Abstract
Schistosomes are the causative agents of schistosomiasis, a neglected tropical disease affecting over 230 million people worldwide. Additionally to their major impact on human health, they are also models of choice in evolutionary biology. These parasitic flatworms are unique among the common hermaphroditic trematodes as they have separate sexes. This so-called “evolutionary scandal” displays a female heterogametic genetic sex-determination system (ZZ males and ZW females), as well as a pronounced adult sexual dimorphism. These phenotypic differences are determined by a shared set of genes in both sexes, potentially leading to intralocus sexual conflicts. To resolve these conflicts in sexually selected traits, molecular mechanisms such as sex-biased gene expression could occur, but parent-of-origin gene expression also provides an alternative. In this work we investigated the latter mechanism, that is, genes expressed preferentially from either the maternal or the paternal allele, in Schistosoma mansoni species. To this end, transcriptomes from male and female hybrid adults obtained by strain crosses were sequenced. Strain-specific single nucleotide polymorphism (SNP) markers allowed us to discriminate the parental origin, while reciprocal crosses helped to differentiate parental expression from strain-specific expression. We identified genes containing SNPs expressed in a parent-of-origin manner consistent with paternal and maternal imprints. Although the majority of the SNPs was identified in mitochondrial and Z-specific loci, the remaining SNPs found in male and female transcriptomes were situated in genes that have the potential to explain sexual differences in schistosome parasites. Furthermore, we identified and validated four new Z-specific scaffolds.
Keywords: parent-of-origin gene expression, sexual dimorphism, intralocus sexual conflict, male–female coevolution, Schistosoma mansoni
Introduction
Schistosomiasis, also known as bilharzia, is a highly prevalent tropical disease affecting over 200 million people worldwide (Engels et al. 2002). It mainly occurs in developing countries and ranks second in term of parasite morbidity and mortality after malaria (King 2010). This chronic infection is caused by different species of the Schistosoma genus, blood flukes, with a complex lifecycle involving two obligatory hosts. Schistosoma mansoni responsible of intestinal schistosomiasis uses Biomphalaria genus as mollusk intermediate host, and humans or rodents as vertebrate definitive hosts. In the vertebrate host, males and females form monogamous couples, and sexually reproduce (Beltran and Boissier 2008). The produced eggs are released out of the host via the feces and, in contact with freshwater, hatch to liberate free-swimming larvae called miracidium. These miracidia actively search and infect their intermediate hosts, transform into two successive intramolluskan stages (i.e., sporocysts), which produce by asexual multiplication thousands of vertebrate-infecting larvae (i.e., cercaria).
Besides the medical importance of this parasite for human health, it is also a very interesting model in terms of evolutionary biology due to its original sexual features. Among the 18,000–24,000 hermaphroditic species recorded in Trematodes, only the Schistosomatidae family (∼100 species) has evolved separate sexes and has thus been qualified as an “evolutionary scandal” by Claude Combes in 1991 (Combes 1991). This gonochorism is accompanied by a pronounced dimorphism between sexes (Basch 1990; Loker and Brant 2006). These differences between males and females exist on different scales ranging from a molecular, to a behavioral level, and have been documented in several studies (see Moné and Boissier 2004 for review). Larval stages are morphologically indistinguishable and one striking difference at the adult stage is the worm musculature (muscular adult male vs. a thin adult female), as well as the presence of a gynaecophorous canal, and a large oral sucker in males allowing them to carry and shelter their female mating partner (Beltran and Boissier 2008, 2009). Sexes are genetically determined during egg fertilization with a ZZ/ZW chromosomal system where females are heterogametic (Grossman et al. 1981). In S. mansoni, females have a W chromosome characterized by large pseudoautosomal regions as well as W-specific sequences almost entirely composed of heterochromatic satellite-type repeats (Lepesant et al. 2012; Protasio et al. 2012). No W-specific protein-coding genes were identified so far (Criscione et al. 2009). Thus, in spite of the observed sexual dimorphism at the adult stage, males and females share a common set of protein-coding genes. There are 782 genes specific to the Z chromosome and are thus found twice in the ZZ males and only once in the ZW females. Studies on ZW female heterogametic systems frequently report a lack of dosage compensation that equalizes Z-linked transcript levels in males and females (Graves 2016). It is also the case in S. mansoni adults where Z-linked expression is reduced relative to autosomal expression in females but not in males (Vicoso and Bachtrog 2011). However, it is not known whether specific genes may be escaping the dosage compensation mechanism in schistosome males, therefore being potentially responsible for male-biased gene expression and accentuating phenotypic differences between sexes. If selection for a particular trait at a particular locus favors different alleles in males versus females (i.e., sexual antagonism), one would expect to find a mechanism to resolve these intralocus sexual conflicts. Gonochoric species usually “solve” these conflicts through sex-biased expression of the sexually antagonistic genes (Ingleby et al. 2015; Lipinska et al. 2015). In schistosomes, sex-biased gene expression was intensively studied at the whole transcriptome level (Fitzpatrick et al. 2005, 2008; Anderson et al. 2015; Lu et al. 2016; Picard et al. 2016), but other molecular mechanisms such as parent-of-origin gene expression have never been investigated.
In diploid gonochoric organisms, sexual reproduction leads to the presence of two copies of each gene in each somatic cell of an individual, one inherited from the mother (i.e., matrigenes) and the other from the father (i.e., patrigenes). Usually both copies of a gene cooperate and are expressed at equal levels allowing compensation of the function in case of deleterious mutation, but in some cases gene expression is restricted to one of the parental alleles leading to paternally versus maternally preferential or exclusive gene expression (aka parent-of-origin gene expression, or genomic imprinting) (Reik and Walter 2001). Different theories have been proposed to explain the evolution of genomic imprinting and the effects of such gene expression in the offspring. The sexual antagonism (Day and Bonduriansky 2004; Bonduriansky 2007; Patten et al. 2014) or maternal–offspring coadaptation (Wolf and Hager 2006) theories suggest that genomic imprinting has the potential to modify resemblance of an individual to its parents (Patten et al. 2014). The sexual antagonism theory proposes that as fathers and mothers have passed the filter of sex-specific selection (alleles successfully transmitted to progeny), it is thus more likely that male offspring will benefit from paternally expressed alleles for male traits, whereas female offspring will benefit from maternally expressed alleles for female traits (Day and Bonduriansky 2004). It is therefore predicted that phenotypic traits benefiting males or females, as for example muscular bodies for males, would be preferentially expressed from the paternal alleles. In schistosomes we therefore expect paternal genes coding for growth enhancers or muscle development to be preferentially expressed in males, and maternal expression in females for traits under sexually antagonistic selection.
On the basis of the maternal–offspring coadaptation theory, maternal alleles may be selected for imprinted expression to provide the greatest combined fitness between the mother and progeny (Wolf and Hager 2006). Therefore in organisms providing maternal investment (e.g., most mammals) we expect maternal expression in both sexes for genes involved in maternal–offspring interactions and more specifically maternal care. In schistosome species, it has been proposed that prezygotic paternal investment (transport of the female to the oviposition site, female maturation, and feeding) is higher than maternal investment (Beltran and Boissier 2008). Therefore, paternal expression in both sexes may be expected.
The evolutionary roles of parent-of-origin expression nevertheless remain controversial and the patterns predicted by those models may be very contrasted but also non-exclusive. Indeed, most of the studies have been carried in plants, mammals and some insects such as coccids and bees (da Rocha and Ferguson-Smith 2004; Ferguson-Smith 2011; Macdonald 2012; Kocher et al. 2015) and support an alternative, but not exclusive conjecture: The kinship theory (Moore and Haig 1991; Haig 2000), which links parent-of-origin gene expression not to sexual, but more generally to parental conflicts. It assumes that parental genomes are not functionally equivalent and do not have the same reproductive interests. In a polyandrous mating system, maternal alleles tend to preserve mother and progeny by a restriction of resources, while the paternal genome favors growth of offspring. An interesting example for such a “battle of sexes” was shown by the paternal expression of the insulin-like growth factor 2 (IGF2) inducing growth, while the maternal expression of its antagonist receptor (IGF2R) tends to reduce growth in mammals (Wilkins and Haig 2003). The kinship model therefore predicts that maternal expression will be favored if a gene has a positive effect when maternally expressed but a negative effect when paternally derived and vice versa for paternal expression. Therefore, we would expect paternal expression in both sexes for genes involved in extraction of maternal resources by offspring. Schistosomes are oviparous organisms that shed hundreds of eggs daily per female, and resource is already present in the egg before fertilization (maternal allocation of energy) but once released the subsequent development is independent of parental resources. The expression patterns will therefore depend on the phenotypic effect of the genes.
Differences in gene expression according to parental origin in schistosomes may be important to identify candidates for sex-specific phenotypes. To this end, we screened male and female transcriptomes in reciprocal hybrids of S. mansoni for parent-of-origin expressed genes, considering both known sex-linked and autosomal genes. This approach relied on strain-specific single nucleotide polymorphic (SNP) markers allowing us to identify the potential allelic expression profile of each transcript, while reciprocal crosses allowed us to discriminate parent-of-origin effects from strain-of-origin effects. We show that most SNPs detected in genes with monoallelic expression are located in mitochondrial and Z-specific loci, thus validating our experimental design and analysis pipeline. We also identified a number of SNPs with preferential expression from one parental allele in the transcriptome of male and female schistosomes, suggesting paternal and maternal imprints. As the genes concerned by these parent-of-origin type SNPs are closely related to development and sex-specific functions, we suggest that parent-of-origin gene expression needs to be explored in depth because it may be used to resolve intralocus sexual conflicts underlying sexual dimorphism in this species. Finally, we discuss our results in regard to current knowledge on schistosome parasites life-history traits and the evolutionary theories supporting imprinting in other organisms.
Materials and Methods
Parasite Origin, Crossing Protocol, and Sequencing
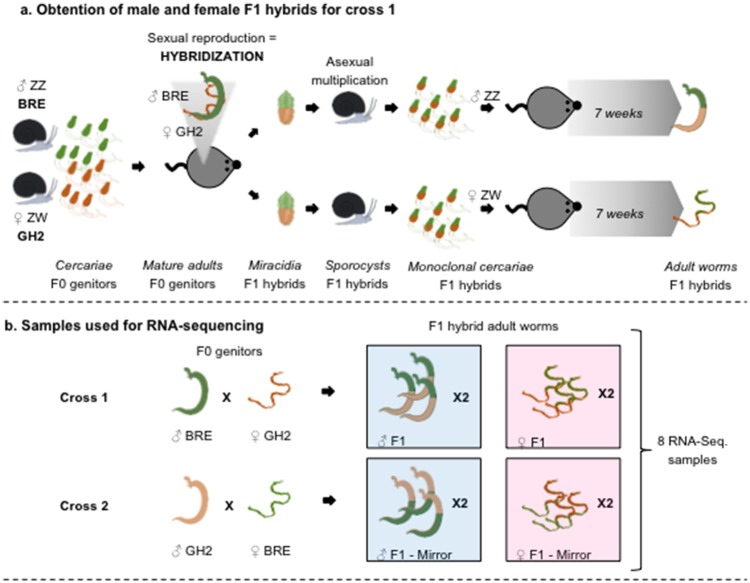
Hybrid adult parasites were obtained by reciprocally crossing two different geographic isolates of the S.mansoni species: The Guadeloupian strain GH2 and the Brazilian strain BRE. We performed biological duplicates of two independent crosses, F1 male and female hybrids (GH2 mother × BRE father) and the reciprocal male and female crosses (BRE mother × GH2 father) as shown in figure 1 and table 1. Each cross was performed in mouse (Mus musculus) definitive hosts, by using Biomphalaria mollusks that were individually exposed to a single miracidium of the BRE or GH2 strain (releasing either male or female clonal populations of BRE or GH2 cercariae). The resulting hybrid miracidiae were individually used to infect mollusks, and adult worms were recovered after mouse unisexual infestation (fig. 1a). Total RNA was extracted from the eight samples separately (fig. 1b) and global sequencing was performed on four lanes (double multiplexing) using paired-end (2 × 125 nt) Hiseq Illumina technology.
Fig. 1.
—(a) Crossing protocol example for Cross 1 and (b) samples used for RNA-sequencing. (a) Mollusks releasing clonal populations of male BRE (ZZ) and female GH2 (ZW) cercariae were used to infect M. musculus definitive hosts and produce F1 eggs releasing hybrid miracidiae. These male or female miracidiae were used to infect mollusks and produce male of female cercariae. Unisexual infection of M. musculus allowed us to recover after 7 weeks, male and female hybrid adult worms. Notice that Cross 2 (male GH2 x female BRE), which is not presented in this figure, was performed the same way.
Table 1.
F1 and Mirror Individuals from Two Reciprocal Crosses
| F1 Male | F1 Female | F1-Mirror Male | F1-Mirror Female | |
|---|---|---|---|---|
| Biological duplicates | ♀ GH2 × ♂ BRE | ♀ GH2 × ♂ BRE | ♀ BRE × ♂ GH2 | ♀ BRE × ♂ GH2 |
| ♀ GH2 × ♂ BRE | ♀ GH2 × ♂ BRE | ♀ BRE × ♂ GH2 | ♀ BRE × ♂ GH2 |
Total RNA Isolation
For each sex and cross, experiments were performed in two biological replicates. RNA extractions were performed alternatively from 20 adult males or 100 adult females. Briefly, parasites were ground in liquid nitrogen and solubilized in TRIzol (Thermo Fisher Scientific). Total RNA was then extracted by adding chloroform. PureLink RNA Mini kit (Ambion) was used for further purification following the manufacturer’s protocol. Total RNA was eluted in 30 µl RNAsecure (Ambion) and incubated at 65 °C for 10 min. Samples were then treated with TURBO DNase (TURBO DNA-free, Ambion) and the reaction was stopped by cooling down on ice for 2 min. RNA was finally purified on a column (RNeasy mini kit, QIAGEN) and eluted in 30 µl RNase-free water. Quality and concentration were assessed by spectrophotometry with the Agilent 2100 Bioanalyzer system. Further details are available at Environmental and Evolutionary Epigenetics Webpage (http://methdb.univ-perp.fr/epievo/; last accessed February 16, 2018).
Illumina Libraries Construction and High-Throughput Sequencing
cDNA library construction and sequencing were performed at the sequencing facility of Montpellier GenomiX (MGX, France). The TruSeq stranded mRNA library construction kit (Illumina Inc., USA) was used according to the manufacturer’s recommendations on 300 ng of total RNA per condition. Briefly, poly-A RNAs were purified using oligo-d(T) magnetic beads. The poly-A+ RNAs were fragmented and reverse transcribed using random hexamers, Super Script II (Life Technologies, ref. 18064-014) and Actinomycin D. During the second-strand generation step, dUTP substitued dTTP to prevent the second strand to be used as a matrix during the final PCR amplification. Double-stranded cDNAs were adenylated at their 3ʹ-ends before ligation was performed using Illumina’s indexed adapters. Ligated cDNAs were amplified following 15 cycles PCR and PCR products were purified using AMPure XP Beads (Beckman Coulter Genomics, ref.A63881). The quantitative and qualitative analysis of the library was carried on Agilent_DNA 1000 chip and qPCR (Applied Biosystems 7500, SYBR Green). The sequencing was performed on a HiSeq2500 in paired-end 2x125nt mode. RNA-Seq reads are available at the NCBI-SRA under the BioProject accession number PRJNA378178.
RNA-Seq Data Processing
Quality Control
Quality control and initial cleaning of the reads was performed with the filter by quality program (version 1.0.0) based on FASTX-toolkit (Blankenberg et al. 2010). Reads with less than 90% of bases with Phred quality score inferior or equal to 30 were discarded (probability of 1 incorrect base call out of 1,000, and a base call accuracy of 99, 9%). Adaptors used for sequencing were removed using the cutadapt program version 1.6 (Martin 2011).
Mapping of the Reads against the Reference Genome
Each sample’s paired-end sequencing reads were independently mapped (i.e., unique reads) to the S.mansoni’s reference genome version 5.2 (Berriman et al. 2009; Protasio et al. 2012) using TopHat2 software (Kim et al. 2013). The alignment was performed in single reads with intron length parameter set between 20 and 50,00 bp. We authorized one mismatch in the anchor region of spliced alignment, applied a microexon search because of the presence of microexon genes in schistosome genomes (DeMarco et al. 2010), and used the very sensitive Bowtie2 option (Langmead et al. 2009). The GTF annotation file (S.mansoni sex-specific transcriptome) produced in a previous work (Picard et al. 2016) served as the reference (http://ihpe.univ-perp.fr/acces-aux-donnees/; last accessed February 16, 2018). In order to avoid false positives in SNP calling, we filtered PCR duplicates by eliminating exact reads found more than eight times (option “Tolerated Duplicates” = four) using “Remduplicates” (Althammer et al. 2011).
Parent-of-Origin Gene Identification
The approach is based on the analysis of the frequency of SNP markers presenting allelic imbalance within the transcriptomic data of the hybrids. The identity of the base can then be used to distinguish allelic origin, and the reciprocal cross helps to discriminate parent-of origin from strain-of-origin biases in allelic expression (fig. 1).
Selecting Strain-Specific SNPs in the Genomic Data of BRE and GH2
SNPs in the genome of the BRE and GH2 strains were previously described (Clément et al. 2013). We used the Freebayes generated BAM files available at http://methdb.univ-perp.fr/downloads/, last accessed February 16, 2018 to assess strain-specific SNPs. We considered informative, any SNP showing fixed alternative allele frequency (AF 0.9) in one strain, and absent or having an alternative allele frequency inferior or equal to 0.1 in the other (AF 0.1) compared with the reference genome (NMRI strain, Puerto Rican Origin).
SNPs Calling in the Transcriptomic Data
Using SAMtools (Li et al. 2009), we produced mpileup files with read coverage information and call quality from the TopHat2 alignment BAM files. We then performed SNPs calling by using VarScan software (version 0.1) on the mpileup files (Koboldt et al. 2012). The minimum depth at position to make a call was set to ten reads, the minimum supporting reads for alternative allele at position were set to two. The minimum base quality to count a read was set to 15, minimum variant allele frequency threshold was set to 1%, and the p-value threshold for calling variants was set to 5%. The number of reads, the number of variant allele, and the alternative base identity were visually checked for ten randomly selected SNPs in each sample with the Integrative Genomics Viewer (IGV) software (Thorvaldsdóttir et al. 2013).
Parental Origin of the Transcripts Analysis
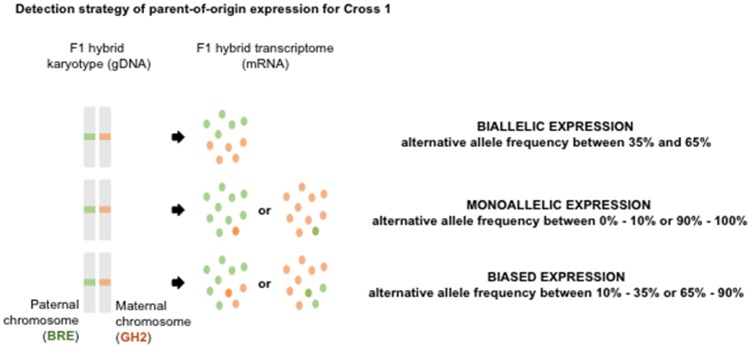
In order to discriminate parent-of-origin from strain-of-origin SNP expression, each sample’s transcriptome variant files were compared with the previous selected strain-specific SNPs (BRE or GH2) and that according to variant positions. We only kept SNPs in the transcriptome for which we had parental information in order to identify the origin from which the SNP is expressed. Three categories were defined based on the expression profiles of the SNPs. Those with alternative allele frequency between 0% and 10% or 90% and 100% were defined as monoallelic (or imprinted). Those with alternative allele frequency between 10% and 35% or 65% and 90% were defined as biased (preferential expression), and those with alternative allele frequency between 35% and 65% were considered as biallelic (fig. 2). Among these, SNPs were conserved only if found in the same category in both replicates. We further added mpileup coverage information to assess whether the SNPs were not detected, or whether the regions were not transcribed. When coverage information indicated reads in the region we considered the allele similar to the reference genome and set allele frequency to 0% (minimum coverage of ten reads). If the region was not covered and therefore not transcribed, we considered position as noninformative.
Fig. 2.
—Detection strategy of parent-of-origin expression: example for Cross 1. Cross 1 was performed using males from the S. mansoni Brazilian strain (BRE) and females from the Guadeloupian strain (GH2). Each hybrid contains a chromosome from the mother (GH2) and from the father (BRE). The trancriptomic analysis of hybrid offspring allowed identifying genes 1) expressed in equal proportions between the mother’s (GH2) and father’s (BRE) allele (i.e., biallelic expression), 2) expressed exclusively from the mother’s (GH2) or father’s (BRE) allele (i.e., monoallelic expression), or 3) expressed preferentially from the mother’s (GH2) or the father’s (BRE) allele (i.e., biased expression).
Comparisons between the Reciprocal Crosses
SNPs with biased and monoallelic expression profiles were compared between reciprocal crosses. This allowed us to identify genes potentially expressed in a parent-of-origin manner when reciprocal crosses agreed with each other on parent-of-origin bias.
Annotation of the Transcriptome
The transcriptome file (extracted from the GTF reference [Picard et al. 2016]) was de novo reannotated on the local server of the laboratory by searching protein databases using a translated nucleotide query (BlastX) against the nonredundant protein sequence database (nr). The 25 best hits were then used to search for gene ontology terms using the Blast2Go program (Conesa et al. 2005). Correspondence between transcript and current version of genome annotation was manually checked using IGV software (Thorvaldsdóttir et al. 2013).
Quantitative PCR Validation of Z-Specific Scaffolds
Simple or double copy of genomic DNA was assessed with male (ZZ) and female (ZW) DNA extracted from adult worms to verify the Z-specific status of unplaced scaffolds containing genes with monoallelic paternal expression. DNA of single adult worms of each sex was extracted as recommended from the QIAGEN QIAamp DNA Micro Kit protocol for isolation of genomic DNA from tissues. Final elution was performed using 40 µl of buffer AE (10 mM Tris·Cl, 0.5 mM ethylenediaminetetraacetic acid, pH 9.0). Primers were designed using the online Primer3plus program (Rozen and Skaletsky 2000) and aligning the primers against S.mansoni’s reference genome v5.2 to verify for high specificity. We used a known Z-specific (double copy in ZZ males and single in ZW females) and an autosomal (double copy in each sex) genomic region as internal control. Primer efficiencies were assessed with serial dilutions of genomic DNA in DNAse free water (dilutions ¼, 1/16, 1/64, and 1/256). Primer efficiency was calculated using the slope of the standard curve with efficiency = −1 + 10 (−1/slope). Quantitative PCR reactions were carried out in a total volume of 10 µl, containing 5 µl of the Takyon 2X reaction mix (Eurogentec), 2 µl of 10 mM primers (forward and reverse), 2 µl of DNA template, and 1 µl of nuclease free water. The qPCR program was set for 40 cycles containing a 10-s denaturation phase at 95 °C, annealing–extension phase at 59 °C for 20 s and 72 °C for 25 s.
Results
RNA-Seq Data Processing
Between 197 and 320 million reads were produced after RNA sequencing for each of the eight samples, representing male and female adult progenies of biological duplicate crosses and mirror crosses of a Brazilian (BRE) and Guadeloupian (GH2) strain of S.mansoni (table 2 and fig. 1). After filtering reads according to their quality Phred score, between 160 and 239 million (∼80%) of them were kept for subsequent read mapping against S.mansoni’s reference genome v5.2 (Berriman et al. 2009; Protasio et al. 2012). At least 90% of the filtered reads were successfully mapped to the reference genome. Because SNPs frequency reliability was essential to detect allele-specific transcripts, potential PCR duplicates (∼50% of reads) were removed.
Table 2.
Number of Sequences Remaining after Each Bioinformatic Steps
| Quality Filtering Q30 |
TopHat2 Mapping | PCR Duplicate Removal |
||||
|---|---|---|---|---|---|---|
| Crosses | Sex | Input | Discarded | Input | Mapped | Output |
| ♀ GH2 × ♂ BRE | M | 203,732,076 | 43,590,513 | 160,141,563 | 144,201,576 | 64,795,806 |
| (90%) | ||||||
| ♀ GH2 × ♂ BRE | M | 278,553,568 | 62,102,488 | 216,451,080 | 197,048,330 | 84,039,096 |
| (91.0%) | ||||||
| ♀ BRE × ♂ GH2 | M | 249,198,708 | 54,496,632 | 194,702,076 | 179,194,594 | 81,242,489 |
| (92.0%) | ||||||
| ♀ BRE × ♂ GH2 | M | 237,406,392 | 52,577,939 | 184,828,453 | 169,678,849 | 79,231,052 |
| (91.8%) | ||||||
| ♀ GH2 × ♂ BRE | F | 197,048,594 | 34,868,591 | 162,180,003 | 147,618,963 | 76,799,603 |
| (91.0%) | ||||||
| ♀ GH2 × ♂ BRE | F | 226,038,046 | 42,007,698 | 184,030,348 | 167,729,763 | 86,451,322 |
| (91.1%) | ||||||
| ♀ BRE × ♂ GH2 | F | 231,467,876 | 57,694,047 | 173,773,829 | 159,153,208 | 80,775,575 |
| (91.6%) | ||||||
| ♀ BRE × ♂ GH2 | F | 320,963,252 | 81,771,733 | 239,191,519 | 220,417,355 | 103,756,971 |
| (92.2%) | ||||||
Identification of Parent-of-Origin Expressed Genes
SNPs Discriminating Parents
In this work, we benefited from 708,898 SNPs identified in the Brazilian (BRE) and Guadeloupian (GH2) strains (Clément et al. 2013). These two inbred strains display low nucleotide diversity within each strain, but a high differentiation level between them (mean Fst = 0.73) (Clément et al. 2013). We selected 230,425 of those SNPs (32%) that were fixed and discriminant between the two strains (“private” SNPs) (genome browser available at http://genome.univ-perp.fr; last accessed February 16, 2018). Among these, 29,796 SNPs (13%) were found across the transcriptome of the hybrid progenies. The majority (56%) of these SNPs was situated in exons of known genes (16,587 exons from 6,612 genes), while 13,209 (44%) were situated in introns or nonannotated regions.
SNPs Expression between Replicates and Reciprocal Crosses
We compared SNP expression patterns between replicates. Between 70% and 77% of the SNPs had the same expression patterns in biological replicates when attributed to one of the three defined categories (i.e., biallelic, biased or monoallelic, see description in Materials and Methods) (table 3). We thus analyzed 10,665 SNPs for males and 14,412 SNPs for females and compared their allele expression profiles between reciprocal crosses to distinguish strain-of-origin effects from parent-of-origin effects (table 3). Among them, 5, 422 (51%) and 7,095 (49%) of the SNPs had a biallelic expression pattern for males and females, respectively. In total, 128 (1%) and 1,063 (7%) SNPs showed a parent-of-origin expression pattern for males and females, respectively. Others were either expressed in a strain-of-origin manner or had a nonconcordant expression pattern between reciprocal crosses, representing 6% of all SNPs (e.g., biallelic in one and biased in the other [table 3]), which could be due to mitonuclear incompatibilities in the hybrid crosses (Wolf 2009; Wolf et al. 2014). Indeed, this type of incompatibilities is expected to be manifested and may lead to aberrant gene expression patterns and/or loss of imprinting in one direction of the cross (Wolff et al. 2014).
Table 3.
Summary Information of SNPs Recovered in the Transcriptomic Data and Associated SNPs Expression Patterns
| Parental Discriminating SNPs in Transcriptome | Categorized SNPs between Replicates | Number of SNPs in Reciprocal Crosses | Biallelic SNPs | Strain-of-OriginSNPs | Parent-of-Origin SNPs | OtherSNPs | ||
|---|---|---|---|---|---|---|---|---|
| M: ♀ GH2 × ♂ BRE | 29,796 | 20,273 | 13,333 | 10,665 | 5,422 | 4,422 | 128 | 693 |
| M: ♀ GH2 × ♂ BRE | 23,509 | |||||||
| (70%) | ||||||||
| M: ♀ BRE × ♂ GH2 | 24,731 | 16,455 | (51%) | (41%) | (1%) | (6%) | ||
| M: ♀ BRE × ♂GH2 | 24,966 | |||||||
| (72%) | ||||||||
| F: ♀ GH2 × ♂ BRE | 25,092 | 17,595 | 14,412 | 7,095 | 5,455 | 1,063 | 799 | |
| F: ♀ GH2 × ♂BRE | 26,286 | |||||||
| (74%) | ||||||||
| F: ♀ BRE × ♂ GH2 | 27,349 | 21,014 | (49%) | (38%) | (7%) | (6%) | ||
| F: ♀ BRE × ♂ GH2 | 28,885 | |||||||
| (77%) | ||||||||
Parent-of-Origin Expressed Genes
We investigated whether SNPs with parent-of-origin patterns were situated in known genes of S. mansoni. In males, 108 SNPs were located in the exons of 15 genes (in addition to one mitochondrial) and expressed from the same parent in both reciprocal crosses. Six different chromosomes were concerned, with three genes on chromosomes 1, 2, and 4, and one gene on the unplaced scaffold_0193 of chromosome 2, the unplaced scaffold_0083 of chromosome 3, chromosome 5, ZW linkage group (Z-specific region), unplaced scaffold_0138 and unplaced scaffold_0164. Most SNPs (n = 86) were located in the same gene (cytochrome c oxidase I) of the mitochondrial genome of maternal inheritance and thus providing an internal control as they had a strictly maternal expression pattern (table 4). Among the 15 genes in males, one displayed monoallelic expressed SNPs from the maternal allele, while 14 genes showed parentally biased SNPs in equal proportion between the mother’s (n = 7) and the father’s allele (n = 7) (table 5).
Table 4.
Functional Annotation of Genes Containing Parent-of-Origin Expressed SNPs
| Chromosomes | Number of Genes with Parental SNPs | Genes or Position | Number of Discriminant SNPs with a Parent-of-Origin Expression | SNPs Positions | Origin | SNPs Expression Pattern | Genes Function |
|---|---|---|---|---|---|---|---|
| Males | |||||||
| Chr_1 | 3 | Smp_151660 | 1 | 2817123 | ♀ | Biased | Putative uncharacterized protein |
| Smp_128980 | 7 |
|
♀ | Monoallelic | Aminomethyltransferase | ||
| Smp_083130 | 2 |
|
♀ | Biased | Beta1, 3-glucuronyltransferase I | ||
| Chr_2 | 3 |
|
1 | 10177131 | ♂ | Biased | Endonuclease-reverse transcriptase |
| Smp_169030 | 1 | 17479792 | ♂ | Biased | Probable asparagine–tRNA mitochondrial | ||
| Smp_147330 | 1 | 19943769 | ♂ | Biased | Probable ATP-dependent RNA helicase dhx34 | ||
| Chr_2.SC_0193 | 1 | Smp_171530 | 1 | 63337 | ♂ | Biased | Beta Parvin related |
| Chr_3.SC_0083 | 1 | Smp_168560 | 1 | 661857 | ♀ | Biased | Steroid dehydrogenase |
| Chr_4 | 3 | Smp_149950 | 1 | 3351765 | ♀ | Biased | Bifunctional coenzyme a synthase |
|
1 | 18631541 | ♂ | Biased | Gag-pol polyprotein | ||
| Smp_131150 | 1 | 28344103 | ♀ | Biased | Exosome component 10 | ||
| Chr_5 | 1 |
|
1 | 538919 | ♀ | Biased | Tpa: endonuclease-reverse transcriptase |
| ZW linkage group | 1 | Smp_171960 | 1 | 15939680 | ♂ | Biased | Dehydrogenase: reductase SDR family 1 |
| SC_0138 | 1 | Smp_125620 | 1 | 248617 | ♂ | Biased | Coiled-coil domain-containing protein 60 |
| SC_0164 | 1 | Smp_094930 | 1 | 60425 | ♀ | Biased | Early growth response protein 1 |
| Mitochondria | 1 |
|
86 | Not shown | ♀ | Monoallelic (mitochondrial) | Cytochrome c oxidase subunit i |
| Females | |||||||
| Chr_1 | 5 | Smp_034860 | 1 | 15501570 | ♀ | Biased | Nuclear receptor 2dbd gamma |
| Smp_128970 | 1 | 21880758 | ♂ | Biased | Endonuclease-reverse transcriptase/Inhibitor of growth protein 3 | ||
| Smp_173620 | 1 | 30834128 | ♀ | Biased | Transmembrane protein C9orf5/Strawberry notch related | ||
|
2 |
|
♀ | Biased | Gag-pol polyprotein | ||
| Smp_154960 | 1 | 44621620 | ♂ | Biased | Putative cop-coated vesicle membrane protein P24 Emp24/gp25l family | ||
| Chr_2 | 2 | Smp_142400 | 1 | 4664882 | ♂ | Biased | Bhlhzip transcription factor Bigmax |
| Smp_122810 | 2 |
|
♂ | Biased | Mechanosensory protein 2/MEChanosensory abnormality family member | ||
| Mitochondria | 1 |
|
24 | Not shown | ♀ | Monoallelic (mitochondrial) | Cytochrome c oxidase subunit i |
Note.—The localization of the genes in known chromosomes or unassembled portion of S. mansoni’s genome for male and female adults is represented in this table. Number of SNPs, parental origin, expression pattern, and functional annotation are also presented. Expressed genes detected in this study that were not in the current annotated regions of S. mansoni’s genome version (v5.2) are mentioned as XLOCs as identified in the GTF transcriptome reference (Picard et al. 2016). Notice that for those genes, the genomic position has been provided.
Table 5.
Number of Genes Containing Parent-of-Origin SNPs
| SNP Patterns in Reciprocal Crosses | Male Progeny | Female Progeny | Total |
|---|---|---|---|
| Genes with paternal monoallelic SNPs | 0 | 0 | 0 |
| Genes with paternal biased SNPs | 7 | 4 | 11 |
| Genes with maternal monoallelic SNPs | 1 (+1 mitochondrial) | 0 (+1 mitochondrial) | 1 |
| Genes with maternal biased SNPs | 7 | 3 | 10 |
| Total monoallelic | 1 | 0 | 1 |
| Total biased | 14 | 7 | 21 |
| Total genes with parent-of-origin expressed SNPs | 15 | 7 | 22 |
Note.—Genes with parent-of-origin SNPs are categorized 1) according to their expression in male and female progenies and 2) according to their expression pattern (i.e., maternal or paternal, and monoallelic or biased).
In females, SNPs showing consistent parent-of-origin expression were found in exons of a total of 378 different genes. Among these, 278 genes had SNPs with a strict paternal expression and were known to be located in Z-specific regions (i.e., ZW linkage group: position 3550000–13340000, 13860000–19650000, and 23230000–30820000 of the v5.2 assembly [Protasio et al. 2012]) (supplementary table S1, Supplementary Material online), which is consistent with the heterogametic status of female schistosomes. Indeed, heterogametic female schistosomes inherit their W chromosome from the mother and the Z from the father: Such patterns provided again an internal control validating the accuracy of our method for the detection of parent-of-origin genes. Z-specific genes were therefore not considered in further analysis.
Additionally, 92 genes located on 22 individual unplaced scaffolds (including Chr_1.unplaced. SC_0034 and Chr_3.unplaced. SC_0192) contained SNPs with a paternal expression pattern (supplementary table S1, Supplementary Material online). We thus considered them as potentially Z-specific and verified their status in the four biggest unplaced scaffolds containing the larger amount of genes, for unique (ZW) or double copy (ZZ) of genomic DNA in males and females using a quantitative PCR approach (table 6). Unplaced scaffold_0115 of the ZW linkage group (Chr_ZW.unplaced.SC_0115), as well as the unplaced scaffolds number 0111 (SC_0111), 0129 (SC_0129), and 0136 (SC_0136), were thereafter classed as Z-specific (table 6). The other scaffolds were not validated by qPCR but were considered as potentially Z-specific and excluded from further analysis (supplementary table S1, Supplementary Material online).
Table 6.
Quantitative PCR Results: Validation of Four New Z-Specific Scaffolds
| Tested Scaffolds or Controls | Target Positions | Primer Sequences (5ʹ–3ʹ) | Expected Product Sizes (bp) | Primer Efficiencies | Fold Changes (female/male) |
|---|---|---|---|---|---|
| Chr_ZW (Z-specific control) | 28071666–28072040 | −TGTTATCAAACGCCCAGTGA- | 375 | 1.8 | 0.47 |
| −CGTTGAAAAGCCGAGTTTGT- | |||||
| Chr_1 (autosome) | 32740790–32741111 | −CCTCACGAGGTACTCGAAGC- | 322 | 1.8 | 1 |
| −TATGGGACCTGCAACCTTTC- | |||||
| Chr_ZW.unplaced.SC_0115 | 23140–23512 | −CCTGCTTAGACCGCCTGTAG- | 373 | 2 | 0.45 |
| −ACTGTTTCGGCCGTAATGTC- | |||||
| 198332–198645 | −TCGGTTGGTGTCTGATGGTA- | 314 | 2 | 0.48 | |
| −CCACTGACCAATTTCCTCAAA- | |||||
| 729203–729462 | −TCATCTGTCTCCCAGGCATT- | 260 | 2 | 0.42 | |
| −GGCAAGAACATGACCGAGAT- | |||||
| SC_0111 | 155978–156340 | −GCTCCTCCATGTCCAACTCT- | 363 | 1.8 | 0.44 |
| −ACGCATTCGTAGCCGAGATA- | |||||
| 626009–626295 | −GGCACCCTGTAAATTCATCC- | 287 | 1.8 | 0.55 | |
| −CCTGCTTTTAGTTGCCCTGA- | |||||
| 1059389–1059655 | −TGGATCCGAAAATTGTTTGTC- | 267 | 1.8 | 0.46 | |
| −GTACCGCTTTCAAAACATGC- | |||||
| SC_0129 | 99114–99482 | −GATGTCAATGTGAGGCCAAA- | 369 | 1.8 | 0.52 |
| −GGCTACTCGTGTCCCGTAAG- | |||||
| 320296–320629 | −GCTTAGGAATAAGCGGTTCG- | 334 | 1.8 | 0.42 | |
| −AACGGCATAAATGGGTGAAT- | |||||
| SC_0136 | 134667–134949 | −TCGATAATCCCATGCACTCA- | 283 | 1.8 | 0.41 |
| −CCTTCATGAAAAACAGGGAAA- | |||||
| 23063–23394 | −AAAAGAACGCTTCACCGAAA- | 332 | 2 | 0.45 | |
| −TGAATCGTGCTGATTCTCCA- |
Note.—Two control regions were used with one in a known Z-specific region (see Protasio et al. 2012) and one in an autosomal region. Three primers were taken arbitrary in the unplaced scaffolds to be tested. The target positions, primer sequences, as well as the expected product sizes, primer efficiencies and female/male fold changes are presented in the table.
The remaining seven genes in females (in addition to one mitochondrial) presented biased SNPs consistent with parent-of-origin expression. Two distinct regions in the genome were concerned, with SNPs located in five genes on chromosome 1, and two genes on chromosome 2. These genes contained biased SNPs expressed from the maternal (n = 3) or paternal (n = 4) alleles. And last, the “cytochrome c oxidase” was identified again with 24 SNPs located in the gene and with a maternal expression (tables 4 and 5).
Function of the Parent-of-Origin Expressed Genes
The majority of genes identified with a parent-of-origin SNP expression in male and female schistosomes (table 4) had biological processes involved in metabolism, growth, and development. We furthermore identified putative genetic mobile elements of unknown functions. Two types of expression patterns were observed with 1) either the expression of paternal SNPs in males and maternal SNPs in females, or 2) the opposite feature with paternal SNPs expressed in females or maternal SNPs expressed in males. Interestingly, most of the genes concerned with these SNPs expression patterns were related to developmental pathways and therefore potentially related to sex-specific and dimorphic phenotypes.
Is first described in this section the function of genes presenting SNPs expressed from the parent of the same sex in progeny. In males, seven genes with paternally expressed SNPs were identified. Among those, we pinpointed the putative ATP-dependent RNA helicase dhx34 (Smp_147330) protein-coding gene, which is part of the DEAD box proteins family, conserved in metazoan they are known for their implication in embryogenesis, spermatogenesis, cellular growth, and division (Godbout and Squire 1993; Johnstone et al. 2005; Matsumoto et al. 2005). Its ortholog in Caenorhabditis elegans (smgl-2) is critical for muscle development (Williams and Waterston 1994). Furthermore, such helicases are part of a complex called the “compensasome” because they mediate dosage compensation mechanism (Sanjuán and Marín 2001) in other organisms. The Beta-Parvin related gene (Smp_171530) is essential during embryogenesis in other organisms (Zhang et al. 2004; Montanez et al. 2009) and its ortholog in C. elegans is required for muscle assembly and function, while mutation of the gene leads to embryonic lethality. The coiled-coil domain-containing protein 60 (Smp_125620) potentially involved in protein demethylation and protein–protein interactions is also thought to be important for embryonic development in vertebrates, with an identified role in the dorsoventral axial establishment of zebrafish (Wei et al. 2016). A dehydrogenase reductase sdr family member gene (Smp_171960) has also been identified and interestingly a previous association study has shown a paternal parent-of-origin effect of that gene on language impairment (Nudel et al. 2014). At last three genes were identified with paternally expressed SNPs, one being a probable asparagine tRNA mitochondrial coding gene (Smp_169030) and has been related to developmental diseases in humans (Sofou et al. 2015; Vanlander et al. 2015), while two are genetic mobile elements with an endonuclease or endopeptidase activity (XLOC_009689 and XLOC_015537).
In females, the genes with maternally expressed SNPs identified had functions linked to development and female specificities. The most relevant gene that can directly be related to female-specific phenotypes was the nuclear receptor 2dbd gamma gene (Smp_034860). It contains a domain similar to the ligand-binding domain of C. elegans nuclear hormone receptor Sex-1 protein. This transcription factor plays pivotal role in sex fate of C. elegans by regulating the transcription of the sex-determination gene xol-1, which specifies male fate when active and hermaphrodite fate when inactive (Carmi et al. 1998; Farboud and Meyer 2006). Other genes related to developmental pathways were identified such as the “Strawberry Notch related” gene (Smp_173620). Notch genes encode for transmembrane proteins and have conserved functions in developmental pathways. They are required during embryogenesis and oogenesis in Drosophila and zebrafish (Coyle-Thompson and Banerjee 1993; Majumdar et al. 1997; Takano et al. 2010).
At last, a genetic mobile element (XLOC_001902), identified as a gag-pol polyprotein, was maternally expressed in schistosome females.
The analysis of SNPs with opposite expression patterns in males and females (i.e., from the parent of opposite sex) also revealed genes potentially involved in male and female developmental pathways but may also be relevant in a coevolution context between sexes. Concerning maternally expressed SNPs in males, eight genes were identified including one with monoallelic expressed SNPs (Smp_128980). This aminomethyltransferase gene is nuclear-encoded but confined to the mitochondria and is involved in glycine metabolism. Also, mainly present in the mitochondrial matrix, we identified the Bifunctional coenzyme A synthase gene (Smp_149950). This last gene predicted to be involved in ATP-binding activity and dephospho-CoA kinase activity has an ortholog in C. elegans (Y65B4A.8), which seems involved in embryo development, feminization of hermaphroditic germ-line, germ cell development, nematode larval development, regulation of cell proliferation, and regulation of meiotic nuclear division (Kerins et al. 2010; Waters et al. 2010).
Other genes with maternally biased SNPs in males were identified. The “putative Beta1-3 glucuronyltransferase” (Smp_083130) which is involved in carbohydrate metabolic process is mainly expressed in the brain of Drosophila and is involved in the growth of peripheral nerves during larval development (Pandey et al. 2011). In C. elegans sqv-8 (homologous to three distinct glucuronyl transferases [GlcAT-I, GlcAT-P, and GlcAT-D]), which encodes a glucuronyl transferase, is required for one-cell embryos and for vulval morphogenesis. Sqv mutants have a vulval defect, but also an oocyte and somatic gonad defect, resulting in hermaphrodite sterility. Moreover, some sqv mutations cause maternal-effect lethality (Herman et al. 1999; Bulik et al. 2000) It has also been shown that glct-6 (another Beta1-3 glucuronyltransferase ortholog in C. elegans) is involved in determination of adult lifespan (Kim and Sun 2007). The “exosome component 10” gene (Smp_131150), also known as “polymyositis scleroderma autoantigen” in humans, is a cell death-related nuclease, that is required for DNA degradation during apoptosis. It is also thought to participate in dosage compensation mechanism by inactivation of the X chromosome. Indeed, in mouse, a specific nuclear component of the exosome (Exosc10) gene involved in mRNA degradation pathways leads to downregulation of spliced Xist transcript production and blocks the onset of the X-inactivation process (Ciaudo et al. 2006). The transcription factor gene “Early growth response protein 1” (Smp_094930) targeting important genes for normal development and differentiation (Silverman et al. 1998; Pagel and Deindl 2011) may also play an important role in Schistosoma blood flukes development. Finally, a Steroid dehydrogenase gene (Smp_168560) was detected with a maternal SNPs expression in males. Interestingly, steroid dehydrogenase pathway was also found overrepresented in male schistosomes from cercariae to adult stages (Picard et al. 2016). In C. elegans, some ortholog genes to Smp_168560 were found (stdh-4, stdh-1, and let-767) they encode for a putative steroid dehydrogenase and expressed not only in larval and adult pharynx (stdh-4) but also in larval intestine, and in both larval and adult body wall muscle and neurons (stdh-1) and is required for normally short lifespan. The ortholog gene let-767 is particularly important for C. elegans development, as it is required for embryogenesis, and female reproduction. This gene is zygotically expressed in the intestine, but a maternal-effect lethal allele (let-767 [s2464]) also exists, indicating that LET-767 is probably provided maternally which is consistent with our maternal expressed SNPs identified. Mutations in this gene cause abnormal embryonic development, slow growth and small adult body size as well as a failure to mature gonads and last an hypersensitivity to low cholesterol (Kuervers et al. 2003). Other genes with opposite expression patterns in males and females could not be associated to any known functions (Smp_151660) or were predicted as a genetic mobile element with an endonuclease activity (XLOC_017885).
In females, five genes were found containing SNPs expressed from the paternal allele. Among them, one gene exhibited a monoallelic expressed SNP, while the four others contained biased SNPs. In this last situation we identified an uncharacterized protein-coding gene (Smp_128970), containing PHD finger domains, related to the “inhibitor of growth protein family” which has been implicated in chromatin-mediated transcriptional regulation in C. elegans embryo and is therefore potentially important for schistosome development (Luo et al. 2009). Other genes were involved in not only protein and vesicle-mediated transport (Smp_154960) but also transcriptional regulation of developmental pathways (Smp_142400) (Steingrímsson et al. 1998; Hallsson et al. 2004; Hsu et al. 2004) and mechanosensory response (Smp_122810) (Huang et al. 1995).
Discussion
Parent-of-origin gene expression has been studied in mammals, plants, and some invertebrates (da Rocha and Ferguson-Smith 2004; Arico et al. 2011; Ferguson-Smith 2011; Macdonald 2012; Kocher et al. 2015), and aberrant expression in gene subject to imprinting has severe consequences, mainly on development and growth (Tycko and Morison 2002). The term imprinting was first used to describe the elimination of an entire paternal chromosome in sciara flies (Crouse 1960) and refers to the entire heterochromatization (silencing) of the paternal genome in the soccid mealybug, which is involved in maleness determination (Khosla et al. 2006). In addition to the kinship theory of genomic imprinting (Haig 2000), another interesting alternative explanation proposes that imprinting may be related to a mechanism for the resolution of intralocus sexual conflict and may be important for traits under sex-specific selection (Day and Bonduriansky 2004; Bonduriansky 2007). This sexual antagonism theory involving male and female coevolution has the potential to act differently for each sex (sex-specific imprinting) and predicts that imprinting may affect organisms with sexual dimorphism (Bonduriansky 2007) as it is the case in schistosomes adult parasites. For that reason, we have investigated S. mansoni transcriptome for parent-of-origin gene expression considering intralocus sexual conflicts and sexual antagonism. We have identified genes containing SNPs expressed in a parent-of-origin manner and even if larger scale parent-of-origin gene expression in this species could not be addressed here we argue that regarding to the function of the genes concerned by these pattern, they have the potential to explain sexual differences in schistosome parasites.
A total of 1,191 SNPs were identified as expressed in a parent-of origin manner in male (128 SNPs) and female (1,063 SNPs) adult schistosomes (table 3). These parental SNPs were located in 15 genes in males (in addition to the cytochrome c oxidase) and in 378 genes in females. As expected, the mitochondrial cytochrome c oxidase gene was also detected with a maternal expression in both sexes, thus providing validation of our methodology. In females, these parental expressed SNPs also revealed as expected 278 known Z-specific genes expressed exclusively from the paternal allele (monoallelic SNPs expression), which is consistent with the schistosomes female heterogametic situation. Furthermore, in females, 92 genes situated on 22 unplaced scaffolds were found with a paternal expression pattern. We hypothesized that they might be related to Z-specific loci and tested the four biggest unplaced scaffolds containing the majority of paternal expressed genes using a quantitative PCR approach. This allowed us to identify four new Z-specific genomic regions (unplaced scaffold 0115 of the ZW linkage group, unplaced scaffold 0111, 0129 and 0136, table 6). We believe that other unplaced scaffolds are potentially Z-specific and need further validation (supplementary table S1, Supplementary Material online).
A total of 22 genes showing expressed SNPs consistent with parent-of-origin expression were identified in reciprocal cross duplicates of female (seven genes) and male (15 genes) adult schistosomes. These genes only contained few SNPs consistent with parental expression and the majority of them was biased rather than monoallelic. Notwithstanding, in males gene “Smp_128980” contained seven monoallelic SNPs expressed from the maternal allele and gene “Smp_083130” contained two biased SNPs expressed from the maternal allele, while in females two biased SNPs were identified in XLOC_001902 and Smp_122810 expressed from the maternal and paternal alleles, respectively (table 4). This is probably the consequence of the poor amount of discriminant SNPs from which we have based our analysis and therefore hampers our ability to confirm any existing imprinting at the whole gene level in this species. Another consideration comes from the fact that imprinting may be tissue specific and that working with whole adult worms may mitigate the resulting expression patterns found in this work. Moreover, in the present experimental design the adult worms recovered from single-sex infections present an immature phenotype (especially for female worms that rely on males for their maturation), which may have biased the expression and hamper the detection of further imprinted genes. Nevertheless, the genes identified with these parent-of-origin expressed SNPs in males and females had interesting functions related to sex-specific phenotypes and sexual dimorphism in this species and share interesting common relations with parent-of-origin genes expression theories.
If we base our reflection on the kinship theory (Haig 2000) defined by parental conflict over resource allocation and offspring fitness, we would of expected to find an impact of gene dosage on the fitness of matrilineal or patrilineal relatives. Moreover, usually schistosome couples are monogamous, but mate change can occur among these parasites (Tchuem Tchuenté et al. 1996; Beltran and Boissier 2008, 2009) potentially causing a source of relatedness asymmetry for matrigenes and patrigenes in offspring. For example, maternally expressed alleles would be selected to reduce the extraction of resources and therefore reduce organism size, while paternally expressed genes should favor taking more resources from the mother and therefore induce growth. Indeed males are much more muscular than females (i.e., sexual dimorphism), and sexual selection may favor large body in males and the opposite in females, thus paternal genes should be selected in males to extract resources at a greater rate than in females. Schistosomes are oviparous organisms therefore neither the paternal or maternal genome can directly influence resource allocation after spawning. As observed in this study for schistosomes, it is difficult to reconcile these predictions to the function of the genes identified in this work except for genes that may influence growth and development of offspring, but this may overlap with the functions predicted by other models and in particular the sexual antagonism theory.
A clear dimorphic trait found in adult males is their strong musculature compared with the thin females. Interestingly we identified genes containing SNPs expressed from the paternal alleles that seem important for muscle development (Smp_147330, Smp_171530, Smp_125620), which is clearly predicted by the sexual antagonism theory. In some case, mutations of orthologous genes in C. elegans lead to embryonic lethality (Smp_171530). Moreover, some of those genes have been related to paternal parent-of-origin effects (Smp_171960) (Nudel et al. 2014) and developmental diseases in humans (Smp_169030) (Sofou et al. 2015; Vanlander et al. 2015), which often find their cause in imprinting defaults. Another interesting point may concern the relationship with dosage compensation mechanism (Smp_147330) as such helicases genes mediate dosage compensation mechanism in other organisms (Sanjuán and Marín 2001). In schistosomes, heterogametic (ZW) females need to balance Z-linked transcript levels relative to homogametic (ZZ) males (Vicoso and Bachtrog 2011; Graves 2016). These previous observations are consistent with the sexual antagonism theory proposed by Day and Bonduriansky (Day and Bonduriansky 2004; Bonduriansky 2007).
Maternal expressed SNPs found in male genes were involved in development (Smp_149950, Smp_094930), with embryogenesis and reproduction (Smp_168560) but also growth (Smp_083130). In C. elegans let-767 (orthologous to Smp_168560) is zygotically expressed in the intestines, and mutations in this gene cause abnormal embryonic development, slow growth and small adult body size as well as a failure to mature gonads and last a hypersensitivity to low cholesterol (Kuervers et al. 2003). Other mutations of C. elegans orthologous genes (sqv genes orthologous to Smp_083130; let-767 orthologous to SMP_168560) cause maternal-effect lethality (Herman et al. 1999; Bulik et al. 2000) indicating that they are probably provided maternally in C. elegans, which is similar to our observations in schistosomes. This may suggest that the maternal genome largely contributes to male phenotypes and is probably also indispensable for proper schistosome development. Moreover, we identified a probable candidate gene (Smp_131150, exosome component 10) known to be involved in dosage compensation mechanism in mouse (Exosc10) as it leads to downregulation of spliced Xist transcript production and blocks the onset of the X-inactivation process (Ciaudo et al. 2006). This seems to indicate a strong coadaptation between sexes in schistosomes. Moreover, consistent with such coadaptation we identified in males two genes (Smp_128980, aminomethyltransferase, containing maternal monoallelic SNPs together with the Bifunctional coenzyme A synthase gene [Smp_149950] containing maternal biased SNPs) that are mainly confined to the mitochondria. This may underlie coevolved complexes between the maternal inherited mitochondrial genome and nuclear maternally expressed genes in males, and therefore male and female coadaptation. Cytonuclear interacting genes seem important in the evolution of genomic imprinting (Wolf 2009). Such genes are important in a coevolution context between sexes because it may allow stronger coadaptation between the maternal inherited mitochondrial genome and the expression of maternal nuclear alleles in males (Beekman et al. 2014).
In females, we identified genes involved in developmental pathways (Smp_173620), embryogenesis and oogenesis and also in transcriptional regulation of embryo development and animal reproduction (Smp_142400). Others were related not only to protein and vesicle-mediated transport (Smp_154960) but also to mechanosensory response (Smp_122810) (Huang et al. 1995). Interestingly, we identified an uncharacterized protein-coding gene (Smp_128970), which seems to play a role in chromatin-mediated transcriptional regulation, and also contain a PHD finger domain, related to the “inhibitor of growth protein family.” This gene found in females has SNPs expressed from the paternal allele and together with other genes expressed from the paternal allele in females may once more allow a better coadaptation between sexes. Indeed, the paternal genome may influence the transcriptomic profile of females in order to increase the adaptive integration of offspring and the paternal genome leading to higher female’s fitness. This observation is also consistent with the sexual antagonism theory, but rather than expressing male- or female-specific coding genes in the same sexes, it shows that the opposite parent may influence the phenotype of the progeny in order to stick with its own sexual features (e.g., Smp_128970; inhibitor of growth protein family expressed from the paternal allele in females potentially tends to reduce females size which is one important dimorphic trait representative of schistosome sexual features). This again shows how difficult the patterns observed in our organism can be reconciled with any particular model on the evolution of parent-of-origin gene expression and be nonexclusive.
Importantly, this also suggests that parent-of-origin gene expression or imprinting may occur in systems that have previously not been the focus of research because of the absence of conflict over maternal investment, but that imprinting shall be present in organisms where coadapted traits may have fitness effects for progeny (Wolf and Hager 2006). This theory described in the maternal–offspring coadaptation theory (Wolf and Hager 2006), in any case does not reject the possibility of having coadaptation between paternal and offspring traits (Wolf and Hager 2006). Therefore, we may have stronger coadaptation between the paternal genome and offspring in schistosomes rather than relative to the maternal genome. Thus, the maternal–offspring coadaptation theory may not be obvious in schistosomes and this may be a reason why we do not observe any particular features predicted by this model. Furthermore, this is plausible considering the strong influence that male schistosomes have on their female’s fitness (Popiel 1986; Boissier and Moné 2001; Lu et al. 2016). An example of such coevolution at the phenotypic level can be observed from the fact that the male schistosomes continuously feed the female via the tegument and that female cannot reach sexual maturity without pairing with a male (Popiel 1986; Beltran and Boissier 2008). In order to explain the monogamous mating system of schistosomes it has been proposed that prezygotic paternal investment (transport of the female to the oviposition site, female maturation, and feeding) is higher than maternal investment (Beltran and Boissier 2008) in contrast with other models on which imprinting has been studied. Second, it is well established that the sex ratio is male-based making the male the more competitive sex (Boissier and Moné 2000; Moné and Boissier 2004; Beltran and Boissier 2010).
At last in females the genes expressed from the maternal allele once more provide interesting elements concordant with the sexual antagonism theory. Indeed, some had particular conserved functions in developmental pathway (Smp_173620; Strawberry Notch related) and required during embryogenesis and oogenesis in other organisms (Coyle-Thompson and Banerjee 1993; Majumdar et al. 1997; Takano et al. 2010). Moreover, we identified a gene with a maternal biased SNP in females (Smp_034860) containing a domain similar to the ligand-binding domain of C.elegans nuclear hormone receptor Sex-1 protein. It is used as an X-chromosome signal that determines nematode sex and may also play a role in dosage compensation mechanisms (Carmi et al. 1998; Farboud and Meyer 2006). As the molecular determinants of sex determination are not yet known in schistosomes besides that it is genetically determined during egg fertilization, this gene seems to be an interesting candidate to explore.
Why Is Parent-of-Origin Expression an Interesting Phenomenon to Explain Gonochorism and Sexual Dimorphism in Schistosomes and May Need Further Consideration?
As seen, most of the expression patterns observed in this work are difficult to reconcile with all predictions made by the different models of the evolution of genomic imprinting, namely the kinship theory (Moore and Haig 1991; Haig 2000), the maternal–offspring coadaptation theory (Wolf and Hager 2006), and the sexual antagonism theory (Day and Bonduriansky 2004; Bonduriansky 2007). Nevertheless regarding sexual dimorphism, we have mainly focused here on patterns predicted by the sexual antagonism theory. Day and Bonduriansky predicted in their sexual antagonism theory (Day and Bonduriansky 2004) that sex-specific imprinting may occur, and in combination with sexually antagonistic selection result in sexual dimorphism. Studies have clearly shown that some parentally expressed genes are expressed in a sex-specific manner and might be at the base of sexual dimorphism and differences in behavior like, for example, maternal care (Gregg et al. 2010), but also have effects on complex traits (Hager et al. 2008). Sex-specific parent-of-origin expression in adults can thus contribute to transcriptional differences leading to morphological and or physiological changes at the base of male and female adult phenotypes in contrast with the schistosomes morphologically indistinguishable larval stages. Therefore, sex-specific imprinting would allow males and females to approach their phenotypic optima and may contribute to the evolution of sexual dimorphism (Bonduriansky 2007). In this work, we highlighted interesting elements concordant with such predictions.
First, most of the genes identified have functions related to development and sex-specific functions. As we know that schistosomes have evolved from hermaphroditic ancestors, both sexes have therefore obligatorily coevolved and parent-of-origin gene expression may allow stronger coadaptation between sexes as seen previously. Second, gonochorism is thought to have appeared with the colonization of warm-blooded species (Loker and Brant 2006). The diversity generated by sexual reproduction was proposed to allow the parasite to counter vertebrate’s immune system. Moreover, sexual dimorphism is essential for schistosomes and allows male and female schistosomes to have complementary roles in the definitive hosts (Basch 1990). Indeed, males are much more muscular than females (i.e., sexual dimorphism), and sexual selection may favor large body in males and the opposite in females. It is interesting to consider that parent-of-origin expression may have allowed sexual reproduction maintenance in order to generate diversity in these species, as imprinting defaults can have detrimental effects in other organisms (Swales and Spears 2005) and cause embryonic lethality. Furthermore, it might have contributed to the evolution of sexual dimorphism as proposed by Day and Bonduriansky’s predictions (Day and Bonduriansky 2004). At last, the identification of genetic mobile elements with parent-of-origin expressed SNPs is also an interesting point as it has been shown that imprinted regions tend to show significantly more transposon insertions or more generally, retroviral repeats, than other regions (Pignatta et al. 2014). Thus, imprinting in schistosomes could also be the side effect of host defense mechanism against foreign DNA (McDonald et al. 2005) that was thereafter selected because of its adaptive significance. It has recently been shown that “captured” genes of retroviral origin (Syncytins) have sex-dependent effects in mice, and that they are involved in the development of the placenta in females, and have a role in muscle development in males (Redelsperger et al. 2016). This again underlies a possible contribution of imprinted genes to sexual dimorphism in schistosome parasites.
Conclusion
Schistosomes are diploid metazoan parasites with separate sexes, thus the maternal and paternal genomes are thought to contribute equally to the fitness of their offspring. In this work, we explored the transcriptome of male and female adult worms to search for parent-of-origin expressed genes. It allowed us to not only identify new Z-Specific loci but also detect genes containing SNPs consistent with parent-of-origin patterns. Because of the poor amount of SNP markers available, we cannot conclusively affirm that imprinting exists at a larger scale in this species, but suggest that it needs further considerations. Interestingly, in males and females an important number of genes identified with parent-of-origin SNPs are related to developmental processes such as embryogenesis, spermatogenesis, and oogenesis and may act as fundamental contributors to male and female phenotypes and more specifically to sexual dimorphism. Other genes related to dosage compensation mechanisms also seem important for sex-specific phenotypes, especially in males, and the identification of the Sex-1 protein gene ortholog found in females is an important candidate to explore the molecular bases of sexual determination in schistosomes. We propose that parent-of-origin gene expression may be a mechanism allowing to mitigate conflict linked to parenthood and potentially solve intralocus sexual conflict in this species as it has been proposed from Day and Bonduriansky’s sexual conflict theory, defining imprinting as a mechanism for the evolution of sex specific traits. In schistosomes, it is appealing to think that if imprinting exists in this species, it may coincide with the shift from hermaphroditism to gonochorism and thus may be at the base sexual reproduction and sexual dimorphism maintenance. Importantly, future studies on parent-of-origin expression in schistosomes should address the question of the underlying molecular mechanisms. Indeed, the imprinting by the usual mechanism of DNA methylation could be controversial in this species for which such mechanism is still under debate (Raddatz et al. 2013); therefore other epigenetic actors will have to be explored, such as histone modifications. Growing evidences show that chromatin modifications are indeed involved in schistosomes development (Roquis et al. 2015), moreover in a sex-specific manner (Picard et al. 2016). Another interesting perspective would be to look for parental biased expression in the nondifferentiated cercarial stage. Finally, by detecting genes with strain-specific expressions, our unprecedented analysis opens new perspectives to understand hybridization mechanisms in schistosomes and thus on the disease dynamics, including interspecific interactions in the context of the modification of species geographical distribution due to climate change (Boissier et al. 2016).
Ethic Statement
All experiments were carried out according to national ethical standards established in the writ of February 1, 2013 (NOR: AGRG1238753A), setting the conditions for approval, planning, and operation of establishments, breeders and suppliers of animals used for scientific purposes and controls. The French Ministère de l’Agriculture et de la Pêche and the French Ministère de l’Education Nationale de la Recherche et de la Technologie provided permit A66040 to the laboratory for animal experiments and certificate to the experimenters (authorization 007083, decree 87–848).
Supplementary Material
Acknowledgments
We would like to thank Anne Rognon and Nathalie Arancibia for rodent and snail maintenance, and Jean-Francois Allienne for technical assistance. We also acknowledge the contribution of the MGX—Montpellier GenomiX sequencing platform. This work was funded by the region Languedoc Roussillon (Chercheur d’avenir Schistosex, recipient: JB).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Literature Cited
- Althammer S, et al. , 2011. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 27(24):3333–3340. p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, et al. , 2015. Schistosoma mansoni egg, adult male and female comparative gene expression analysis and identification of novel genes by RNA-seq. PLoS Negl Trop Dis. 9(12):e0004334–e0004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico JK, et al. , 2011. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 7(6):e1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch PF. 1990. Why do schistosomes have separate sexes? Parasitol Today. 6(5):160–163. [DOI] [PubMed] [Google Scholar]
- Beekman M, Dowling DK, Aanen DK.. 2014. The costs of being male: are there sex-specific effects of uniparental mitochondrial inheritance? Philos Trans R Soc B. 369(1646):20130440.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran S, Boissier J.. 2008. Schistosome monogamy: who, how, and why? Trends Parasitol. 24(9):386–391. [DOI] [PubMed] [Google Scholar]
- Beltran S, Boissier J.. 2009. Are schistosomes socially and genetically monogamous? Parasitol Res. 104(2):481–483. [DOI] [PubMed] [Google Scholar]
- Beltran S, Boissier J.. 2010. Male-biased sex ratio: why and what consequences for the genus Schistosoma? Trends Parasitol. 26(2):63–69. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. , 2009. The genome of the blood fluke Schistosoma mansoni. Nature 460(7253):352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, et al. , 2010. Manipulation of FASTQ data with galaxy. Bioinformatics 26(14):1783–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissier J, et al. , 2016. Outbreak of urogenital schistosomiasis in Corsica (France): an epidemiological case study. Lancet Infect Dis. 16(8):971–979. [DOI] [PubMed] [Google Scholar]
- Boissier J, Moné H.. 2000. Experimental observations on the sex ratio of adult Schistosoma mansoni, with comments on the natural male bias. Parasitology. 121(Pt 4):379–383. [DOI] [PubMed] [Google Scholar]
- Boissier J, Moné H.. 2001. Relationship between worm burden and male proportion in Schistosoma mansoni experimentally infected rodents and primates. A meta-analytical approach. Int J Parasitol. 31(14):1597–1599. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R. 2007. The genetic architecture of sexual dimorphism: the potential roles of genomic imprinting and condition dependence In: Fairbairn DJ, Blanckenhorn WU, Szekely T, editors. Sex, Size and Gender Roles: Evolution- ary Studies of Sexual Size Dimorphism. Oxford: Oxford University Press; pp. 176–184. [Google Scholar]
- Bulik DA, et al. , 2000. sqv-3,-7, and-8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 97(20):10838–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ.. 1998. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature 396(6707):168–173. [DOI] [PubMed] [Google Scholar]
- Ciaudo C, et al. , 2006. Nuclear mRNA degradation pathway (s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2(6):e94.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément JAJ, et al. , 2013. Private selective sweeps identified from next-generation pool-sequencing reveal convergent pathways under selection in two inbred Schistosoma mansoni strains. PLoS Negl Trop Dis. 7(12):e2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes C. 1991. The schistosome scandal. Acta Oecol. 12(1):165–173. [Google Scholar]
- Conesa A, et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Coyle-Thompson CA, Banerjee U.. 1993. The strawberry notch gene functions with Notch in common developmental pathways. Development (Cambridge, England) 119(2):377–395. [DOI] [PubMed] [Google Scholar]
- Criscione CD, et al. , 2009. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 10(6):R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse HV. 1960. The controlling element in sex chromosome behavior in Sciara. Genetics 45(10):1429–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, Bonduriansky R.. 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167(4):1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco R, et al. , 2010. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 20(8):1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels D, et al. , 2002. The global epidemiological situation of schistosomiasis and new approaches to control and research. Acta Trop. 82(2):139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Meyer B.. 2006. Mechanism of sex-1 function in sex determination and dosage compensation In: Development and Evolution Meeting. http://www.citeulike.org/group/6190/article/3326016. [Google Scholar]
- Ferguson-Smith AC. 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 12(8):565–575. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, et al. , 2005. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Mol Biochem Parasitol. 141(1):1–13. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, et al. , 2008. Use of genomic DNA as an indirect reference for identifying gender-associated transcripts in morphologically identical, but chromosomally distinct, Schistosoma mansoni cercariae. PLoS Negl Trop Dis. 2(10):e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R, Squire J.. 1993. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci U S A. 90(16):7578–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 17(1):33–46. [DOI] [PubMed] [Google Scholar]
- Gregg C, et al. , 2010. Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329(5992):682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AI, Short RB, Cain GD.. 1981. Karyotype evolution and sex chromosome differentiation in Schistosomes (Trematoda, Schistosomatidae). Chromosoma 84(3):413–430. [DOI] [PubMed] [Google Scholar]
- Hager R, et al. , 2008. Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol. 8:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. 2000. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 31(1):9–32. [Google Scholar]
- Hallsson JH, et al. , 2004. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167(1):233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman T, Hartwieg E, Horvitz HR.. 1999. sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination. Proc Natl Acad Sci U S A. 96(3):968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-H, Hsieh-Li H-M, Li H.. 2004. Dysfunctional spermatogenesis in transgenic mice overexpressing bHLH-Zip transcription factor, Spz1. Exp Cell Res. 294(1):185–198. [DOI] [PubMed] [Google Scholar]
- Huang M, et al. , 1995. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 378(6554):292–295. [DOI] [PubMed] [Google Scholar]
- Ingleby FC, Flis I, Morrow EH.. 2015. Sex-biased gene expression and sexual conflict throughout development. Cold Spring Harbor Perspect Biol. 7 (1):a017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, et al. , 2005. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 277(1):92–101. [DOI] [PubMed] [Google Scholar]
- Kerins JA, et al. , 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn. 239(5):1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S, Mendiratta G, Brahmachari V.. 2006. Genomic imprinting in the mealybugs. Cytogenet Genome Res. 113(1–4):41–52. p [DOI] [PubMed] [Google Scholar]
- Kim D, et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sun H.. 2007. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell 6(4):489–503. [DOI] [PubMed] [Google Scholar]
- King CH. 2010. Parasites and poverty: the case of schistosomiasis. Acta Trop. 113(2):95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22(3):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, et al. , 2015. A search for parent-of-origin effects on honey bee gene expression. G3 5(8):1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuervers LM, et al. , 2003. The sterol modifying enzyme LET-767 is essential for growth, reproduction and development in Caenorhabditis elegans. Mol Genet Genomics. 270(2):121–131. [DOI] [PubMed] [Google Scholar]
- Langmead B, et al. , 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant JM, et al. , 2012. Chromatin structural changes around satellite repeats on the female sex chromosome in Schistosoma mansoni and their possible role in sex chromosome emergence. Genome Biol. 13(2):R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska AA, et al. , 2015. Sexual dimorphism and the evolution of sex-biased gene expression in the brown alga ectocarpus. Mol Biol Evol. 32(6):1581–1597. [DOI] [PubMed] [Google Scholar]
- Loker ES, Brant SV.. 2006. Diversification, dioecy and dimorphism in schistosomes. Trends Parasitol. 22(11):521–528. [DOI] [PubMed] [Google Scholar]
- Lu Z, et al. , 2016. Schistosome sex matters: a deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci Rep. 6:31150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shah S, Riabowol K, Mains PE.. 2009. The Caenorhabditis elegans ing-3 gene regulates ionizing radiation-induced germ-cell apoptosis in a p53-associated pathway. Genetics 181(2):473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald WA. 2012. Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects. Genet Res Int. 2012:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Nagaraj R, Banerjee U.. 1997. Strawberry notch encodes a conserved nuclear protein that functions downstream of notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev. 11(10):1341–1353. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10. [Google Scholar]
- Matsumoto K, et al. , 2005. Expression of rck/p54, a DEAD-box RNA helicase, in gametogenesis and early embryogenesis of mice. Dev Dyn. 233(3):1149–1156. [DOI] [PubMed] [Google Scholar]
- McDonald JF, Matzke MA, Matzke AJ.. 2005. Host defenses to transposable elements and the evolution of genomic imprinting. Cytogenet Genome Res. 110(1–4):242–249. [DOI] [PubMed] [Google Scholar]
- Moné H, Boissier J.. 2004. Sexual biology of schistosomes. Adv Parasitol. 57(4):89–189. [DOI] [PubMed] [Google Scholar]
- Montanez E, et al. , 2009. Alpha-parvin controls vascular mural cell recruitment to vessel wall by regulating RhoA/ROCK signalling. EMBO J. 28(20):3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D.. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7(2):45–49. [DOI] [PubMed] [Google Scholar]
- Nudel R, et al. , 2014. Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav. 13(4):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel J-I, Deindl E.. 2011. Early growth response 1—a transcription factor in the crossfire of signal transduction cascades. Indian J Biochem Biophys. 48(4):226–235. [PubMed] [Google Scholar]
- Pandey R, Blanco J, Udolph G.. 2011. The glucuronyltransferase GlcAT-P is required for stretch growth of peripheral nerves in Drosophila. PLoS ONE 6(11):e28106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten MM, et al. , 2014. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity 113(2):119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard MAL, et al. , 2016. Sex-biased transcriptome of Schistosoma mansoni: host-parasite interaction, genetic determinants and epigenetic regulators are associated with sexual differentiation. PLoS Negl Trop Dis. 10(9):e0004930–e0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, et al. , 2014. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3:e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popiel I. 1986. Male-stimulated female maturation in Schistosoma: a review. J Chem Ecol. 12(8):1745–1754. [DOI] [PubMed] [Google Scholar]
- Protasio AV, et al. , 2012. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis. 6(1):e1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, et al. , 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 110(21):8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelsperger F, et al. , 2016. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet. 12(9):e1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Walter J.. 2001. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2(1):21–32. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Ferguson-Smith AC.. 2004. Genomic imprinting. Curr Biol. 14(16):R646–R649. [DOI] [PubMed] [Google Scholar]
- Roquis D, et al. , 2015. The epigenome of Schistosoma mansoni provides insight about how cercariae poise transcription until infection. PLoS Negl Trop Dis. 9(8):e0003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H.. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132:365–386. [DOI] [PubMed] [Google Scholar]
- Sanjuán R, Marín I.. 2001. Tracing the origin of the compensasome: evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol Biol Evol. 18(3):330–343. [DOI] [PubMed] [Google Scholar]
- Silverman ES, et al. , 1998. cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1). Biochem J. 336(1):183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofou K, et al. , 2015. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genomic Med 3(1):59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG.. 1998. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125(23):4607–4616. [DOI] [PubMed] [Google Scholar]
- Swales AKE, Spears N.. 2005. Genomic imprinting and reproduction. Reproduction 130(4):389–399. p [DOI] [PubMed] [Google Scholar]
- Tchuem Tchuenté LA, et al. , 1996. Mating behaviour in schistosomes: are paired worms always faithful? Parasitol Today. 12(6):231–236. [DOI] [PubMed] [Google Scholar]
- Takano A, et al. , 2010. Expression of strawberry notch family genes during zebrafish embryogenesis. Dev Dyn. 239(6):1789–1796. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B, Morison IM.. 2002. Physiological functions of imprinted genes. J Cell Physiol. 192(3):245–258. [DOI] [PubMed] [Google Scholar]
- Vanlander AV, et al. , 2015. Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2). Hum Mutat. 36(2):222–231. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 3:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters K, Yang AZ, Reinke V.. 2010. Genome-wide analysis of germ cell proliferation in C. elegans identifies VRK-1 as a key regulator of CEP-1/p53. Dev Biol. 344(2):1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, et al. , 2016. The coiled-coil domain containing protein Ccdc136b antagonizes maternal Wnt/β-catenin activity during zebrafish dorsoventral axial patterning. J Genet Genomics. 43(7):431–438. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D.. 2003. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 4(5):359.. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH.. 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 124(4):475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. 2009. Cytonuclear interactions can favor the evolution of genomic imprinting. Evolution 63(5):1364–1371. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R.. 2006. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4(12):e380–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Oakey RJ, Feil R.. 2014. Imprinted gene expression in hybrids: perturbed mechanisms and evolutionary implications. Heredity (Edinb). 113(2):167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JN, et al. , 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 369(1646):20130443. [DOI] [PMC free article] [PubMed]
- Zhang Y, et al. , 2004. Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J Biol Chem. 279(40):41695–41705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.