Abstract
Genetic switches must alternate between states whose probabilities are dependent on regulatory signals. Classical examples of transcriptional control in bacteria depend on repressive DNA loops anchored by proteins whose structures are sensitive to small molecule inducers or co-repressors. We are interested in exploiting these natural principles to engineer artificial switches for transcriptional control of bacterial genes. Here, we implement designed homodimeric DNA looping proteins (‘Transcription Activator-Like Effector Dimers’; TALEDs) for this purpose in living bacteria. Using well-studied FKBP dimerization domains, we build switches that mimic regulatory characteristics of classical Escherichia coli lactose, galactose and tryptophan operon promoters, including induction or co-repression by small molecules. Engineered DNA looping using TALEDs is thus a new approach to tuning gene expression in bacteria. Similar principles may also be applicable for gene control in eukaryotes.
INTRODUCTION
Regulated control of gene expression is a key feature of living cells. Classical examples include the Escherichia coli lactose, galactose and tryptophan operons (1–3). These operons are regulated by genetic ‘off’ or ‘on’ switches that prevent or enhance RNA polymerase access to the gene promoter, respectively. Regulatory control typically does not involve changing the concentration of regulatory proteins, but rather changing concentrations of small molecules that induce allosteric changes in these regulatory proteins. We are interested in engineering new gene regulatory switches in bacteria based on natural principles.
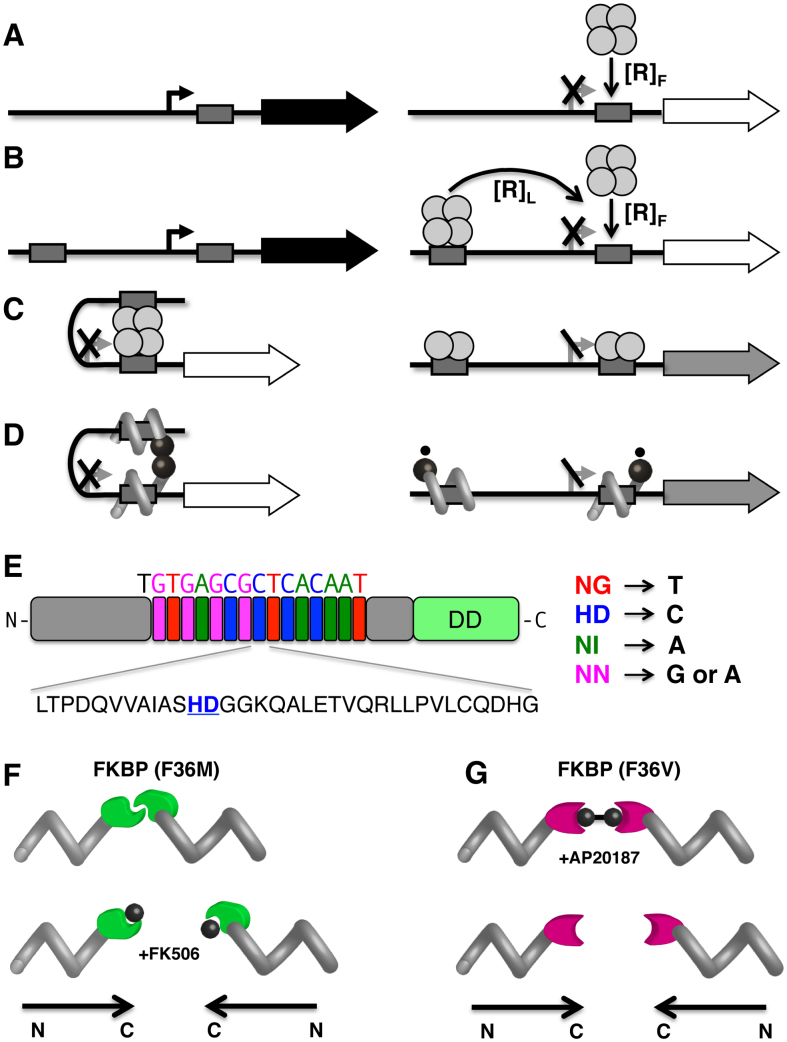
DNA looping is exploited as a fundamental principle in bacterial gene repression (4–12). DNA looping represses transcription initiation by at least two mechanisms. First, for bidentate repressor proteins like Lac repressor (LacI), protein saturation of an operator DNA sequence overlapping the RNA polymerase binding site can be increased by increasing the total local repressor concentration by contributions from repressors looping from a distant site (5,11,13,14) compare Figure 1A and B). Second, trapping of a promoter within a strained DNA loop may be intrinsically repressive (15–18). While small molecule-induced allosteric affects in natural gene control switches typically alter the affinity of regulatory proteins for DNA (Figure 1C), here we explore switches where it is protein dimerization that is regulated by the small molecule (Figure 1D).
Figure 1.
Engineering DNA regulatory loops by analogy with natural bacterial repressors. (A) Constitutive ‘off’ switch. ‘On’ state (left) involves an unoccupied operator (grey box) at the promoter (broken arrow) allowing transcription of the downstream gene (filled arrow). ‘Off" state (right) involves a repressor protein (shown as a tetramer) binding at the operator, preventing promoter access by RNA polymerase. (B) Enhancing effective repressor concentration by DNA looping. Two unoccupied operators (grey boxes) flank the promoter to be regulated (left). The effective concentration of repressor (shown as a bidentate tetramer) at the proximal regulatory operator is the sum of contributions due to free, [R]F, and tethered, [R]L (distal) repressors (right). (C) Inducible ‘off’ switch. ‘Off’ state (left) involves DNA looping anchored by a bidentate tetrameric repressor protein. ‘On’ state (right) is induced by tetramer destabilization into dimers, removing the contribution of tethered (distal) repressors. Residual repression is indicated by grey arrow. (D) Analogous inducible ‘off’ switch involving a designed TALED. A small molecule (small circles) decreases dimer stability. (E) TALED design. TALE protein (gray) with sequence-specific RVD cassettes highlighted and the amino acid sequence of one cassette illustrated (below) with specificity diresidue underlined. DD: dimerization domain. (F) Controlled TALED dimerization. TALE-FKBP(F36M) dimerization variant (left) is broken by FK506 (small circle). Polypeptide polarity is shown below. (G) TALE-FKBP(F36V) dimerization variant (right) is induced by AP20187.
We describe the design and testing of a regulated gene repression system controlled by dimerization of engineered sequence-specific Transcription Activator-Like Effector Dimers (TALEDs) expressed in living bacteria (Figure 1D). Transcription Activator-Like Effectors (TALEs) are remarkable protein products of plant pathogenic bacteria (19). Bacterial injection of these proteins into hosts is believed to foster infection through transcriptional activation of host genes. TALEs are composed of repeats of 34-amino acid domains carrying a repeat variable diresidue (RVD) encoding specificity for a target base pair. TALE proteins engage the DNA major groove, forming a right-handed protein spiral that binds at a specific DNA target sequence (20,21), bringing a C-terminal transcription activation domain to the protein binding site in chromatin. TALE proteins have previously been fused to nucleases (‘TALENs’) for genetic engineering. Here we replace the TALE activation domain with a dimerization domain [Figure 1E; (19,22)]. In the present work we create new gene control switches in living bacteria by implementing TALEDs to mimic features of the natural lac, gal and trp gene control systems of E. coli. In particular, we borrow from the gal operon switch the concept that repression can be partly dependent on repressor protein dimerization.
MATERIALS AND METHODS
DNA looping reporter constructs
DNA looping constructs (Supplemental Table S1) were based on plasmid pJ992 (23), created by modifications of pFW11-null (24,25). See supplemental methods for full details.
TALE-FKBP protein expression
Assembly and cloning of genes encoding TALEs has been facilitated by semi-automated methods (26). TALE-FKBP protein expression plasmids were created using modified versions of pJ1034 (low expression promoter) and pJ1035 (moderate expression promoter) (25). Plasmid pJ1035 contains the bacterial UV5 promoter with complete –10 and –35 boxes. The low expression plasmid pJ1034 contains the –10 box of the UV5 promoter with a non-ideal –35 box (24). See supplemental methods for full details.
E. coli β-galactosidase reporter assay
LacZ expression was measured using a liquid β-galactosidase colorimetric enzyme assay (27). Repression quantitated in terms of repression ratio (RR):
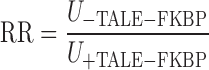 |
with the contribution to the repression ratio due to free repressor defined as RRF:
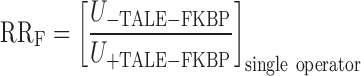 |
the overall contributions to the repression ratio due to free repressor and DNA looping defined as RRT:
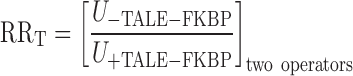 |
and the contributions to the repression ratio due to DNA looping defined as RRL:
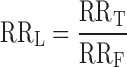 |
See supplemental methods for full details.
Chemically induced dimerization of FKBP(F36V) variants
Bacterial stains containing TALE-FKBP(F36V) variants were subcultured in the presence or absence of 1 μM dimerizing agent, AP20187 (Clontech), dissolved in methanol, followed by standard β-galactosidase assay. See supplemental methods for full details.
Chemical disruption of FKBP(F36M) variant dimerization
Bacterial strains containing the TALE-FKBP expression plasmids pJ2307 (low protein expression) or pJ2309 (moderate protein expression) were grown in LB medium containing ampicillin. For reporter assay, ∼1.2 × 105 bacterial cells were subcultured into 1.1 ml LB medium in the presence or absence of 3 μg/ml PMBN (Sigma). Also included at the time of subculture was dimer disruptor FK506 (Sigma), dissolved in DMSO to yield either 1 or 5 μM final concentration. See supplemental methods for full details.
RESULTS
TALED design
Designed TALEDs (Figure 1E and Supplemental Figure S1) were created by modification of a semi-automated method where clusters of modules with proper RVDs are encoded in DNA segments assembled by a Golden Gate procedure (26). In the original assembly system, RVDs are cloned into a plasmid with a C-terminal nuclease in place of the natural transcriptional activation domain. For production of TALEDs, the nuclease domain was replaced with various FKBP modules to facilitate dimerization. Fusion to a C-terminal FKBP(F36M) mutant domain allows constitutive dimerization (Figure 1F) that can be disrupted by small molecule FK506. FKBP(F36M) dimer affinity has been reported to be 30 μM in vitro (28). In contrast, fusion to a C-terminal FKBP(F36V) domain creates monomers whose homodimerization depends on the additional of a small molecule chemical dimerizer such as AP20187 (Figure 1G). In this case, dimer affinity has been reported to be in the nM range in vitro (29). Details are provided in Supplemental Figure S1.
lac looping model systems
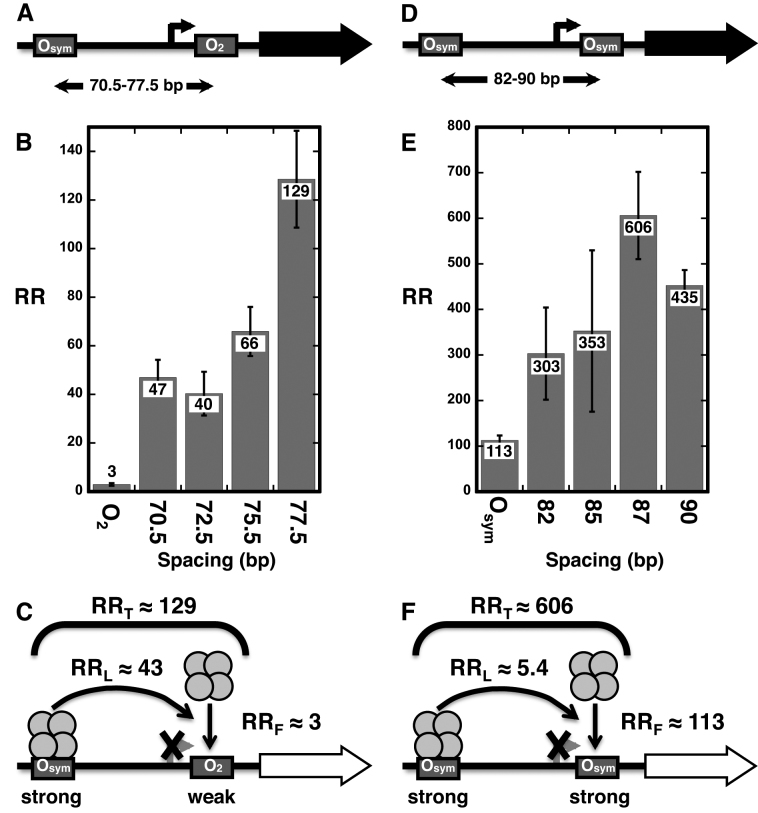
We and others have been studying engineered repression by DNA looping using elements of the natural E. coli lac operon (17,18,23,30–32). To create an in vivo model for emulation using designed TALEDs, we assembled components of the lac control switch and analyzed quantitatively their behavior. This design is different from our previous published experiments because gene induction is accomplished by elimination of functional LacI rather than by addition of IPTG inducer. This scheme allows the results to be directly compared to an analogous system where TALEDs substitute for LacI. In one series (Figure 2A), a promoter-proximal pseudo-palindromic operator (O2) controls RNA polymerase access to the promoter, but is recognized weakly by LacI, while a palindromic operator that strongly binds LacI (Osym) is placed at four distances (70.5, 72.5, 75.5, 77.5 bp) upstream. Design details are shown in Supplemental Figure S2A. Resulting measurements of DNA repression in vivo are shown in Figure 2B and interpreted in Figure 2C. The repression ratio (RR; see methods) is defined as the level of expression of the lacZ reporter gene in the absence of any functional LacI (full ‘on’ state), divided by the corresponding level of lacZ expression when LacI is expressed at normal physiological levels (full ‘off’ state). As shown in Figure 2C, repression attributable to free repressor binding at O2 (RRF) has a value of 3, while increased local repressor concentration due to looping from Osym gives RRL as high as 43, increasing repression by a factor of as much as 14, for a total RRT of up to 129. For comparison we also studied a lac looping switch where both proximal and distal palindromic Osym operators bind LacI strongly (Figure 2D and Supplemental Figure S2B). Here, repression is much more complete and is dominated by the strong binding of free repressor at the proximal operator. Nonetheless, increasing local repressor concentration by DNA looping still contributes a factor of up to 5.4 to overall repression (Figure 2E and F). Thus, these model systems illustrate that optimal operator spacing of natural components of the lac control switch allow DNA looping contributions to repression (RRL) in the range of 5–40 (Supplemental Table S2).
Figure 2.
DNA repression looping paradigm: Tetrameric LacI repression loop. (A) Strong distal Osym operator and weak proximal O2 operator flanking test promoter. (B) Measured repression ratios (RR) comparing repression by a single proximal O2 operator with loops of four lengths between distal Osym and proximal O2. (C) Interpretation of results showing respective contributions of free (RRF) and tethered (RRL) repressors, whose product is RRT. Here, the contribution of distal tethered repressor to effective repressor concentration at the proximal site is more than 14-fold higher than for free repressor. (D–F) Comparable data for loops involving two strong Osym operators. Data for all experiments reflect the mean of at least six determinations with standard deviation indicated.
TALED-based lac repression in vivo
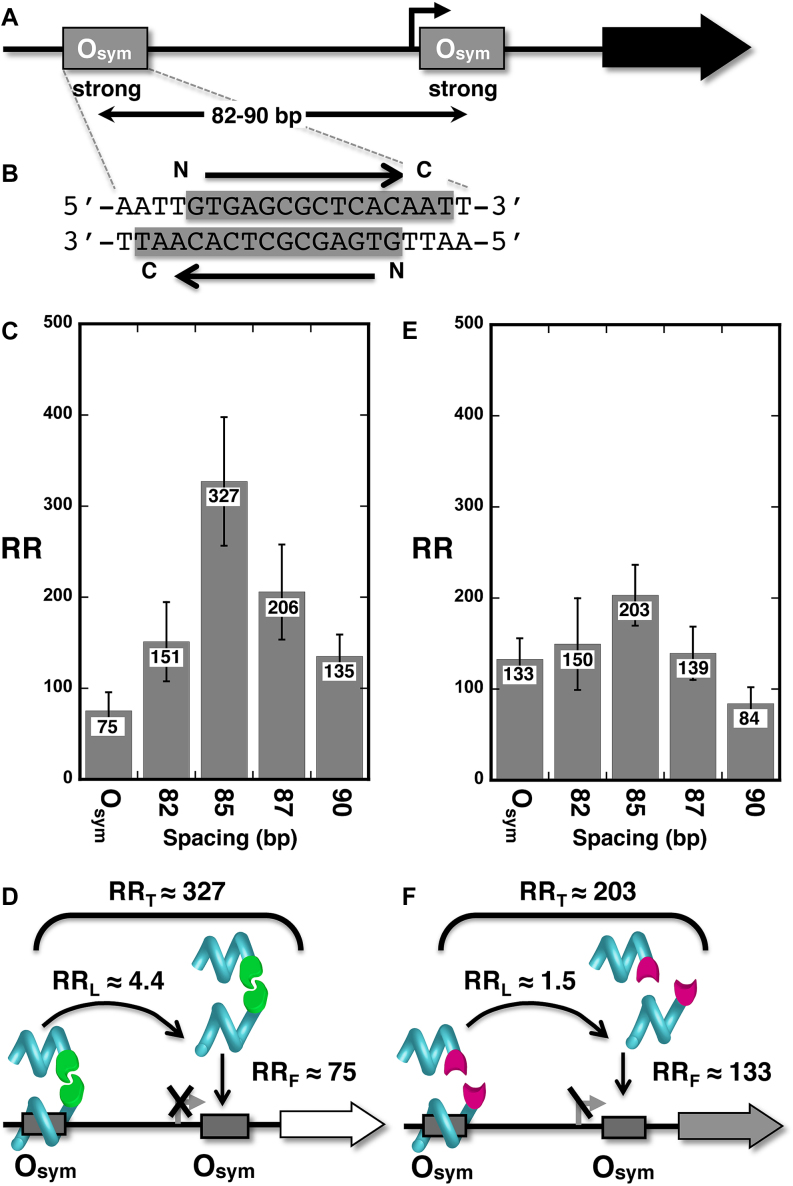
To test whether DNA looping by designed TALEDs could lead to an enhancement of gene repression comparable to that caused by looping by LacI, we designed a TALE protein against a 15-bp sequence within the Osym operators present in the test construct (Figure 3A). Operator symmetry dictates that this TALE protein can bind in either of two orientations (Figure 3B), unavoidably creating four potential competing DNA repression loops with different geometries (Supplemental Figure S3). Four operator spacing constructs and a control bearing only a proximal operator were tested in living E. coli cells after expression of the Osym-specific TALED with FKBP(F36M) constitutive dimerization domain from a moderate promoter. The results (Figure 3C and D) demonstrate that enhancement of repression due to DNA looping anchored by the TALED (RRL ∼ 4.4) is comparable to repression enhancement for LacI in this system (compare Figures 2F and 3D). This is true despite the potential for four competing DNA–protein loops of different geometries for each operator spacing. Importantly, significant enhancement of repression is not observed for TALE fusions involving FKBP(F36V) domains, which do not dimerize (Figure 3E and F). Qualitatively similar results were obtained when TALEDs were expressed at lower levels (Supplemental Table S3). Importantly, we also showed that TALED binding to the distal Osym operator at various distances from a promoter lacking a proximal operator had no effect on gene expression (Supplemental Table S4). This result confirms that repression enhancement is due to DNA looping, as designed. Together, these results demonstrate for the first time engineered in vivo transcriptional regulation by DNA looping anchored by a designed TALED.
Figure 3.
A designed TALED represses gene expression by DNA looping. (A) Test system involving Osym operators at four spacings. (B) Designed TALE protein binding sites (gray) and orientations within each symmetrical Osym operator. (C) Measured repression ratios (RR) comparing repression by a single proximal Osym operator with DNA loops of four lengths between distal Osym and proximal Osym. (D) Interpretation of results showing respective contributions of free (RRF) and tethered (RRL) TALEs, whose product is RRT. Here TALED-induced DNA looping enhances repression by 4.4-fold. (E and F) Comparable data for experiments with TALE monomers.
Tuning TALED-controlled gene repression
As shown in Figure 2, quantitative enhancement of gene repression by DNA looping with LacI is a function of operator affinity and loop geometry. To test this in the context of TALEDs, we designed a series of variant operators and tested repression from these operators when they were placed alone in the proximal position in cells expressing the Osym-specific TALED fused to FKBP(F36M). Unlike recognition of palindromic Osym, most of the variant operators are asymmetric and are recognized in only a single orientation. TALED binding to these isolated variant operators indeed led to weaker repression, as designed (Supplemental Figure S4).
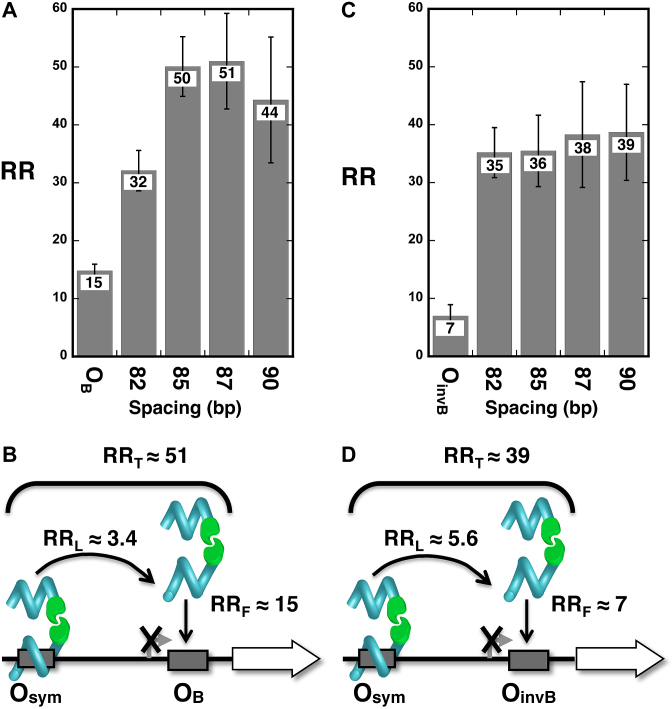
We then tested TALED-mediated enhancement of gene repression by DNA looping between distal Osym and either proximal OB (Figure 4A and B) or proximal OinvB (Figure 4C and D) operators. These configurations involve weaker proximal operators and probably two (rather than four) competing DNA loops (Supplemental Figure S3). Maximal RRL values ranged from 3.4–5.6 in these cases. Qualitatively similar results were obtained when TALEDs were expressed at lower levels (Supplemental Table S5).
Figure 4.
Tuning operator affinity and TALED repression looping. Data (A, C) and interpretation (B, D) for gene repression by designed TALED loops between strong (Osym) distal operators and weak OB (A, B) and OinvB (C, D) operators.
Regulation of gene repression by TALEDs: co-repression or induction by small molecules
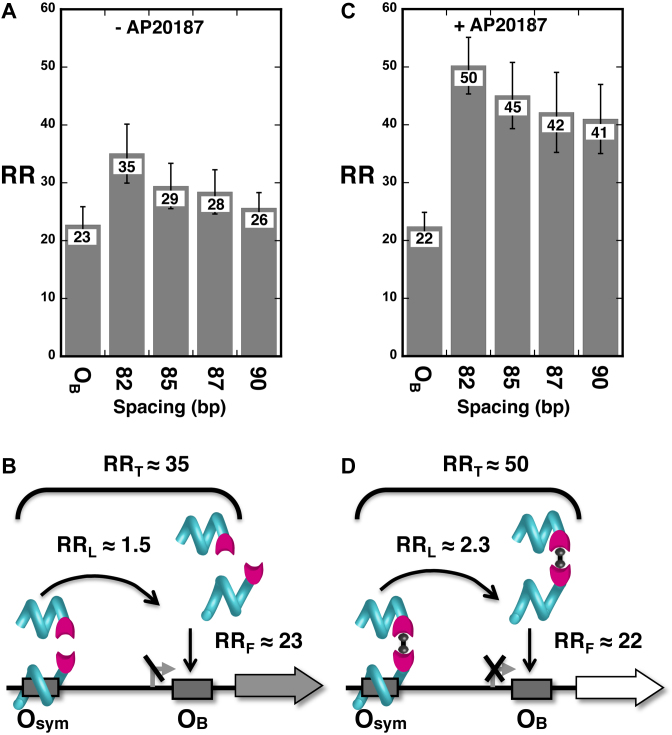
A hallmark of natural gene regulation is triggering by small molecules that increase or decrease repression of gene transcription. We sought to mimic this regulation with TALEDs whose FKBP domain dimerization is sensitive to small molecules. We began with a model of co-repression where partial gene repression by an TALE protein monomer bearing FKBP(F36V) binding at a weak OB proximal operator (Figure 5A and B) yields an RRF value of ∼23 and a very low RRL value of ∼1.5. This anticipated result is consistent with the lack of FKBP(F36V) dimerization in the absence of a chemical dimerizer (Figure 1G). Addition of the cell-permeable chemical dimerizer AP20187 at a concentration found to be effective in E. coli (Supplemental Figure S5) revealed engineered co-repression as designed (Figure 5C and D). The RRL value in the presence of co-repressor was ∼2.3, raising overall repression (RRT) to ∼50 for the optimal operator spacing. Qualitatively similar results were obtained when TALEDs were expressed at lower levels (Supplemental Table S6). We cannot conclusively explain the difference between the RRL value of ∼2.3 obtained for chemical dimerization (Figure 5D) vs. the value of ∼4.4 obtained for protein-protein dimerization (Figure 3D). Based on the reported affinity of the chemical dimerizer, it might have been anticipated that repression by chemical dimerization might be tighter. It seems likely that this discrepancy is due to two non-optimal features of the chemical dimerizer AP20187. These are (i) intracellular AP20187 dose may not be optimal (Supplemental Figure S5) and (ii) there will always be a competition between one dimerizer molecule bridging two proteins (desired) and one dimerizer molecule capping each protein (undesired).
Figure 5.
Chemically-induced dimerization triggers TALED repression by DNA looping. Data (A, C) and interpretation (B, D) for gene repression involving designed TALE-FKBP(F36V) repressors without (A, B) or with (C, D) chemically-induced dimerization by 1 μM AP20187.
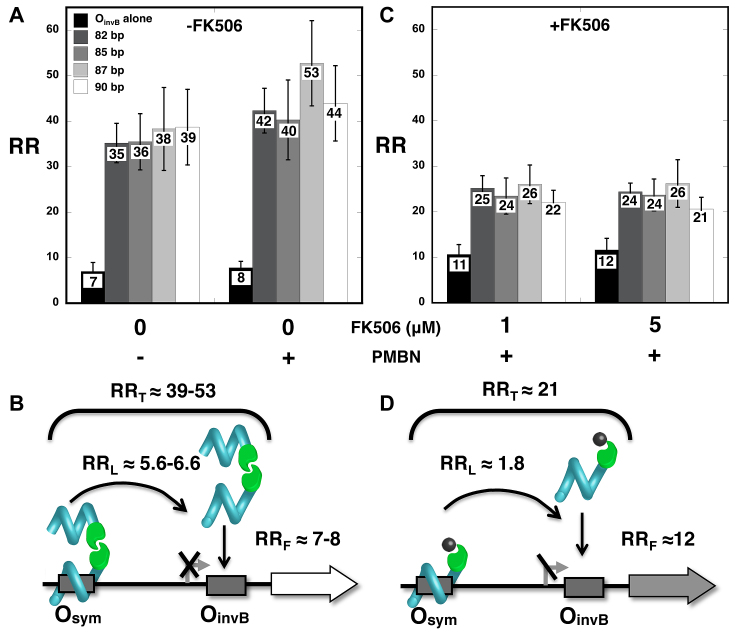
We likewise sought to model gene induction with a small molecule by artificially triggering disruption of TALED dimerization in E. coli. In this case, E. coli cells carrying the previously-studied combination of distal Osym and proximal OinvB operators and expressing a dimerizing TALED fusion to FKBP(F36M) (Figure 4C and D) were tested in the absence or presence of the permeabilizing agent polymyxin B nonapeptide [PMBN; (33,34)]. PMBN concentration was optimized by measuring uptake of small molecule antibiotics and FK506 (Supplemental Figure S6). Addition of permeabilizer preserves DNA looping and gene repression (Figure 6A and B). Strong RRL values of ∼5.6–6.6 are observed for an overall RRT of ∼39–53. In contrast, with addition of the small molecule FK506 (1 or 5 μM), TALED dimerization is inhibited, and derepression is observed (RRL value ∼1.8) as intended (Figure 6C and D). Qualitatively similar results were obtained when TALEDs were expressed at lower levels (Supplemental Table S7).
Figure 6.
Chemically-induced TALED disruption of dimerization induces gene expression by DNA un-looping. Data (A, C) and interpretation (B, D) for gene repression involving designed TALE-FKBP(F36M) repressors without (A, B) or with (C, D) chemically-induced disruption of dimerization by FK506.
DISCUSSION
Our focus in this work has been engineered approaches to artificial control of transcription initiation in E. coli. We based our strategy on the DNA looping paradigm common in natural gene control. Here we engineer homodimeric TALEDs for this purpose, demonstrating that DNA looping increases promoter repression to extents comparable to switches involving LacI protein. Exploiting dimerization domains based on FKBP variants, we further show how small molecules can act as co-repressors or inducers in these engineered switches by regulating TALE protein dimerization. This approach is distinct from two elegant prior in vitro studies that did not explore gene control in living cells. Gowetski et al. engineered DNA loops through designed coiled-coil peptides (32). Praetorius and Dietz have recently shown in vitro assembly of complex duplex DNA shapes by DNA looping anchored by designed constitutive dimeric TALE proteins (35). In contrast, our work implements TALEDs in living cells with controlled TALE dimerization regulating gene expression.
In the present study, we have intentionally limited ourselves to adapting cis regulatory elements from the E. coli lac operon where much is known about DNA looping between natural and artificial sequence variants of the palindromic lac operator sequence. Building lac operator-specific TALEDs has the advantage that it allows the engineered system to be directly compared to lac regulation by LacI. On the other hand, the palindromic nature of natural lac operators complicates designed DNA loops anchored by asymmetric TALEDs because multiple competing loop geometries always result. Our data show that this complexity does not prevent the desired functions, but it typically obscures the expected sinusoidal dependence of repression on distance between operators (phasing). We thus attribute the absence of sinusoidal repression dependence on operator phasing to the presence of competing loops (where one loop need not dominate), together with the flexibility of the TALED proteins themselves. An additional complication to interpretation of preferred operator phasings is that the FKBP dimer interface and protein orientations for the FKBP F36M dimer are likely to differ from FKBP F36V after chemical dimerization (28). Nonetheless, for general application the profound advantage of designed sequence-specific TALE-based protein domains will easily permit specification of unique DNA loops by selecting non-palindromic operator sequences.
Future implementations of the TALED strategy described here can be envisioned to exploit the formation of heterodimeric repressor proteins. When combined with selective heterodimer stabilization or destabilization by available small molecules, such approaches will further generalize microbial applications. Likewise, further development of small molecule triggers to which bacterial cells are highly permeable will further advance application.
It is important to emphasize that regulation of our engineered switches is achieved by controlling the probability of repressor protein dimerization rather than the allosteric control of DNA binding affinity that is pervasive in biological systems. Thus, controlled elimination of DNA looping reduces, but does not eliminate, TALE protein monomer binding at the proximal operator. Overcoming this resulting weak residual repression would involve devising TALE proteins that mimic natural repressors whose DNA affinity can be allosterically controlled by small molecules.
In summary, the present work engineers control of bacterial gene expression by mimicry of natural DNA looping mechanisms using designed repressors whose dimerization can be controlled. It is plausible that similar approaches can be implemented in the context of transcription initiation from eukaryotic promoters to create regulated transcriptional repression by TALED-directed looping of chromatin via arbitrary targeted sequences. Such approaches would greatly extend the previously-recognized principle of eukaryotic gene regulation by DNA looping (36,37).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Justin Peters for helpful discussions.
Authors contributions: Nicole A. Becker, Karl J. Clark and L. James Maher: Project conception and manuscript preparation. Nicole A. Becker, Tanya L. Schwab: Experimental work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Mayo Foundation; National Institutes of Health [GM75965 to L.J.M.]. The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Müller-Hill B. The Lac Operon: A Short History of a Genetic Paradigm. 1996; Berlin; NY: Walter de Gruyter. [Google Scholar]
- 2. Weickert M.J., Adhya S.. The galactose regulon of Escherichia coli. Mol. Microbiol. 1993; 10:245–251. [DOI] [PubMed] [Google Scholar]
- 3. Yanofsky C. Tryptophan biosynthesis in Escherichia coli. Genetic determination of the proteins involved. JAMA. 1971; 218:1026–1035. [PubMed] [Google Scholar]
- 4. Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu. Rev. Genet. 1989; 23:227–250. [DOI] [PubMed] [Google Scholar]
- 5. Bellomy G., Mossing M., Record M.. Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988; 27:3900–3906. [DOI] [PubMed] [Google Scholar]
- 6. Matthews K. DNA looping. Microbiol. Rev. 1992; 56:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schleif R. DNA looping. Ann. Rev. Biochem. 1992; 61:199–223. [DOI] [PubMed] [Google Scholar]
- 8. Rippe K., von Hippel P.H., Langowski J.. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci. 1995; 20:500–506. [DOI] [PubMed] [Google Scholar]
- 9. Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 2001; 26:733–740. [DOI] [PubMed] [Google Scholar]
- 10. Vilar J.M., Saiz L.. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 2005; 15:136–144. [DOI] [PubMed] [Google Scholar]
- 11. Oehler S., Müller-Hill B.. High local concentration: a fundamental strategy of life. J. Mol. Biol. 2009; 395:242–253. [DOI] [PubMed] [Google Scholar]
- 12. Cournac A., Plumbridge J.. DNA looping in prokaryotes: experimental and theoretical approaches. J. Bacteriol. 2013; 195:1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Law S.M., Bellomy G.R., Schlax P.J., Record M.T. Jr. In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local Lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J. Mol. Biol. 1993; 230:161–173. [DOI] [PubMed] [Google Scholar]
- 14. Mossing M.C., Record M.T. Jr. Upstream operators enhance repression of the lac promoter. Science. 1986; 233:889–892. [DOI] [PubMed] [Google Scholar]
- 15. Mandal N., Su W., Haber R., Adhya S., Echols H.. DNA looping in cellular repression of transcription of the galactose operon. Genes Dev. 1990; 4:410–418. [DOI] [PubMed] [Google Scholar]
- 16. Lionberger T., Meyhofer E.. Highly bent DNA: A novel repressor of T7 RNA polymerase. Biophys. J. 2010; 98:69a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker N.A., Peters J.P., Lionberger T.A., Maher L.J.. Mechanism of promoter repression by Lac repressor-DNA loops. Nucleic Acids Res. 2012; 41:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Becker N.A., Greiner A.M., Peters J.P., Maher L.J.. Bacterial promoter repression by DNA looping without protein-protein binding competition. Nucleic Acids Res. 2014; 42:5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bogdanove A.J., Voytas D.F.. TAL effectors: customizable proteins for DNA targeting. Science. 2011; 333:1843–1846. [DOI] [PubMed] [Google Scholar]
- 20. Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.-K., Shi Y., Yan N.. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012; 335:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mak A.N.-S., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L.. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012; 335:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim Y., Kweon J., Kim A., Chon J.K., Yoo J.Y., Kim H.J., Kim S., Lee C., Jeong E., Chung E. et al. . A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013; 31:251–258. [DOI] [PubMed] [Google Scholar]
- 23. Becker N.A., Kahn J.D., Maher L.J. 3rd. Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005; 349:716–730. [DOI] [PubMed] [Google Scholar]
- 24. Whipple F.W. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998; 26:3700–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peters J.P., Becker N.A., Rueter E.M., Bajzer Z., Kahn J.D., Maher L.J.. Quantitative methods for measuring DNA flexibility in vitro and in vivo. Meth. Enzymol. 2011; 488:287–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma A.C., McNulty M.S., Poshusta T.L., Campbell J.M., Martinez-Galvez G., Argue D.P., Lee H.B., Urban M.D., Bullard C.E., Blackburn P.R. et al. . FusX: a rapid one-step transcription activator-like effector assembly system for genome science. Hum. Gene Ther. 2016; 27:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller J. A Short Course in Bacterial Genetics. 1992; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 28. Rollins C.T., Rivera V.M., Woolfson D.N., Keenan T., Hatada M., Adams S.E., Andrade L.J., Yaeger D., van Schravendijk M.R., Holt D.A. et al. . A ligand-reversible dimerization system for controlling protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clackson T., Yang W., Rozamus L.W., Hatada M., Amara J.F., Rollins C.T., Stevenson L.F., Magari S.R., Wood S.A., Courage N.L. et al. . Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bond L.M., Peters J.P., Becker N.A., Kahn J.D., Maher L.J.. Gene repression by minimal lac loops in vivo. Nucleic Acids Res. 2010; 38:8072–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haeusler A.R., Goodson K.A., Lillian T.D., Wang X., Goyal S.S., Perkins N.C., Kahn J.D.. FRET studies of a landscape of Lac repressor-mediated DNA loops. Nucleic Acids Res. 2012; 40:4432–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gowetski D.B., Kodis E.J., Kahn J.D.. Rationally designed coiled-coil DNA looping peptides control DNA topology. Nucleic Acids Res. 2013; 41:8253–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992; 56:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Viljanen P., Vaara M.. Susceptibility of gram-negative bacteria to polymyxin B nonapeptide. Antimicrob Agents Chemother. 1984; 25:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Praetorius F., Dietz H.. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science. 2017; 355: [DOI] [PubMed] [Google Scholar]
- 36. Vilar J.M., Saiz L.. Control of gene expression by modulated self-assembly. Nucleic Acids Res. 2011; 39:6854–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yasmin R., Yeung K.T., Chung R.H., Gaczynska M.E., Osmulski P.A., Noy N.. DNA-looping by RXR tetramers permits transcriptional regulation “at a distance”. J. Mol. Biol. 2004; 343:327–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.