Abstract
Small interfering RNA (siRNA)-based drugs require chemical modifications or formulation to promote stability, minimize innate immunity, and enable delivery to target tissues. Partially modified siRNAs (up to 70% of the nucleotides) provide significant stabilization in vitro and are commercially available; thus are commonly used to evaluate efficacy of bio-conjugates for in vivo delivery. In contrast, most clinically-advanced non-formulated compounds, using conjugation as a delivery strategy, are fully chemically modified (100% of nucleotides). Here, we compare partially and fully chemically modified siRNAs in conjugate mediated delivery. We show that fully modified siRNAs are retained at 100x greater levels in various tissues, independently of the nature of the conjugate or siRNA sequence, and support productive mRNA silencing. Thus, fully chemically stabilized siRNAs may provide a better platform to identify novel moieties (peptides, aptamers, small molecules) for targeted RNAi delivery.
INTRODUCTION
A variety of chemical modification patterns have been explored to improve siRNA stability (1), ranging from the simple introduction of a dTdT overhang to highly complicated patterns that remove the chemical nature of RNA (2). Commonly used, commercially available scaffolds include modification of every other nucleotide (3) or all pyrimidines in a given sequence (4). These ‘conventional’ modification patterns substantially enhance siRNA stability in vitro (5) and block innate immune activation in vivo (6).
Partially modified siRNAs have been extensively used to study the impact of conjugates on siRNA distribution and in vivo efficacy. A wide range of conjugate modalities have been tested: steroids (7), lipids (8), folate (9), vitamins (10), aptamers (11) and antibodies (12) all demonstrating only marginal efficacy. All of these conjugates were evaluated in the context of naked or partially modified siRNA, miRNAs and antisense oligonucleotides.
A recent breakthrough in conjugate-mediated delivery was the development of the triple N-acetylgalactosamine (GalNAc) conjugated siRNA (13), which drives efficient, receptor-mediated uptake into hepatocytes. The GalNAc-conjugated siRNAs were fully modified using an advanced version of an alternating 2′-fluoro, 2′-O-methyl pattern first described in 2005 (3). Additionally, in the context of single stranded RISC entering oligonucleotides, chemical stabilization was shown to be absolutely essential for in vivo efficacy (14,15). Recently, fine-tuning of the chemical stabilization pattern, including increases in the 2′-O-methyl content, incorporation of additional phosphorothioates (14) and 5′ phosphate stabilization (16–19) have been shown to even further enhance long-term efficacy of conjugated siRNAs.
Here, we systematically compare the distribution, tissue accumulation, and efficacy of partially and fully modified siRNA scaffolds and show that full chemical stabilization of siRNA is preferred for in vivo applications, independently of the siRNA sequence or the nature of the conjugate used.
MATERIALS AND METHODS
Oligonucleotide synthesis, deprotection and purification
Oligonucleotides were synthesized using standard and modified (2′-fluoro, 2′-O-methyl) phosphoramidite, solid-phase synthesis conditions using a MerMade 12 (BioAutomation, Irving, TX, USA) and Expedite DNA/RNA synthesizer (ABI 8909). Unconjugated oligonucleotide strands were grown on controlled pore glass functionalized with a long-chain alkyl amine and Unylinker® terminus (Chemgenes, #N-4000-10) and cholesterol-conjugated oligonucleotides were synthesized on modified solid support (Chemgenes, # N-9166-05). Oligonucleotides were removed from CPG, deprotected, and purified by HPLC as described previously (20). Purified oligonucleotides were passed over a Hi-Trap cation exchange column to exchange the counter-ion with sodium. Oligonucleotides identity were confirmed by HPLC–mass spectrometry.
In vitro RISC loading and cleavage assay
S100 cell extract from ago2−/− +Ago2 MEF cells were incubated with 50 nM chemically modified or unmodified let-7a siRNA for 30 min at 37°C. After 30 min, 100 nM of 32P capped labeled RNA was incubated at 23°C for 10 min to measure RISC concentration. After 10 min, temperature was shifted to 37°C (multi-turnover condition). Cell extract, RNA labeling, and cleavage assay were performed as previously described by Salomon et al. (21).
Cell culture (HeLa and CTB cells)
HeLa cells (ATCC, #CCL-2) were maintained in DMEM (Cellgro, #10–013CV) supplemented with 10% fetal bovine serum (FBS) (Gibco, #26140) and 100 U/ml penicillin/streptomycin (Invitrogen, #15140) and grown at 37°C and 5% CO2. Cells were split every 2–5 days, and discarded after fifteen passages. Primary cytotrophoblast cells (CTB) from human placenta were provided by Dr. S Ananth Karumanchi. CTBs were maintained in Medium 119 (Gibco, #11043) supplemented with 10% FBS (Gibco, #26140) and 100 U/ml penicillin/streptomycin (Invitrogen, #15140) at 37°C and 5% CO2.
Preparation of primary neurons
Mouse primary cortical neurons were prepared as previously described (22). In brief, pregnant C57BL6 females were anesthetized with Avertin (Sigma, #T48402), followed by cervical dislocation. Brains of E15.5 day embryos were removed, and meninges were carefully detached. Cortices were isolated and transferred into a 1.5-ml tube with pre-warmed papain solution for 25 min at 37°C, 5% CO2, to dissolve tissue. Cortices were dissociated by repeated pipetting and plated on poly-l-lysine coated plates. Anti-mitotics, 0.484 μl/ml of UTP Na3 (Sigma, #U6625) and 0.2402 μl/ml of FdUMP (Sigma, #F3503), were added to neuronal cultures to prevent growth of non-neuronal cells. Protocols for oligonucleotide treatment have been described in detail in Alterman et al. bioprotocols(22).
Hepatocyte cell culture
Human primary hepatocytes (Gibco, #HMCPUS) were thawed and plated in Cryopreserved Hepatocytes Recovery Medium (Gibco, #CM7000), consisting of Williams E Medium (Gibco, #A12176-01) supplemented with 5% fetal bovine serum, 1 μM dexamethasone in DMSO, 100 U/ml penicillin and streptomycin, 4 μg/ml human recombinant insulin, 2 mM GlutaMAX and 15 mM HEPES pH 7.4. Plated hepatocytes were allowed to recover for 6 h at 37°C and 5% CO2 and subsequently maintained in Williams E Medium supplemented with the Hepatocyte Maintenance Supplement Pack (Gibco, #CM4000), containing 0.1 μM dexamethasone in DMSO, 50 U/ml penicillin and streptomycin, 6.25 μg/ml human recombinant insulin, 6.25 μg/ml human transferrin, 6.25 μg/ml selenous acid, 6.25 μg/ml bovine serum albumin, 6.25 μg/ml linoleic acid, 2 mM GlutaMAX,15 mM HEPES pH 7.4 and 5 mM CaCl2.
Passive uptake of oligonucleotides (HeLa and CTB cells)
Cells were plated in DMEM containing 6% FBS at 10 000 cells per well in 96-well tissue culture plates. CTB cells were plated in Medium 199 containing 6% FBS at 25 000 cells per well in 96-well plates. Hydrophobically modified siRNA (hsiRNA) was diluted in OptiMEM (Gibco, #31985-088), and added to cells resulting in 3% FBS final. Cells were incubated for 72 h at 37°C and 5% CO2. Cells were lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix).
Passive uptake of oligonucleotides (hepatocytes)
Cells were plated in Williams E Medium supplemented with the Hepatocyte Maintenance Supplement Pack (Gibco, #CM4000) at 25 000 cells per well in 96-well tissue culture plates. hsiRNA, hydrophobically modified siRNA, was diluted in OptiMEM (Gibco, #31985-088) to a 2× stock concentration and 50 μl of this solution was combined with 50 μl of hepatocyte maintenance medium for a final volume of 100 μl. Cells were incubated for 24 h at 37°C and 5% CO2. Cells were lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix).
Lipid-mediated delivery (HeLa cells)
Cells were plated in DMEM with 6% FBS at 10,000 cells per well in 96-well tissue culture treated plates. siRNA was diluted in OptiMEM, and mixed 1:1 with Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, #13778150) (final transfection reagent concentration = 0.3 μl/25 μl/well). siRNA and transfection reagent was added to cells resulting in 3% FBS final. Cells were incubated for 72 h at 37°C and 5% CO2.
mRNA quantification from cells and tissue punches
mRNA was quantified using the QuantiGene 2.0 Assay (Affymetrix, #QS0011). Cells were lysed in 250 μl diluted lysis mixture with proteinase K (Affymetrix, #13228) for 30 min at 55°C. Probe sets were diluted as specified in the manufacturer's protocol. Cell lysates and diluted probe sets were added to the capture plate and amplification and luminescent detection were carried out as specified in the manufacturer's protocol.
Tissue punches (∼5 mg) were homogenized in 300 μl of Homogenizing Buffer (Affymetrix, #10642) with proteinase K in a 96-well plate format on a QIAGEN TissueLyser II (Qiagen, #85300). Lysates and diluted probe sets were added to the capture plate and signal was amplified and detected according to the manufacturer's protocol. This method is described in detail in Coles et al. (23).
Luminescence was detected on either a Veritas Luminometer (Promega, #998-9100) or a Tecan M1000 (Tecan, Morrisville, NC, USA). The specific mRNAs detected are specified in each graph. Catalog numbers are as follows: human HTT (Affymetrix, #SA-50339), mouse HTT (Affymetrix, #SB-14150), human PPIB (Affymetrix, #SA-10003), human sFLT1-i13 (Affymetrix, #SA-50459), mouse sFlt1 (Affymetrix, #SB-50049). Data sets were normalized to housekeeping genes specified in figure legends. Catalog numbers are as follows: human PPIB (Affymetrix, #SA-10003), mouse Ppib (Affymetrix, #SB-10002), human YWHAZ (Affymetrix, #SA-13271), mouse Flt1 (Affymetrix, #SB-14012).
PNA (peptide nucleic acid) based assay for detection of hsiRNA in mouse tissues (24,25)
hsiRNA guide strand in tissues were quantified using PNA hybridization assay. Tissues were lysed in MasterPure tissue lysis Solution (EpiCentre, #MC85200) in the presence of proteinase K (2 mg/ml) (Invitrogen, #25530-049) in TissueLyser II (Quiagen) (10mg tissue in 100 μl lysis solution). Sodium dodecyl sulphate (SDS) from lysate was precipitated with KCl (3 M) and pelleted at 5000 g. hsiRNA in cleared supernatant was hybridized to a Cy3-labeled PNA fully complementary to guide strand (PNABio, Thousand Oaks, CA, USA) and injected into HPLC DNAPac PA100 anion exchange column (Thermo Fisher, #043010) and Cy3 fluorescence was monitored and peaks integrated. Mobile phase for HPLC was 50% water–50% acetonitrile, 25 mM Tris–HCl (pH 8.5), 1 mM EDTA and salt gradient was 0–800 mM NaClO4. For the calibration curve known amounts of hsiRNAsFLT1 duplexes were spiked into the tissue lysis solution.
Animal experiments
Animal experiments were performed in accordance with animal care ethics approval and guidelines of University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411). Mice were 6–10 weeks of age at the time of experiments. All animals were kept on 12-h light/dark cycle in pathogen-free facility with food and water ad libitum. Before all studies all animals were euthanized as follows: animals were deeply anesthetized with 0.1% Avertin and transcardially perfused with a 4% paraformaldehyde solution in phosphate buffered saline, pH 7.2.
Injection of hsiRNA and FM-hsiRNA
For systemic administration of oligonucleotides C57BL/6 or FVB/NJ (both form Jackson Laboratory) mice were injected either with phosphate buffered saline (PBS) or with different amounts of hsiRNA, resuspended in PBS, through the tail vein (IV) or SC. For in vivo tissue distribution C57BL/6 were injected with 10 mg/kg of hsiRNAsFLT1 and euthanized 24 h after injection. For in vivo efficacy FVB/NJ mice were injected with 20 mg/kg using the same protocol. Animals were euthanized 7 days after injection and tissues were taken for microscopy, hsiRNA quantification (using PNA hybridization assay), and for mRNA quantification using QuantiGene 2.0 assay (23).
Imaging
C57BL/6 mice were injected intravenously (IV, via tail vein) or subcutaneously (SC, interscapular, between shoulders) with 10 mg/kg of Cy3-labeled hsiRNA (Cy3-hsiRNAsFLT1 or Cy3-FM-hsiRNAsFLT1). After 24 h mice were sacrificed and liver, kidney, spleen, intestine, muscle (quadriceps), pancreas, lung tissues, as well as fat, skin and tail from the site of injection, were removed and fixed in 10% formalin (Sigma, #HT501320) overnight at 4°C. Fixed tissues were embedded in paraffin, and sliced into 4 μm sections that were mounted on glass slides. Sections were deparaffinized by incubating in Xylene twice for 8 min. Sections were rehydrated in serial ethanol solutions (100%, 95%, 80% dilutions in water) for 4 min each, then washed twice for two minutes with PBS. Washed slides were incubated with 0.25 μg/ml DAPI (Molecular Probes, #D3571) in PBS for 1 min then washed twice for 2 min with PBS. Slides were mounted with PermaFluor mounting medium (Thermo, #TA030FM) and coverslips and dried overnight before imaging on a Leica DM5500 microscope fitted with a DFC365 FX fluorescence camera.
Statistical analysis
Data were analyzed using GraphPad Prism 6 software. IC50 curves were fitted using log(inhibitor) versus response—variable slope (four parameters). For Figure 4A–D, statistics were calculated using one-way ANOVA with Tukey's test for multiple comparisons, with significance calculated relative to PBS control injected animals.
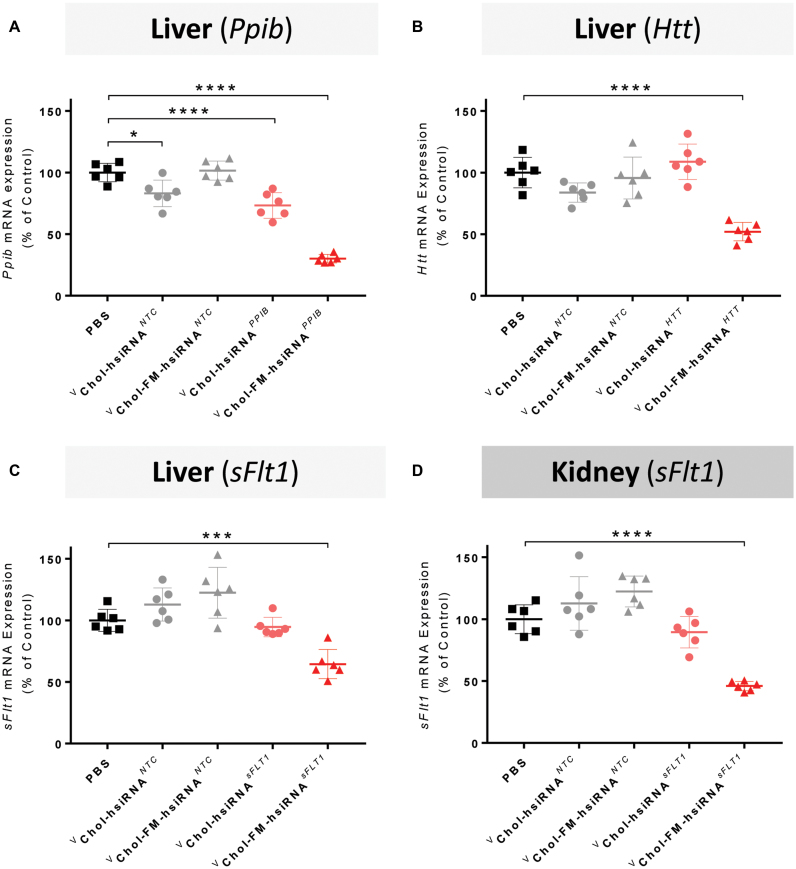
Figure 4.
Fully modified hsiRNAs are more efficacious than partially modified hsiRNAs following systemic administration. (A–C) Quantification of target (Ppib, Htt, sFlt1, respectively) mRNA silencing in liver 7 days after SC administration of cholesterol-conjugated vhsiRNANTC, vFM-hsiRNANTC, vhsiRNATarget, vFM-hsiRNATarget at 20 mg/kg. (D) Quantification of Ppib mRNA silencing in kidney 7 days after SC administration of cholesterol-conjugated vhsiRNANTC, vFM-hsiRNANTC, *hsiRNA, *FM-hsiRNAsFlt1 at 20 mg/kg. FVBNj mice (n = 6 per group). mRNA levels were measured with QuantiGene 2.0 (Affymetrix) assay. Htt, Ppib, and sFlt1 mRNA levels normalized to housekeeping gene, Hprt. All data presented as percent of PBS treated control. All error bars represent mean ± SD. *P< 0.05, ***P< 0.001; ****P< 0.0001 as calculated by one-way ANOVA with Tukey's test for multiple comparisons. v = 5′-vinyl phosphonate, NTC = non-targeting control.
RESULTS
Full chemical stabilization enables efficient conjugate-mediated siRNA efficacy in vitro
To compare the impact of partially or fully modified scaffolds on conjugate-mediated distribution and silencing in vivo, we used asymmetric, cholesterol-modified siRNAs as a model (Figure 1). The asymmetric design, termed hsiRNAs (15-nucleotide passenger strand with a 3′-conjugate and 20-nucleotide guide strand) lowers the melting temperature of the double-strand region, to facilitate the dissociation of the non-cleavable modified guide strand from the RNA-Induced Silencing Complex (RISC) during loading. RISC loading can happened through passenger strand cleavage (26) or dissociation. Full chemical modification of the passenger strand blocks the cleavage pathway, leaving dissociation as the only functional alternative for RISC loading. This makes the thermal melt (Tm) of the fully chemically stabilized duplex one of the primary activity-limiting factors. The relative impact of asymmetric duplex configuration on compound efficacy is sequence dependent. Supplementary Figure S1A and B shows the activity of two siRNA sequences in the context of either the conventional symmetric (20-mer-20-mer) or asymmetric (15-mer-20-mer) duplex configuration. For the siRNA targeting HTT there is no difference between the two configurations, whereas for the siRNA targeting sFLT1, reduction in duplex length substantially improves compound efficacy. The single-strand region of the guide strand contains phosphorothioate linkages (27), providing additional stabilization and enhancing cellular internalization (Ly. et al, prepared for publication). This chemical scaffold has been extensively characterized including mechanism of trafficking, clearance kinetics, and systemic and local delivery (19,24,28–33).
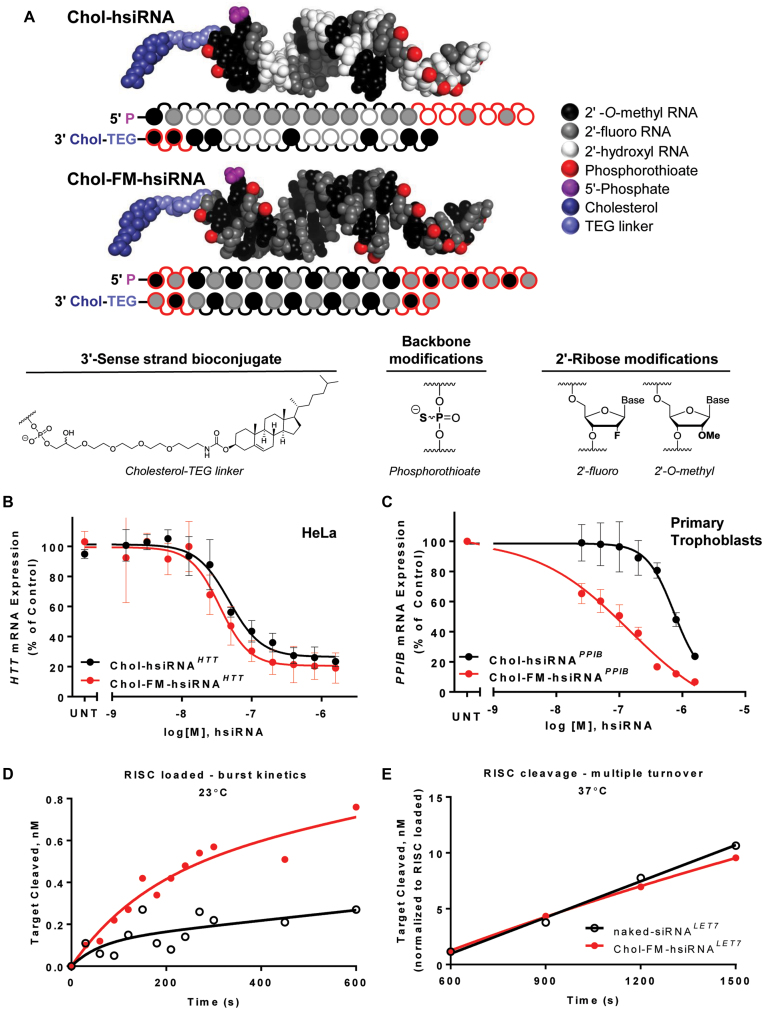
Figure 1.
Chemical composition and cellular efficacy of fully modified hsiRNAs. (A) Chemical structure, modification pattern, and molecular model of partially (hsiRNA) and fully modified (FM-hsiRNA) hydrophobic siRNA. (B and C) Comparison of hsiRNA and FM-hsiRNA activity in vitro. HeLa (B) or primary cytotrophoblasts (C) were incubated with hsiRNA or FM-hsiRNA at concentrations shown for 72h. mRNA levels were measured using QuantiGene (Affymetrix) normalized to housekeeping gene (human PPIB for (B) and human YWHAZ for (C)), and presented as percent of untreated control (n = 3, mean ± SD). UNT—untreated cells. (D, E) RISC loading and cleavage comparison of unmodified, naked-siRNALET7, or FM-hsiRNALET7 (D) FM-hsiRNA LET7 shows an increase in loaded RISC as evident by single turnover target cleavage. (E) Multiple turnover cleavage rate (normalized to loaded RISC) is similar between naked-siRNA, or FM-hsiRNA.
Cholesterol conjugation to partially modified hsiRNAs results in robust cellular uptake in vitro and potent local silencing in vivo (28,30), but marginal systemic efficacy. Thus, cholesterol-conjugated siRNAs provide a good starting point to evaluate the impact of extensive chemical stabilization on conjugate-mediated hsiRNA delivery.
Using a previously identified, partially modified hsiRNA sequence targeting huntingtin (HTT) mRNA (30), we synthesized and tested a panel of fully modified hsiRNAs (FM-hsiRNAs) based on previously reported patterns (2,3). Though several configurations were functional, an alternating 2′-O-methyl, 2′–fluoro pattern, with a chemically monophosphorylated, 2′-O-methyl-modified uridine (U) at position 1, and a 2′-fluoro modified nucleotide at position 14 of the guide strand performed the best (Figure 1A, Supplementary Figure S1C and Supplementary Table S1). An alternating pattern with a 2′–fluoro in position 1 of the guide strand reduced silencing activity (data not shown), likely due to placement of 2′-O-methyl groups in positions 2 and 14, which negatively affects potency in the context of heavily modified duplexes (34). The guide strand was also 5′ -chemically phosphorylated, as terminal 2′-O-methylated U is not a good a substrate for intracellular kinases as natural RNA. We also added two phosphorothioate linkages to both 5′ and 3′ ends of the passenger and guide strands to provide additional resistance to exonucleases. Figure 1A shows the modification patterns and PyMol structure models of the conventionally modified hsiRNA and the most active fully modified hsiRNA (FM-hsiRNA).
The FM-hsiRNA pattern supported similar or improved silencing when applied to previously identified functional siRNAs targeting HTT, PPIB, sFLT1, TIE2 (35), PLK1 and SOD1 mRNAs (Figure 1B and C, Supplemental Figures S1 and S2 and data not shown). Moreover, FM-hsiRNAs improved silencing in cultured primary neurons and primary trophoblasts, adherent and suspension cell types, respectively, both of which can be difficult to transfect using conventional methods (36,37). The activity improvement provided by full chemical stabilization was more pronounced in the non-adherent primary trophoblasts where cholesterol mediated uptake is generally slower (Figure 1C). When directly compared to an unmodified, unconjugated siRNA, Chol-FM-hsiRNA showed significantly enhanced efficacy following passive uptake (IC50 of 33.5 nM) while naked siRNA is inactive. However, both compounds showed comparable potency (IC50 of 3.5 nM for siRNA and 0.9 nM FM-hsiRNA) following lipid transfection, further confirming that full modification is compatible with RISC function in cells (Supplementary Figure S3). Similarly, fully modified GalNAc-conjugated siRNAs were more active in primary hepatocytes than partially modified GalNAc-conjugated siRNAs (Supplementary Figure S4), indicating that improvement in potency is not specific to the nature of the conjugate or the cell type treated.
Fully chemically modified siRNAs are efficiently loaded into RISC complex
The loading and cleavage rates of FM-hsiRNA were compared to a completely non-modified Let-7 siRNA in a loading and cleavage assay described earlier (38). The FM-hsiRNA loaded almost three times more RISC complex compared to the naked siRNA (Figure 1D), which is likely a result of the enhanced stability of FM-hsiRNA. Once the loaded RISC was under multiple turnover conditions (37°C), the in vitro cleavage rates of both the FM-hsiRNA and the non-modified oligo were similar (after normalizing to equal numbers of loaded RISC) (Figure 1E). Thus, full modification does not interfere with the ability of the tested oligonucleotide to be recognized by, and functionally load into the RISC complex.
Comparison of partially and fully chemically modified siRNAs in conjugate-mediated systemic delivery
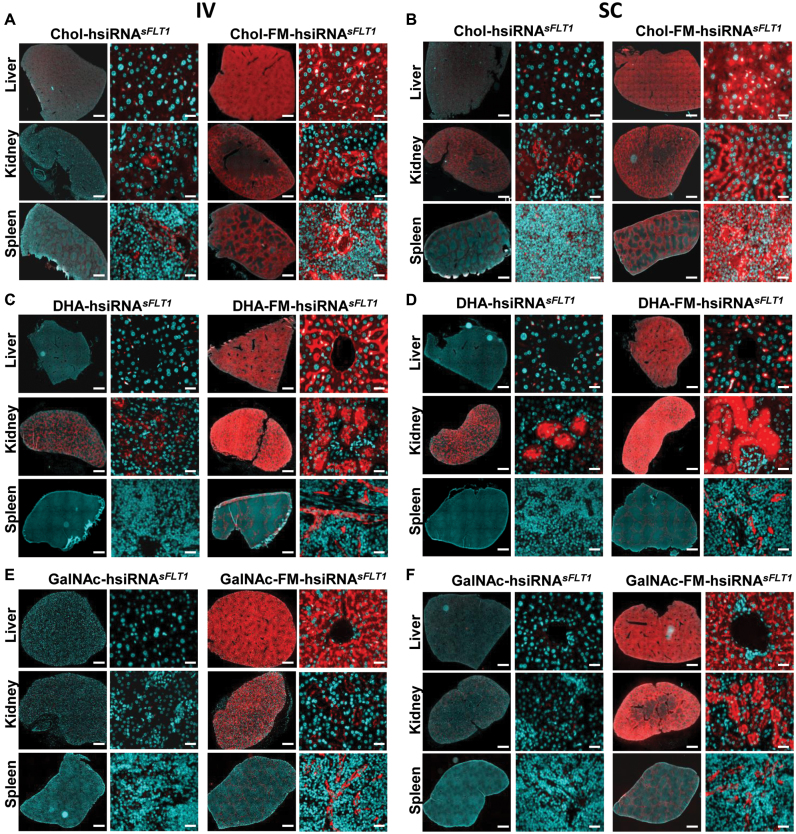
Both partially modified (e.g. all pyrimidines) and fully modified siRNAs have increased stability in vitro (from minutes to days in 50% serum) over non-modified siRNAs (3,4). In vivo, however, oligonucleotides are exposed to an aggressive nuclease environment that cannot be adequately mimicked in vitro. Thus the stability of partially and fully modified siRNAs might be quite different from one another in vivo. To compare the distribution of partially modified and fully modified hsiRNAs in vivo, we administered 10 mg/kg of Cy3-labeled Chol (cholesterol)-hsiRNA and Chol-FM-hsiRNA by intravenous (IV) or subcutaneous (SC) injection (Figure 2A and B). Twenty-four hours after injection, we harvested tissues from mice and visualized the siRNA distribution by fluorescence microscopy. Injection of partially modified Cy3-Chol-hsiRNA resulted in minimal levels of fluorescence, observed only in liver and kidney. In contrast, injection of Cy3-Chol-FM-hsiRNA resulted in intense accumulation of fluorescence in tissues throughout the body, including liver, kidney, spleen, fat, and skin (Figure 2A and B and data not shown).
Figure 2.
Systemically administered fully modified hsiRNA shows enhanced tissue distribution. Tissue distribution of Cy3-Chol-hsiRNA sFLT1 and Cy3-Chol-FM-hsiRNA sFLT after 10 mg/kg intravenous (IV) tail vein injection (A) or 10 mg/kg subcutaneous (SC) injection (B). Tissue distribution of Cy3-DHA-hsiRNA sFLT1 and Cy3-DHA-FM-hsiRNA sFLT after 10 mg/kg intravenous (IV) tail vein injection (C) or 10 mg/kg subcutaneous (SC) injection (D). Tissue distribution of Cy3-GalNAc-hsiRNA sFLT1 and Cy3-GalNAc-FM-hsiRNA sFLT after 10 mg/kg intravenous (IV) tail vein injection (E) or 10 mg/kg subcutaneous (SC) injection (F). Cy3-hsiRNA (red), nuclei stained with DAPI (blue). For every conjugate the left panels are tiled tissue images (scale bar = 1 mm) and the right panels are higher magnification tissue images (scale bar = 25 μm). Image is representative.
To evaluate whether a significant enhancement in retention upon full chemical stabilization is specific to the cholesterol conjugate, we evaluated two other conjugates with potential for tissue delivery: Docosahexaenoic acid (DHA) (26), and GalNAc (39). The fully modified and partially modified GalNAc and DHA conjugate compounds were synthesized and their tissue distribution evaluated 24 h post IV and SC injection (Figure 2C–F). As with cholesterol, we observed a substantial increase in compound tissue accumulation with the FM-hsiRNA, independently of the nature of the conjugate. It is important to notice that changing the conjugate resulted in a change in tissue distribution profile, which could only be visualized in the context of fully modified hsiRNA.
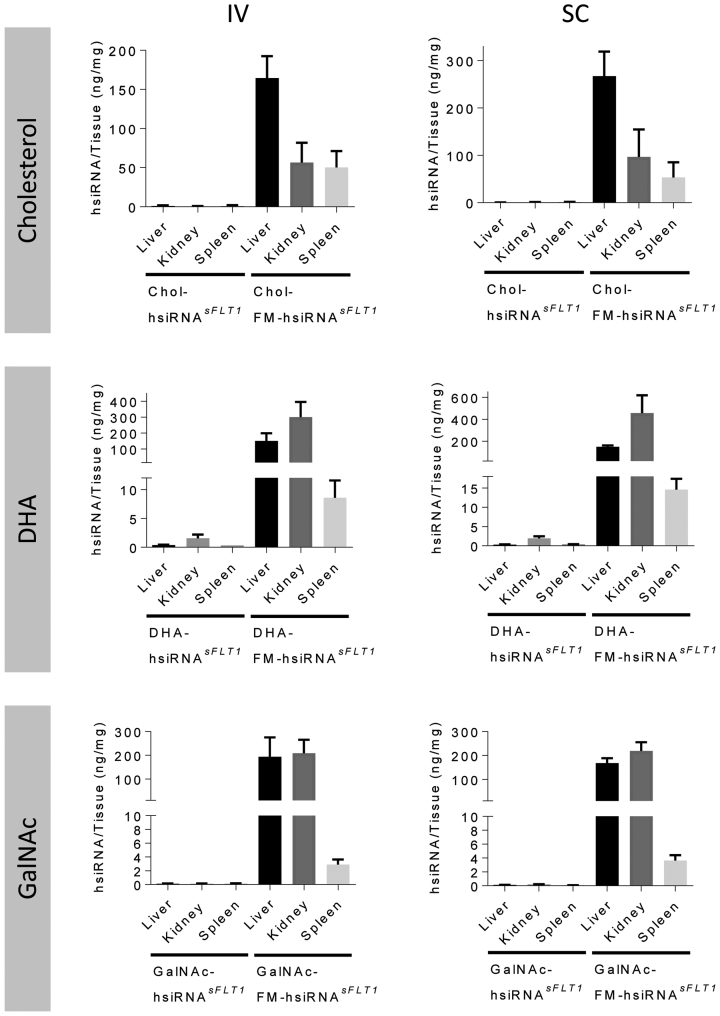
To insure that the qualitative robust difference observed in fluorescence distribution between partially and fully modified hsiRNAs is not an artifact of tissue processing or presence of the fluorescent label, we used a PNA hybridization-based assay (24,25) to evaluate the accumulation of these compounds quantitatively. This method allows for the detection of intact guide strands present in tissue biopsies and is not dependent on the presence of the fluorescent label. (Supplementary Figure S5). Figure 3 shows the quantification data for the three different conjugates (Cholesterol, DHA, and GalNAc) attached to fully and partially modified hsiRNA scaffolds injected SC and IV. In all cases, independently of the route of administration, or the type of conjugate, over 100x tissue accumulation was observed in the context of fully chemically modified siRNAs. FM-hsiRNA guide strands accumulated to relatively high levels, 20–200 ng/mg in liver, kidney, and spleen (Figure 2B and C). Consistent with the imaging data, the tissue distribution profile was effected by the type of conjugate used, with cholesterol preferentially accumulating in liver (∼150 ng/mg), DHA in kidney (300–400 ng/mg) and GalNAc in both liver and kidney (∼200 ng/mg). While cholesterol and DHA show distribution to tissues beyond the liver and kidney, such as the spleen, skin, and fat (data not shown), GalNAc distribution was effectively exclusive to liver and kidney (Figure 2E and F). Hence, full chemical stabilization significantly enhances conjugate-mediated tissue accumulation independent of the conjugate nature, or route of administration, and is heavily reliant on the full chemical modification of the siRNA.
Figure 3.
Systemic administration of fully modified hsiRNAs shows enhanced tissue accumulation. Guide strand tissue quantification by PNA hybridization-based assay in tissues from Figure 2. Guide strand quantification of Cy3-Chol-hsiRNA sFLT1 and Cy3-Chol-FM-hsiRNAsFLT after 10 mg/kg intravenous (IV) tail vein injection (A) or 10 mg/kg subcutaneous (SC) injection (B). Guide strand quantification of Cy3-DHA-hsiRNA sFLT1 and Cy3-DHA-FM-hsiRNA sFLT after 10 mg/kg intravenous (IV) tail vein injection (C) or 10 mg/kg subcutaneous (SC) injection (D). Guide strand quantification of Cy3-GalNAc-hsiRNA sFLT1 and Cy3-GalNAc-FM-hsiRNA sFLT after 10 mg/kg intravenous (IV) tail vein injection (E) or 10 mg/kg subcutaneous (SC) injection (F). Data presented as mean ± SD (n = 3 mice).
Full chemical stabilization enables productive silencing in vivo.
The poor accumulation and retention of partially modified hsiRNAs is consistent with published studies showing that systemic silencing by partially modified siRNA lipophilic conjugates requires repetitive delivery with high doses (50–80 mg/kg) (8). Consistently, when cholesterol-conjugated partially modified hsiRNA was delivered systemically at 2 × 50 mg/kg dose levels we were unable to detect silencing, even in the liver, a primary tissue where cholesterol modified siRNAs distribute.
To confirm that significant FM-hsiRNA tissue accumulation results in functional gene silencing, we evaluated silencing efficiency in the liver and the kidney. For this experiment, we selected the following targets: Ppib, a commonly used housekeeping gene expressed in all cell types, Htt, a key target in Huntington's disease, also expressed in all cell types, and sFlt1 (soluble fms-like tyrosine kinase 1 or VEGFR1), which is primarily expressed in endothelial cells and kidney proximal tubules epithelia, where Chol-FM-hsiRNAs tend to accumulate (Figure 2A and B).
We, and others, have recently demonstrated that chemical stabilization of the 5′ phosphate of the guide strand increases in vivo efficacy of siRNA conjugates (17,19,40). Thus for evaluation of targeted gene silencing in vivo, partially and fully modified siRNA scaffolds were synthesized with a 5′-(E)-vinyl phosphonate (VP). Animals were injected with 20 mg/kg of fully or partially modified hsiRNAs. The level of targeted gene expression was evaluated a week post injection using the QuantiGene 2.0 Assay. We also injected a non-targeting control (NTC) compound of the same chemical composition but not targeting the intended mRNA, and PBS as controls. For all three genes tested, injection of controls resulted in no significant changes in gene expression. In contrast, all three genes were significantly silenced in liver after injection of a fully chemically stabilized variant (Figure 4A–C). The same phenomena was observed in kidney, where sFlt1 was significantly downregulated by Chol-FM-hsiRNA (Figure 4D). In both liver and kidney, partially modified hsiRNA did not support efficient silencing. The degree of silencing varies from target to target and is likely due to differences in cell types and levels of target mRNA expression. While distribution and efficacy are significantly enhanced by full chemical modification, injection of cholesterol, DHA and GalNAc-conjugated, fully chemically modified, siRNAs were well tolerated at the doses injected, with animals showing no observable adverse events or changes in blood chemistry (data not shown).
This confirms that full chemical stabilization enables both siRNA tissue accumulation and productive silencing.
DISCUSSION
Here we have compared partially and fully chemical modified siRNA scaffolds in conjugate mediated delivery in vitro and in vivo. We have utilized a simple siRNA scaffold, utilizing an alternating 2′-fluoro, 2′-O-methyl modification (3) pattern in the context of an asymmetric siRNA. We have shown that full chemical modification does not interfere with RISC assembly and target cleavage in vitro and supports the activity of previously identified, functional siRNA sequences (Figure 1 and Supplementary Table S1) in cell culture. In fact, this chemical configuration supports efficient RISC loading (similar or better than non-modified RNA duplex) and target cleavage in vitro, allows productive RISC assembly and function in cells and in vivo in animals.
Historically, extensive chemical modifications of siRNAs were shown to negatively impact siRNA efficacy, resulting only in a small fraction of pre-identified, functional, naked siRNAs being successfully converted to modified scaffolds (1,41–43). The two major contributors to modification-related negative impact on silencing activity are modifications forcing the nucleic acid into a suboptimal geometry (both for loading and target cleavage) and/or making the duplex too stable, interfering with guide strand loading (26). Indeed, complete modification of the guide strand with 2′-fluoro resulted in almost three fold drop in silencing efficacy in vitro (Supplemental Figure S1C). Both, 2′-fluoro and 2′-O-methyl modifications favor the C3′-endo ribose conformation, but 2′-fluoro modifications slightly over-wind the duplex and 2′-O-methyl slightly under-wind the RNA duplex. In addition, 2′-fluoro modification are more hydrophobic and thus might contribute to enhancement in cellular uptake and in vivo distribution (44). Alternating these two types of modifications supports formation of an A-form helical structure, the geometry required for guide strand-based positioning of the targeted mRNA into the cleavage center of RISC (45). This is consistent with the recently published crystal structure of a fully modified guide strand showing ability to adopt a variety of confirmations (46) Alternating 2′-fluoro and 2′-O-methyl modifications on both strands pairing 2′-fluoro on one strand with 2′-O-methyl on the other weakens the modification-induced thermodynamic gain (2,3) and reduces the sequence related biases. The shorter (15-nucleotide) duplex region will also help to promote release of the non-cleavable passenger strand, alleviating a major thermodynamic limiting step in RISC assembly with fully modified, FM-hsiRNAs (45). Our findings suggest that RISC assembled with the asymmetric fully modified-siRNA is as effective as RISC assembled with non-modified siRNAs (Figure 1D and E). This modification pattern can be applied to previously validated siRNA sequences, as the HTT and PPIB sequences used in this study were originally discovered using a partially modified scaffold. By switching the sequence to the fully modified pattern we effectively reduced the IC50 by almost 2.5× in vitro. We are in the process of evaluating the exact impact of these chemistries on RISC assembly and cleavage kinetics using single molecule approaches (21).
Interestingly, the relative impact of full chemical modification on in vitro efficacy was significantly less pronounced than in vivo. Similar effects have been seen for 5′ phosphate stabilization where in vitro, 5′-vinyl phosphonate and 5′ phosphate hsiRNAs show similar efficacy, while in vivo phosphate stabilization clearly enhances efficacy (19). It is likely that in the harsh biological environment of systemic delivery, chemical stabilization impact on efficacy is much more pronounced than in vitro or upon local delivery.
Systemically administered FM-hsiRNAs accumulate in tissues throughout the body including: liver, kidney, spleen, fat, and skin. As expected, all oligonucleotides, including lipophilic conjugated siRNAs (8,47) preferentially accumulated in the liver where silencing of the target genes was observed. Additionally, significant levels of compound accumulated in the kidneys and spleen (Figures 2A–D and 3), but importantly the substantial differences observed between the fully and partially modified siRNAs were not dependent on the nature of the conjugates themselves but rather the modification pattern of the siRNA.
The increase in tissue accumulation (50–250 ng/mg), was observed with different conjugates, but it was predominantly the nature of conjugate that had an impact on tissue distribution profile. This is consistent with a recently published paper demonstrating that cholesterol modified siRNAs can silence genes in muscle, although dose levels necessary to achieve this effect were high (50mg/kg) (48). DHA-conjugates accumulate to a higher extent in the kidneys, indicating that changing the nature of the conjugate can be used as a strategy to alter tissue distribution. Interestingly, with GalNAc–hsiRNAs we observed similar distribution between kidneys and liver, specifically after SC administration. This was surprising, as GalNAc internalization is believed to be dependent on ASGPR (asialoglycoprotein receptor) overexpressed in hepatocytes. It is possible that partial kidney delivery is due to the presence of the single stranded phosphorothioated tail or the oversaturation of the ASGPR receptors at the dose levels used in this study (10 mg/kg).
With increased accumulation we observed an increase in productive silencing. However, the level of silencing was not directly proportional to the increase in siRNA tissue accumulation. It is well understood that for both antisense oligonucleotides and siRNAs a significant fraction of internalized compound is trapped non-productively and different cells accumulate oligonucleotides to different degrees. This phenomenon is the subject of active investigation.
While the nature of the conjugate clearly effected tissue accumulation, these effects can be observed only in the context of a fully chemically modified scaffold. This demonstrates that some previously discarded or dismissed siRNA conjugates evaluated in the context of partially or non-modified siRNAs might have served as viable delivery strategies and may be worth re-testing in the context of a fully modified pattern to show its true potential.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to especially thank Darryl Conte for help with manuscript writing and editing, all members of the Khvorova Labfor their guidance and Claudio Punzo for the use of his fluorescence microscope.
Author contributions: M.R.H. and J.F.A. designed and M.R.H. and D.E. synthesized all compounds. M.N. and L.R. synthesized DHA and Chol support. A.A.T. and A.H.C. conducted all in vivo experiments and imaging. J.F.A., A.A.T. and M.F.O. conducted all in vitro screening and data analysis. R.A.H. developed and conducted PNA analysis. M.F.O. designed PyMol structures. W.S and P.Z. conceptualized and performed in vitro RISC complex assembly. A.K., N.A., M.J.M., D.V.M., J.F.A. and M.R.H. came up with the concept and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
RO1GM1088030181, RO1NS038194, 5F32NS095508-03 and UH3TR000888; CHDI Foundation [A6367]; Bill and Melissa Gates foundation [OPP1086170];. The compounds synthesis was supported by S10 OD 020012-01. The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. A.K. disclosed ownership of the stock of RXi Pharmaceuticals and Advirna. Other authors have no conflict of interest to report.
REFERENCES
- 1. Watts J.K., Corey D.R.. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012; 226:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deleavey G.F., Watts J.K., Alain T., Robert F., Kalota A., Aishwarya V., Pelletier J., Gewirtz A.M., Sonenberg N., Damha M.J.. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010; 38:4547–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B.. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005; 48:901–904. [DOI] [PubMed] [Google Scholar]
- 4. Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C.. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006; 342:919–927. [DOI] [PubMed] [Google Scholar]
- 5. Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J.. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003; 31:2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Judge A.D., Bola G., Lee A.C., MacLachlan I.. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006; 13:494–505. [DOI] [PubMed] [Google Scholar]
- 7. Chen Q., Butler D., Querbes W., Pandey R.K., Ge P., Maier M.A., Zhang L., Rajeev K.G., Nechev L., Kotelianski V. et al. . Lipophilic siRNAs mediate efficient gene silencing in oligodendrocytes with direct CNS delivery. J. Controlled Release. 2010; 144:227–232. [DOI] [PubMed] [Google Scholar]
- 8. Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. et al. . Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotech. 2007; 25:1149–1157. [DOI] [PubMed] [Google Scholar]
- 9. Dohmen C., Frohlich T., Lachelt U., Rohl I., Vornlocher H.-P., Hadwiger P., Wagner E.. Defined folate-PEG-siRNA conjugates for receptor-specific gene silencing. Mol. Ther. Nucleic Acids. 2012; 1:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T.. Efficient in vivo delivery of siRNA to the liver by conjugation of [alpha]-tocopherol. Mol. Ther. 2008; 16:734–740. [DOI] [PubMed] [Google Scholar]
- 11. McNamara J.O., Andrechek E.R., Wang Y., Viles K.D., Rempel R.E., Gilboa E., Sullenger B.A., Giangrande P.H.. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotech. 2006; 24:1005–1015. [DOI] [PubMed] [Google Scholar]
- 12. Cuellar T.L., Barnes D., Nelson C., Tanguay J., Yu S.-F., Wen X., Scales S.J., Gesch J., Davis D., van Brabant Smith A. et al. . Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB–siRNA conjugates. Nucleic Acids Res. 2015; 43:1189–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. et al. . Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 14. Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S. et al. . Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017; 45:10969–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R.. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012; 150:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lima Walt F., Prakash Thazha P., Murray Heather M., Kinberger Garth A., Li W., Chappell Alfred E., Li Cheryl S., Murray Susan F., Gaus H., Seth Punit P. et al. . Single-stranded siRNAs activate RNAi in animals. Cell. 2012; 150:883–894. [DOI] [PubMed] [Google Scholar]
- 17. Prakash T.P., Kinberger G.A., Murray H.M., Chappell A., Riney S., Graham M.J., Lima W.F., Swayze E.E., Seth P.P.. Synergistic effect of phosphorothioate, 5′-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioorg. Med. Chem. Lett. 2016; 26:2817–2820. [DOI] [PubMed] [Google Scholar]
- 18. Prakash T.P., Lima W.F., Murray H.M., Li W., Kinberger G.A., Chappell A.E., Gaus H., Seth P.P., Bhat B., Crooke S.T. et al. . Identification of metabolically stable 5-phosphate analogs that support single-stranded siRNA activity. Nucleic Acids Res. 2017; 45:6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haraszti R.A., Roux L., Coles A.H., Turanov A.A., Alterman J.F., Echeverria D., Godinho B., Aronin N., Khvorova A.. 5-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017; 45:7581–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Osborn M.F., Alterman J.F., Nikan M., Cao H., Didiot M.C., Hassler M.R., Coles A.H., Khvorova A.. Guanabenz (Wytensin) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res. 2015; 43:8664–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alterman J.F., Coles A.H., Hall L.M., Aronin N., Khvorova A., Didiot M.C.. A High-throughput Assay for mRNA Silencing in Primary Cortical Neurons in vitro with Oligonucleotide Therapeutics. Bio Protoc. 2017; 7:e2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coles A.H., Osborn M.F., Alterman J.F., Turanov A.A., Godinho B.M., Kennington L., Chase K., Aronin N., Khvorova A.. A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 2016; 26:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Godinho B., Gilbert J.W., Haraszti R.A., Coles A.H., Biscans A., Roux L., Nikan M., Echeverria D., Hassler M., Khvorova A.. Pharmacokinetic profiling of conjugated therapeutic oligonucleotides: a high-throughput method based upon serial blood microsampling coupled to peptide nucleic acid hybridization assay. Nucleic Acid Ther. 2017; 27:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roehl I., Schuster M., Seiffert S.. Oligonucleotide Detection Method. 2008; US20110201006.
- 26. Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D.. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005; 123:607–620. [DOI] [PubMed] [Google Scholar]
- 27. Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014; 24:374–387. [DOI] [PubMed] [Google Scholar]
- 28. Byrne M., Tzekov R., Wang Y., Rodgers A., Cardia J., Ford G., Holton K., Pandarinathan L., Lapierre J., Stanney W. et al. . Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocular Pharmacol. Ther. 2013; 29:855–864. [DOI] [PubMed] [Google Scholar]
- 29. Nikan M., Osborn M.F., Coles A.H., Godinho B.M.D.C., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A.. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids. 2016; 5:e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. et al. . Hydrophobically modified siRNAs silence Huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015; 4:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017; 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ly S., Navaroli D.M., Didiot M.C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. et al. . Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017; 45:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nikan M., Osborn M.F., Coles A.H., Biscans A., Godinho B., Haraszti R.A., Sapp E., Echeverria D., DiFiglia M., Aronin N. et al. . Synthesis and evaluation of parenchymal retention and efficacy of a metabolically stable O-phosphocholine-N-docosahexaenoyl-l-serine siRNA conjugate in mouse brain. Bioconjug. Chem. 2017; 28:1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. et al. . Position-specific chemical modification of siRNAs reduces “off-target" transcript silencing. RNA (New York, N.Y.). 2006; 12:1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dahlman J.E., Barnes C., Khan O.F., Thiriot A., Jhunjunwala S., Shaw T.E., Xing Y., Sager H.B., Sahay G., Speciner L. et al. . In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nano. 2014; 9:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dass C.R., Burton M.A.. Modified microplex vector enhances transfection of cells in culture while maintaining tumour-selective gene delivery in-vivo. J. Pharm. Pharmacol. 2002; 55:19–25. [DOI] [PubMed] [Google Scholar]
- 37. Karra D., Dahm R.. Transfection techniques for neuronal cells. J. Neurosci. 2010; 30:6171–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. et al. . Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 40. Parmar R., Willoughby J.L., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. et al. . 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. Chembiochem. 2016; 17:985–989. [DOI] [PubMed] [Google Scholar]
- 41. Rozema D.B., Lewis D.L., Wakefield D.H., Wong S.C., Klein J.J., Roesch P.L., Bertin S.L., Reppen T.W., Chu Q., Blokhin A.V. et al. . Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watts J.K., Deleavey G.F., Damha M.J.. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008; 13:842–855. [DOI] [PubMed] [Google Scholar]
- 43. Deleavey G.F., Frank F., Hassler M., Wisnovsky S., Nagar B., Damha M.J.. The 5′ binding MID domain of human Argonaute2 tolerates chemically modified nucleotide analogues. Nucleic Acid Ther. 2013; 23:81–87. [DOI] [PubMed] [Google Scholar]
- 44. Manoharan M., Akinc A., Pandey R.K., Qin J., Hadwiger P., John M., Mills K., Charisse K., Maier M.A., Nechev L. et al. . Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. Engl. 2011; 50:2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science (New York, N.Y.). 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schirle N.T., Kinberger G.A., Murray H.F., Lima W.F., Prakash T.P., MacRae I.J.. Structural analysis of human argonaute-2 bound to a modified siRNA guide. J. Am. Chem. Soc. 2016; 138:8694–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J. et al. . Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004; 432:173–178. [DOI] [PubMed] [Google Scholar]
- 48. Khan T., Weber H., DiMuzio J., Matter A., Dogdas B., Shah T., Thankappan A., Disa J., Jadhav V., Lubbers L. et al. . Silencing myostatin using cholesterol-conjugated siRNAs induces muscle growth. Mol. Ther. Nucleic Acids. 2016; 5:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.