis of the epigenetic marks that label telomeres might be challenged by the influence of subtelomeres and/or ITSs (5). Whereas telomeres and subtelomeres cannot be differentiated by microscopy techniques, ITSs might interfere with the analyses of telomeric chromatin structure by ChIP. Thus, microscopy and ChIP techniques reveal the chromatin structure of telomeric regions (including telomeres and subtelomeres) or telomeric repeats (including telomeres and ITSs), respectively. Accordingly, hereinafter we will refer to telomeric regions or telomeric repeats.
Euchromatic features have been associated with human telomeric repeats. The levels of H3K9me3 have been mentioned to be relatively low in the telomeric repeats of human fibroblasts (27). In addition, the transcriptional regulator mixed lineage leukemia, a mammalian trithorax-group gene product, binds to telomeric repeats and mono, di and trimethylates H3K4 in HS68 primary fibroblasts (28). In agreement with these results, a study based on genome-wide ChIP-seq experiments has reported low levels of H3K9me3 associated with the telomeric repeats in CD4+ cells (29). The most significant telomeric modifications found in this study were euchromatic marks (H2BK5me1 and H3K4me3). However, it is not clear whether telomeres were analyzed independently of ITSs in this study because the sequence used to identify telomeric reads was not indicated.
Although some studies have reported that human telomeric repeats associate with low levels of heterochromatic marks, other reports have described that they are heterochromatic. TElomeric Repeats containing RNAs (TERRA) are transcribed from subtelomeric regions toward the telomeres (30) and have been proposed to participate in heterochromatin formation at human telomeres (31,32). siRNA depletion of TERRA causes a decrease of H3K9me3 association with the telomeric repeats of HCT116 cells (32,33). In addition, overexpression of telomerase leads to telomeres elongation and higher levels of H3K9me3 at telomeric repeats (31). This and other results have led to the idea that long telomeres are more heterochromatic than short telomeres (16). However, late passage HS68 primary fibroblasts, which have short telomeres, have lower levels of H3K4 methylation at telomeric repeats than early passage fibroblast (28).
The aforementioned data reveal that the heterochromatic nature of human telomeres is controversial. To try to address this issue we have studied the epigenetic modifications of human telomeres by performing statistical analyses of multiple ChIP-seq experiments (19). Our experimental approach involved pooling together data corresponding to different cell lines. Thus, we could integrate multiple data for every epigenetic modification and perform statistical significance analyses. In addition, we minimized the bias associated with the study of repetitive sequences discarding low mappability and coverage experiments. This experimental procedure has allowed us to establish that human telomeres are not enriched in H3K9me3 in many common laboratory cell lines. In turn, telomeres are labeled with H4K20me1 and H3K27ac. Hence, our studies reveal that telomeres exhibit euchromatic features in most commonly studied human cell lines.
H3K27ac is related to enhancer activity and can be established by p300/CBP (23–25). We investigated the presence of these two proteins in human telomeres by analyzing some previously reported ChIP-seq studies that addressed the genome-wide distribution of these proteins (Supplementary Table S3). Interestingly, we found that p300, but not CBP, associates with human telomeres. This result and, in general, our experimental approach is supported by the fact that p300 co-immunocytolocalize with TRF2 and acetylates it, which is important for telomere function (34). Hence, p300 might catalyze the acetylation of H3K27 in human telomeres.
Although we have shown that human telomeres are euchromatic according to their epigenetic marks, telomeres are known to exhibit heterochromatic features according to their compactness and replication time. However, they should not be considered heterochromatin. Telomeres have a short nucleosomal spacing and organize into a compact structure according to cytological observations (35,36). Nevertheless, their overall micrococcal nuclease digestion patterns are similar to the patterns of bulk chromatin. Indeed, telomeric mononucleosomes are more accessible to micrococcal nuclease than bulk mononucleosomes (37). As a consequence, telomeres can eventually render lower amounts of mononucleosomes than bulk chromatin after digestion with micrococal nuclease (35,38). In turn, the heterochromatic nucleosomes present in chromocenters have longer spacing than the telomeric nucleosomes and, after micrococcal nuclease digestion, render mononucleosomes that are more resistant to micrococcal nuclease cleavage (37–39). Hence, the chromatin structure of telomeres is different to that of the heterochromatic pericentromeric chromocenters. In addition, the late replication of telomeres during the cell cycle should be related to subtelomeric heterochromatin. The epigenetic marks of heterochromatin are known to contribute to their late replication. In Schizosaccharomyces pombe, H3K9me3 have been shown to control the replication of late firing origins (40). Similarly, H3K9me3 has been proposed to prime origins for late replication in humans (41). Indeed, HP1 binds to H3K9me3 and to the nuclear lamina, which could delay replication timing (42). Hence, telomeres might replicate late during the cell cycle due, at least in part, to the late firing of the origins present in subtelomeric heterochromatic regions.
Influence of subtelomeric heterochromatin on telomere functions
Subtelomeric regions in humans are heterochromatic and exhibit dense and widespread H3K9me3 and CpG methylation (43–45). Human subtelomeric heterochromatin can silence the expression of nearby located genes and TERRA, which associate with TRF2, HP1 and H3K9me3. This silencing is referred to as telomere position effect (30,32,46–48).
Mutations in different proteins involved in the formation of heterochromatin including DNMT3B, SUV39H1, HP1α, SIRT6 or WRN impair subtelomeric heterochromatin formation and can eventually lead to telomere shortening, to the appearance of telomeric-induced foci, to a p53-dependent DNA damage response and to cellular senescence (31,45,49–53). A direct role of subtelomeric heterochromatin on these processes is argued by the fact that targeting of DNMT3A to subtelomeric regions results in increased levels of subtelomeric DNA methylation and telomere lengthening (54). Moreover, HP1-γ has been shown to bind to the shelterin protein TIN2 and is required for telomere cohesion during S-phase (55). Since we have found that human telomeres are not heterochromatic in most commonly studied human cell lines, our results together with the aforementioned data support that subtelomeric heterochromatin controls human telomere functions. Indeed, the loss of subtelomeric DNA methylation in ICF syndrome cells leads to high levels of TERRA that form telomeric R-loops which, in turn, can cause telomere dysfunction (56).
We have found that U2OS cells, which maintain their telomeres through an aberrant pathway associated with cancer (ALT), exhibit heterochromatic levels of H3K9me3 at telomeres. In turn, H1 embryonic stem cells telomeres do not show enriched levels of H3K9me3 (Figure 4). The absence of H3K9me3 enrichment in H1 telomeres is in apparent contradiction with previously reported immunocytolocalization studies. These studies show that 12–27% of the TRF1 foci co-localize with H3K9me3 in H1 embryonic stem cells. In addition, 1–5% of the TRF2 foci have been shown to co-localize with HP1 (isoforms α or γ) in HT1080 cells (31,57). These results reflect the presence of heterochromatin in human telomeric regions. However, certain levels of co-immunocytolocalization of TRF1 or TRF2 with H3K9me3 or HP1 could be explained by the existence of H3K9me3 mountains, which have a length of 20–200 Kbp and are present in about 30% of the subtelomeric regions in human embryonic stem cells (45).
Telomeres in ALT cells have heterogeneous length and contain degenerated telomeric repeats that might arise through recombination with subtelomeric regions (26,58,59). Hence, ALT telomeres become ‘subtelomeric’ according to their sequence composition. Similarly, our results argue that U2OS telomeres become ‘subtelomeric’ according to their levels of H3K9me3. It will be interesting to address whether heterochromatic levels of H3K9me3 are also found in the telomeres of other cell lines undergoing ALT. The telomeric levels of H3K9me3 have been previously studied by ChIP followed by hybridization with a telomeric probe in ALT positive and telomerase positive fibroblasts (60). This study has reported lower density of H3K9me3 in the telomeric repeats of ALT cells than in the telomeric repeats of telomerase positive cells. Since the ratio of H3K9me3/H3 found in the telomeric repeats of both kinds of fibroblasts was similar, this difference was ascribed to a lower nucleosome density in the ALT fibroblasts. However, the levels of H3K9me3 present in the telomeric repeats of both kinds of fibroblasts were not compared with those of known heterochromatic loci, such as the α-satellite. In addition, we believe that the sensitivity to micrococcal nuclease and nucleosomal spacing of telomeres found for the ALT fibroblast are compatible with a heterochromatic structure. Hence, future studies should further characterize the epigenetic characteristics of telomeric regions in ALT cells.
In summary, we have seen that telomeres in most of the commonly studied human cell lines are not heterochromatic but exhibit euchromatic marks. However, heterochromatin is known to play a major role in telomere biology, which suggests that the integrity of subtelomeric heterochromatin might be important for the proper functioning of telomeres, as previously proposed for A. thaliana (10,14). Interestingly, we have also seen that certain cancer cells that maintain their telomeres through ALT have heterochromatic levels of H3K9me3 in their telomeres. These findings open new perspectives in the study of human telomere epigenetics that might influence the design of strategies to limit undesired cell proliferation. It will be interesting to ascertain whether heterochromatic telomeres are also present in other specific cell types or in cells with mutations that lead to telomere dysfunction and illness.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to NCBI, ENA and Galaxy for sharing their resources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy, Industry and Competitiveness (BIO2016–78955-P) to M.A.V.; European Research Council (ERC2014 AdG669898 TARLOOP); Spanish Ministry of Economy and Competitiveness (BFU2016–75058-P); Worldwide Cancer Research (15–0098) to A.A. Funding for open access charge: Spanish Ministry of Economy, Industry and Competitiveness (BIO2016-78955-P).
Conflict of interest statement. None declared.
REFERENCES
- 1. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 2. Blackburn E.H. Telomeres and telomerase: the means to the end (Nobel lecture). Angew. Chem. 2010; 49:7405–7421. [DOI] [PubMed] [Google Scholar]
- 3. Meyne J., Baker R.J., Hobart H.H., Hsu T.C., Ryder O.A., Ward O.G., Wiley J.E., Wurster-Hill D.H., Yates T.L., Moyzis R.K.. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990; 99:3–10. [DOI] [PubMed] [Google Scholar]
- 4. Richards E.J., Goodman H.M., Ausubel F.M.. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991; 19:3351–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaquero-Sedas M.I., Vega-Palas M.A.. On the chromatin structure of eukaryotic telomeres. Epigenetics. 2011; 6:1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin K.W., Yan J.. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutat. Res. 2008; 658:95–110. [DOI] [PubMed] [Google Scholar]
- 7. Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007; 8:299–309. [DOI] [PubMed] [Google Scholar]
- 8. Galati A., Micheli E., Cacchione S.. Chromatin structure in telomere dynamics. Front. Oncol. 2013; 3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giraud-Panis M.J., Pisano S., Benarroch-Popivker D., Pei B., Le Du M.H., Gilson E.. One identity or more for telomeres?. Front. Oncol. 2013; 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vega-Vaquero A., Bonora G., Morselli M., Vaquero-Sedas M.I., Rubbi L., Pellegrini M., Vega-Palas M.A.. Novel features of telomere biology revealed by the absence of telomeric DNA methylation. Genome Res. 2016; 26:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vaquero-Sedas M.I., Luo C., Vega-Palas M.A.. Analysis of the epigenetic status of telomeres by using ChIP-seq data. Nucleic Acids Res. 2012; 40:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaquero-Sedas M.I., Vega-Palas M.A.. Differential association of Arabidopsis telomeres and centromeres with histone H3 variants. Sci. Rep. 2013; 3:1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gámez-Arjona F.M., López-López C., Vaquero-Sedas M.I., Vega-Palas M.A.. On the organization of the nucleosomes associated with telomeric sequences. Biochim. Biophys. Acta. 2010; 1803:1058–1061. [DOI] [PubMed] [Google Scholar]
- 14. Vaquero-Sedas M.I., Gamez-Arjona F.M., Vega-Palas M.A.. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011; 39:2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K.. siRNA–mediated methylation of Arabidopsis telomeres. PLos Genet. 2010; 6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ichikawa Y., Nishimura Y., Kurumizaka H., Shimizu M.. Nucleosome organization and chromatin dynamics in telomeres. Biomol. Concepts. 2015; 6:67–75. [DOI] [PubMed] [Google Scholar]
- 17. Anchelin M., Murcia L., Alcaraz-Perez F., Garcia-Navarro E.M., Cayuela M.L.. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS One. 2011; 6:e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siomos M., Riha K.. Wendel J. Telomeres and their biology. Plant Genome Diversity. 2012; 1:Wien: Springer; 71–81. [Google Scholar]
- 19. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Capurso D., Xiong H., Segal M.R.. A histone arginine methylation localizes to nucleosomes in satellite II and III DNA sequences in the human genome. BMC Genomics. 2012; 13:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Negishi Y., Kawaji H., Minoda A., Usui K.. Identification of chromatin marks at TERRA promoter and encoding region. Biochem. Biophys. Res. Commun. 2015; 467:1052–1057. [DOI] [PubMed] [Google Scholar]
- 23. Holmqvist P.H., Mannervik M.. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013; 4:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tie F., Banerjee R., Stratton C.A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M.O., Scacheri P.C., Harte P.J.. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009; 136:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K.. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011; 30:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Sullivan R.J., Almouzni G.. Assembly of telomeric chromatin to create ALTernative endings. Trends Cell Biol. 2014; 24:675–685. [DOI] [PubMed] [Google Scholar]
- 27. O'Sullivan R.J., Kubicek S., Schreiber S.L., Karlseder J.. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010; 17:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caslini C., Connelly J.A., Serna A., Broccoli D., Hess J.L.. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol. Cell. Biol. 2009; 29:4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosenfeld J.A., Wang Z., Schones D.E., Zhao K., DeSalle R., Zhang M.Q.. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009; 10:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 31. Arnoult N., Van Beneden A., Decottignies A.. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat. Struct. Mol. Biol. 2012; 19:948–956. [DOI] [PubMed] [Google Scholar]
- 32. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng Z., Wang Z., Stong N., Plasschaert R., Moczan A., Chen H.S., Hu S., Wikramasinghe P., Davuluri R.V., Bartolomei M.S. et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012; 31:4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Her Y.R., Chung I.K.. p300-mediated acetylation of TRF2 is required for maintaining functional telomeres. Nucleic Acids Res. 2013; 41:2267–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makarov V.L., Lejnine S., Bedoyan J., Langmore J.P.. Nucleosomal organization of telomere-specific chromatin in rat. Cell. 1993; 73:775–787. [DOI] [PubMed] [Google Scholar]
- 36. Bandaria J.N., Qin P., Berk V., Chu S., Yildiz A.. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016; 164:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tommerup H., Dousmanis A., de Lange T.. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994; 14:5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inoue A., Hyle J., Lechner M.S., Lahti J.M.. Mammalian ChlR1 has a role in heterochromatin organization. Exp. Cell Res. 2011; 317:2522–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan J., Xu L., Crawford G., Wang Z., Burgess S.M.. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol. Cell. Biol. 2006; 26:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zofall M., Smith D.R., Mizuguchi T., Dhakshnamoorthy J., Grewal S.I.. Taz1-shelterin promotes facultative heterochromatin assembly at chromosome-internal sites containing late replication origins. Mol. Cell. 2016; 62:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Khan A., Marks A.B., Smith O.K., Giri S., Lin Y.C., Creager R., MacAlpine D.M., Prasanth K.V., Aladjem M.I. et al. Temporal association of ORCA/LRWD1 to late-firing origins during G1 dictates heterochromatin replication and organization. Nucleic Acids Res. 2016; 45:2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith O.K., Aladjem M.I.. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J. Mol. Biol. 2014; 426:3330–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ng L.J., Cropley J.E., Pickett H.A., Reddel R.R., Suter C.M.. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009; 37:1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brock G.J., Charlton J., Bird A.. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999; 240:269–277. [DOI] [PubMed] [Google Scholar]
- 45. Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A. et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015; 348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baur J.A., Zou Y., Shay J.W., Wright W.E.. Telomere position effect in human cells. Science. 2001; 292:2075–2077. [DOI] [PubMed] [Google Scholar]
- 47. Arnoult N., Karlseder J.. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015; 22:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A.. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990; 63:751–762. [DOI] [PubMed] [Google Scholar]
- 49. Yehezkel S., Segev Y., Viegas-Pequignot E., Skorecki K., Selig S.. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008; 17:2776–2789. [DOI] [PubMed] [Google Scholar]
- 50. Deng Z., Campbell A.E., Lieberman P.M.. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle. 2010; 9:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sagie S., Ellran E., Katzir H., Shaked R., Yehezkel S., Laevsky I., Ghanayim A., Geiger D., Tzukerman M., Selig S.. Induced pluripotent stem cells as a model for telomeric abnormalities in ICF type I syndrome. Hum. Mol. Genet. 2014; 23:3629–3640. [DOI] [PubMed] [Google Scholar]
- 52. Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008; 452:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tennen R.I., Bua D.J., Wright W.E., Chua K.F.. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011; 2:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choudhury S.R., Cui Y., Narayanan A., Gilley D.P., Huda N., Lo C.L., Zhou F.C., Yernool D., Irudayaraj J.. Optogenetic regulation of site-specific subtelomeric DNA methylation. Oncotarget. 2016; 7:50380–50391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Canudas S., Houghtaling B.R., Bhanot M., Sasa G., Savage S.A., Bertuch A.A., Smith S.. A role for heterochromatin protein 1gamma at human telomeres. Genes Dev. 2011; 25:1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sagie S., Toubiana S., Hartono S.R., Katzir H., Tzur-Gilat A., Havazelet S., Francastel C., Velasco G., Chedin F., Selig S.. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 2017; 8:14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng S., Liu L., Sun Y., Xie P., Hu L., Yuan D., Chen D., Ouyang Q., Lin G., Lu G.. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J. Cell Sci. 2014; 127:752–762. [DOI] [PubMed] [Google Scholar]
- 58. Varley H., Pickett H.A., Foxon J.L., Reddel R.R., Royle N.J.. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 2002; 30:301–305. [DOI] [PubMed] [Google Scholar]
- 59. Conomos D., Stutz M.D., Hills M., Neumann A.A., Bryan T.M., Reddel R.R., Pickett H.A.. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012; 199:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Episkopou H., Draskovic I., Van Beneden A., Tilman G., Mattiussi M., Gobin M., Arnoult N., Londono-Vallejo A., Decottignies A.. Alternative lengthening of telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014; 42:4391–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.