Abstract
Poly(ADP-ribose) polymerases (PARPs) act as DNA break sensors and catalyze the synthesis of polymers of ADP-ribose (PAR) covalently attached to acceptor proteins at DNA damage sites. It has been demonstrated that both mammalian PARP1 and PARP2 PARylate double-strand break termini in DNA oligonucleotide duplexes in vitro. Here, we show that mammalian PARP2 and PARP3 can PARylate and mono(ADP-ribosyl)ate (MARylate), respectively, 5′- and 3′-terminal phosphate residues at double- and single-strand break termini of a DNA molecule containing multiple strand breaks. PARP3-catalyzed DNA MARylation can be considered a new type of reversible post-replicative DNA modification. According to DNA substrate specificity of PARP3 and PARP2, we propose a putative mechanistic model of PARP-catalyzed strand break–oriented ADP-ribosylation of DNA termini. Notably, PARP-mediated DNA ADP-ribosylation can be more effective than PARPs’ auto-ADP-ribosylation depending on the DNA substrates and reaction conditions used. Finally, we show an effective PARP3- or PARP2-catalyzed ADP-ribosylation of high-molecular-weight (∼3-kb) DNA molecules, PARP-mediated DNA PARylation in cell-free extracts and a persisting signal of anti-PAR antibodies in a serially purified genomic DNA from bleomycin-treated poly(ADP-ribose) glycohydrolase-depleted HeLa cells. These results suggest that certain types of complex DNA breaks can be effectively ADP-ribosylated by PARPs in cellular response to DNA damage.
INTRODUCTION
The poly(ADP-ribose) polymerase (PARP) family of proteins, also referred to as diphtheria toxin-like ADP-ribosyltransferases (ARTD), includes 17 known members sharing the conserved ADP-ribosyl transferase (ART) domain. Fifteen of them catalyze the synthesis of monomers or polymers of ADP-ribose (MAR or PAR, respectively) covalently attached to acceptor proteins using nicotinamide adenine dinucleotide (NAD+) as a substrate (1–3). Protein ADP-ribosylation alters the function of the modified proteins or provides a scaffold for the recruitment of other proteins. It regulates a number of biological processes including the DNA damage response, chromatin reorganization, transcriptional regulation, apoptosis, mitosis, cell metabolism and development [for review see (4–7)]. The founding member PARP1 as well as PARP2 and PARP3 play important roles in the repair of DNA strand breaks and are known to be catalytically activated through interaction with DNA damage and catalyze auto-ADP-ribosylation and ADP-ribosylation of other nuclear acceptor proteins [for review see (7–9)]. It was shown that PARP1–3 are recruited to sites of DNA damage induced by laser microirradiation or site-specific nucleases (10–12). PARP1 and PARP2 account for most of cellular PARylation activity (75–95% and 5–15%, respectively) after DNA damage (13–15). In vivo and cellular studies have revealed that depletion of PARP1 or PARP2 results in hypersensitivity to ionizing radiation, to oxidative stress and to alkylating agents (16–18). PARP1 is recruited to single- and double-strand DNA breaks (SSBs and DSBs, respectively) in genomic DNA and is involved in multiple DNA repair pathways, including base excision repair (BER), homologous recombination (HR), non-homologous end-joining (NHEJ) and nucleotide excision repair (NER) (8,19). PARP2 has partially redundant functions with PARP1 that are essential for cell survival. Indeed, double-knockout Parp1−/−Parp2−/− mice show early embryonic mortality (18). Both PARP1 and PARP2 but not PARP3 are required for hydroxyurea-induced HR to ensure cell survival after replication arrest (20) but either PARP1 or PARP2 activity is sufficient for XRCC1 recruitment at an oxidative SSB (21). Nevertheless, there is a growing body of evidence of some specific functions of PARP2 in the maintenance of genomic stability and in development (22). It has been demonstrated that PARP2 (i) has slower kinetics of recruitment to damaged sites (10), (ii) is activated by different types of DNA structures (14,23–25) and (iii) PARylates an overlapping but distinct set of protein targets relative to that of PARP1 (26–28). A recent genetic study revealed that both PARP1 and PARP2 are essential in HR-deficient mice lacking the H2AX histone variant but only PARP1 is required in NHEJ-deficient (Xrcc5−/− or Prkdc−/−) mice (29).
PARP3 is a third member of the PARP family and is characterized by mono(ADP-ribosyl)ation (MARylation) activity at the sites of DNA breaks (30,31). It has been reported that PARP3 interacts with proteins of the canonical NHEJ (C-NHEJ) pathway, including DNA-PKcs, DNA ligase IV, Ku70/80 and APLF; ADP-ribosylates some of them (Ku70/80 and others); facilitates their recruitment to a DSB and consequently stimulates both earlier and later steps of accurate C-NHEJ (32–34). PARP3 and PARP1 act synergistically in response to X-irradiation in human and mouse cells (11). In the absence of PARP3, cells become more sensitive to antitumoral drugs generating DSBs and show a significant delay in the repair of radio-induced DSBs (11,32). PARP3 expression is tightly regulated with preferential expression in well-differentiated cells (35) in contrast to PARP1 and PARP2, which are expressed ubiquitously with a preference for proliferating cells (17). Proteomic screening assays identified considerable overlaps among the protein targets ADP-ribosylated by different DNA-dependent PARPs, where, surprisingly, PARP3 has the largest number of unique targets (26–27,36–37).
All three DNA-dependent PARPs have tryptophan-glycine-arginine (WGR) and C-terminal catalytic (CAT) domains and the overall three-dimensional structures of their CAT domains are similar (38). They share an allosteric regulatory mechanism of DNA-dependent catalytic activation via local destabilization of CAT (39). In contrast to PARP1, PARP2 and PARP3 do not contain the N-terminal zinc finger domains essential for initial DNA binding of PARP1 or the central auto-modification domain containing a BRCT fold (40). Short N-terminal regions (NTRs) of PARP2 and PARP3 (70 and 40 residues, respectively) are not strictly required for DNA-dependent activation but rather contribute to the overall binding affinity and specificity for SSB as shown for PARP2 (24,41). Both PARP2 and PARP3 are preferentially activated by an SSB harboring a 5′ phosphate, indicating selective activation in response to DNA damage or DNA repair intermediates competent for DNA ligation (14,23–25,31).
Recently, we demonstrated that mammalian PARP1 and PARP2 can catalyze covalent addition of ADP-ribose units not only to proteins but also to terminal phosphates and to the 2′-OH terminus of a modified nucleotide at a DNA strand break, producing a covalent PAR–DNA adduct in vitro (25). Furthermore, the PARP-catalyzed DNA PARylation is a reversible process because PAR can be entirely degraded by poly(ADP-ribose) glycohydrolase (PARG) (25). Nevertheless, the detailed molecular mechanisms of PARP-dependent DNA ADP-ribosylation and the biological role of this process are still unclear. At present, the participation of PARP3 in DNA ADP-ribosylation has not been demonstrated.
In this study, we examined DNA ADP-ribosylation activity and the DNA substrate preference of PARP3 as compared with structurally similar PARP2. We show that PARP3 can effectively produce MAR–DNA adducts on terminal phosphate residues at DSB and SSB termini of short and long DNA molecules, sharing its substrate specificity with PARP2. We demonstrate effective ADP-ribosylation of 5′-terminal thiophosphates at DSB termini that makes such MAR–DNA adducts resistant to PARG hydrolysis. We found that depending on configuration of DNA strand breaks, the DNA termini can become preferred acceptor sites for ADP-ribosylation as compared to proteins. According to the data obtained, we propose a putative mechanistic model of DNA break-oriented DNA ADP-ribosylation by PARP3 or PARP2 that could explain our results. Our findings reveal effective PARP3- or PARP2-catalyzed ADP-ribosylation of ∼3-kb DNA plasmid-based substrates and DNA PARylation activity in nuclear extracts from HeLa cells. Finally, immunoblotting of purified genomic DNA from PARG-depleted HeLa cells after genotoxic treatment provides indirect evidence of the presence of PAR–DNA adducts in live cells. The physiological relevance and possible biological outcomes of the PARP-catalyzed ADP-ribosylation of DNA are discussed.
MATERIALS AND METHODS
Proteins, chemicals and reagents
Proteinase K from Tritirachium album and Deoxyribonuclease I from bovine pancreas (DNase I) were purchased from Sigma–Aldrich (France). CIP (alkaline phosphatase, calf intestinal), TdT (terminal deoxynucleotidyl transferase) and PmlI and Nb.BsmI endonucleases were purchased from New England Biolabs France (Evry, France). Human PARP1 (EC 2.4.2.30) and bovine PARG were purchased from Trevigen (Gaithersburg, USA). The plasmids coding for murine PARP2 and human PARP3 were kindly provided by Dr V. Schreiber (ESBS, Illkirch, France). SUMO fusion expression vector pETHSUL (GenBank: EF205333.1) and pSUPER vector coding for the catalytic domain of the Saccharomyces cerevisiae SUMO hydrolase (dtUD1) were kindly provided by Dr P. Loll (Drexel University, Philadelphia, USA). The untagged PARP2 protein was expressed and purified from insect cells as described previously (23). His6 and Strep-II double-tagged dtUD1 hydrolase was expressed and purified as described elsewhere (42). Bleomycin was acquired from Sanofi-Aventis (France), and olaparib (AZD2281, Ku-0059436) from Selleck Chemicals (Houston, USA).
Purification of PARP3
Human PARP3 was cloned into the pETHSUL vector using the overlap extension polymerase chain reaction cloning approach (43). N-terminal His6-tagged SUMO-PARP3 fusion was expressed in Escherichia coli Rosetta 2 (DE3) electro-competent cells (Novagen). The bacterial culture was grown at 37°C in a LB medium (supplemented with 100 μg/ml ampicillin) to OD600 = 0.6–0.8. The protein expression was induced by 0.5 mM isopropyl β-D-galactopyranoside (IPTG; Sigma-Aldrich) during overnight incubation at 18°C. The bacteria were harvested by centrifugation, and cell pellets were lysed using a French press at 18 000 psi in a buffer consisting of 20 mM HEPES-KOH pH 7.6, 40 mM NaCl and 0.1% (w/v) NP-40 supplemented with the cOmplete™ Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland). Lysates were cleared by centrifugation at 40 000 × g for 30 min at 4°C, and the resulting supernatant was adjusted to 500 mM NaCl and 20 mM imidazole and loaded onto a HiTrap Chelating HP column (Amersham Biosciences, GE Healthcare). The column was washed with buffer A (20 mM HEPES-KOH pH 7.6, 500 mM NaCl, 40 mM imidazole) and the bound PARP3 fusion was eluted with a linear 20–500 mM gradient of imidazole in buffer A on Äkta Purifier (GE Healthcare). Eluted fractions of the PARP3 fusion were diluted three times in 20 mM HEPES-KOH pH 7.6, and the His6-SUMO tag was cleaved by incubation with an equimolar concentration of His6 and Strep-II double-tagged dtUD1 SUMO hydrolase for 1 h at 4°C. The protein mixture obtained was loaded onto an additional HisTrap HP column (Amersham Biosciences, GE Healthcare) equilibrated with buffer A. The flow-through fractions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and the fractions containing the homogeneous untagged PARP3 protein were stored at −80°C in 50% glycerol. The concentration of the purified proteins was determined by the method of Bradford.
Oligonucleotides, Dbait molecules and plasmid-based DNA substrates
Sequences of the oligonucleotides, their duplexes and Dbait molecules used in the present work are shown in Supplementary Table S1 and Figure S12, respectively. Regular oligonucleotides, oligonucleotides with thiophosphates and Dbait molecules, containing a hexaethyleneglycol linker [(CH2-CH2-O)6] tethering two complementary DNA strands, were purchased from Eurogentec (Seraing, Belgium). Prior to enzymatic assays, the oligonucleotides were labeled at the 5′-OH end using T4 polynucleotide kinase (Thermo Scientific) in the presence of [γ-32P]ATP (3000 Ci⋅mmol−1) (PerkinElmer) or at the 3′-OH end by means of TdT in the presence of [α-32P]-3′-dATP (cordycepin 5′-triphosphate, 5,000 Ci⋅mmol−1; PerkinElmer). Cold ATP at 1 mM was added to phosphorylate the remaining non-labeled oligonucleotides. After labeling reactions, the radioactively labeled oligonucleotides were desalted on a Sephadex G-25 column, equilibrated with water and then annealed with a corresponding complementary strand for 3 min at 65°C in the following buffer: 20 mM HEPES-KOH (pH 7.6) and 50 mM KCl. Radioactive labeling of duplex DNA was also performed using radioactive [adenylate-32P]NAD+ (800 Ci⋅mmol−1) (PerkinElmer) in the presence of PARPs. To prepare a 5′-[32P]labeled linearized nicked plasmid substrate, 50 μM pML2 plasmid (pBluescript-based plasmid after insertion of a unique PmlI site) was linearized with 30 U of PmlI for 1 h at 37°C in 1 × CutSmart buffer (New England Biolabs, France), then nicked with 15 U of Nb.BsmI for 1 h at 65°C. The 32P label was introduced by reannealing with the 5′-[32P]labeled [5′-d(GTGGTTGTAAAACCTCAGCCAG)-3′] oligonucleotide corresponding to the 22-nt fragment between the 5′ DSB end and the Nb.BsmI-induced nick.
An assay for PARP-dependent DNA ADP-ribosylation
This assay was carried out as described previously (25). Briefly, 40 nM [32P]labeled oligonucleotide duplexes were combined with 50 nM PARP3 or 40 nM PARP2 in the presence of 1 mM NAD+ in ADPR buffer [20 mM HEPES-KOH pH 7.6, 50 mM KCl, 5 mM MgCl2, 1 mM 1,4-dithiothreitol (DTT) and 100 μg/ml bovine serum albumin (BSA)]. The mixture was incubated for 2 min (30 min for PARP2) at 37°C, unless otherwise stated. After the reaction, the samples were incubated in the presence of 50 ng/μl proteinase K and 0.15% SDS for 30 min at 50°C followed by addition of 4 M urea and incubation for 10 s at 95°C. The reaction products were analyzed by electrophoresis in denaturing 20% (w/v) polyacrylamide gels (PAGE; 7 M Urea, 0.5 × Tris-Borate-EDTA buffer, 42°C). The gels were used to expose a Fuji FLA-3000 Phosphor Screen, which was then scanned with Typhoon FLA-9500 and analyzed using the Image Gauge 4.0 software.
Analysis of the efficiency of PARP3- or PARP2-catalyzed auto- and DNA ADP-ribosylation
The efficiency of PARP2-catalyzed auto- and DNA ADP-ribosylation was measured using a cold ExoA•RexTnick duplex phosphorylated at the 5′ end of the nick, with or without a thiophosphate at the 5′ DSB terminus. The assay was performed in ADPR buffer but without BSA. One micromolar PARP2 was added to 1 μM oligonucleotide duplex and incubated in the presence of 0.5 μM [adenylate-32P]NAD+ for 30 min. The reaction products were treated with PARG at 50 pg/μl for 30 min at 30°C and then with 10.5 U DNase I for 30 min at 37°C in the presence of 0.5 mM CaCl2 or treated with 50 ng/μl proteinase K for 30 min at 50°C in the presence of 0.1% SDS. The reactions were terminated by the addition of a stop solution (7.5 M Urea, 0.33% SDS, 10 mM ethylenediaminetetraacetic acid (EDTA), bromophenol blue) at 1:1 (v/v), heated at 95°C for 10 s, and the products of the reactions were analyzed by denaturing PAGE as described above. The efficiency of PARP3-catalyzed auto- and DNA ADP-ribosylation was evaluated as described for PARP2, except that the 5′-terminal thiophosphate at the 5′ DSB terminus of ExoA•RexTnick was replaced by a 5′ phosphate and no additional PARG treatment was carried out.
The cell line, culture conditions and preparation of nuclear extracts
Stable PARG knockdown (shPARG/PARGKD) and control (shCTL/BD650) HeLa cell lines have been described elsewhere (44). The cells were grown in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with penicillin/streptomycin (Gibco, Gaithersburg, USA) and 10% of fetal bovine serum in a humidified atmosphere containing 5% of CO2. After harvesting, the cells were washed twice in cold phosphate-buffered saline (PBS). All the procedures were conducted at 4°C. The cell pellets were resuspended in 3 volumes (w/v) of cytoplasmic extract buffer (10 mM HEPES-KOH pH 7.6, 10 mM KCl, 0.1 mM EDTA, 0.15 mM spermine, 0.7 mM spermidine, 1 × cOmplete protease inhibitors EDTA-free [Roche], 1 mM DTT); 0.1% NP-40 was added immediately after cell resuspension. The cells were allowed to swell on ice for 5 min. Nuclei were collected by centrifugation (500 × g, 5 min), then resuspended in one volume of nuclear extract buffer (20 mM HEPES-KOH pH 7.6, 0.4 M NaCl, 1 mM EDTA, 25% glycerol, 1 × cOmplete protease inhibitors EDTA-free [Roche], 1 mM DTT). After 10-min incubation on ice, the samples were centrifuged at 13 000 × g for 5 min. The nuclear extracts (supernatants) were stored at −20°C if not used immediately.
Identification of PAR–DNA adducts in genomic DNA (gDNA)
To this end, PARylation was stimulated by treating HeLa PARGKD cells with 100 μg/ml bleomycin in PBS for 30 min at 37°C. gDNA from untreated HeLa BD650 cells (control gDNA) or from HeLa PARGKD cells treated with bleomycin was extracted from 2 × 106 cells using a Maxwell® RSC purifier (Promega) and the Maxwell® RSC Blood DNA Kit according to the manufacturer’s protocol, including extended up to 1 h Proteinase K (2.0 mg/ml) treatment at 56°C. Optionally, gDNA from treated PARGKD cells representing the first round of purification was incubated with 1 M hydroxylamine (pH 8.0) for 2 h at room temperature. A free PAR polymer was prepared as follows: PARP1 (38 ng/μl) was auto-PARylated in ADPR buffer in the presence of 1 mM NAD+ and 50 nM non-phosphorylated oligonucleotide duplex Exo40f•RexT (Supplementary Table S1, DNA structure N° 28) for 30 min at 37°C followed by Proteinase K treatment and PAR purification on a Maxwell® RSC purifier as described above for gDNA. PAR concentration was estimated by anti-PAR immunoblotting in comparison with a commercially available free PAR polymer (Trevigen, USA) of known concentration represented as the concentration of ADP-ribose monomers. Next, treated or untreated gDNA from PARGKD cells and control gDNA were mixed or not mixed with free PAR (19 pmol PAR per 1 μg of gDNA), and gDNA was successively purified on Maxwell® RSC purifier several times as described above. In each purification round, 5 μg of gDNA was used per Maxwell® RSC Blood DNA Kit cartridge. To identify PAR–DNA adducts, 1200, 380, 120 or 38 ng of gDNA from each purification round was loaded on a Hybond N+ nylon membrane (GE Healthcare). DNA and PAR were fixed to the membrane by heating at 80°C for 2 h and were analyzed with a mouse monoclonal anti-poly(ADP-ribose) 10H antibody (1:1000, Enzo Lifesciences Inc., USA). Immunodetection was conducted by the ECL method, followed by a scan using an Amersham® Imager 600 (GE Healthcare). Digital images were quantified using the ImageQuant TL v.8.1.0.0 software.
RESULTS
Human PARP3 efficiently catalyzes mono(ADP-ribosyl)ation of DNA oligonucleotide duplexes in vitro
Here, we aimed to characterize the possible DNA ADP-ribosylation activity of the PARP3 enzyme. For this purpose, we used the ExoA•RexTnick duplex containing a 5′-phosphorylated nick and [32P]labeled 5′-phosphate at the DSB terminus of the nicked strand (Supplementary Table S1, DNA substrate N° 1). According to our previous study, it was one of the preferred DNA substrates for PARP2-catalyzed DNA PARylation (25). As shown in Figure 1, incubation of the 5′-[32P]labeled ExoA•RexTnick duplex with recombinant PARP3 in the presence of 1 mM NAD+ resulted in formation of a ∼1-nt-heavier band on a denaturing polyacrylamide gel in a time- and protein concentration-dependent manner (Figure 1A and B). Analysis of the Matrix Assisted Laser Desorption Ionization Time-Of-Flight (MALDI TOF) mass spectrum of the corresponding reaction products (Supplementary Figure S1) revealed that incubation with PARP3 led to the appearance of a mono-charged peak at [M-H]− = 7180.1 Da corresponding to the 5′-phosphorylated 21mer ExoA that contains one ADP-ribose residue (calculated mass 7180.2 Da, Supplementary Figure S1B). These results clearly indicate that PARP3 efficiently catalyzes MARylation of the 5′-phosphorylated ExoA oligonucleotide.
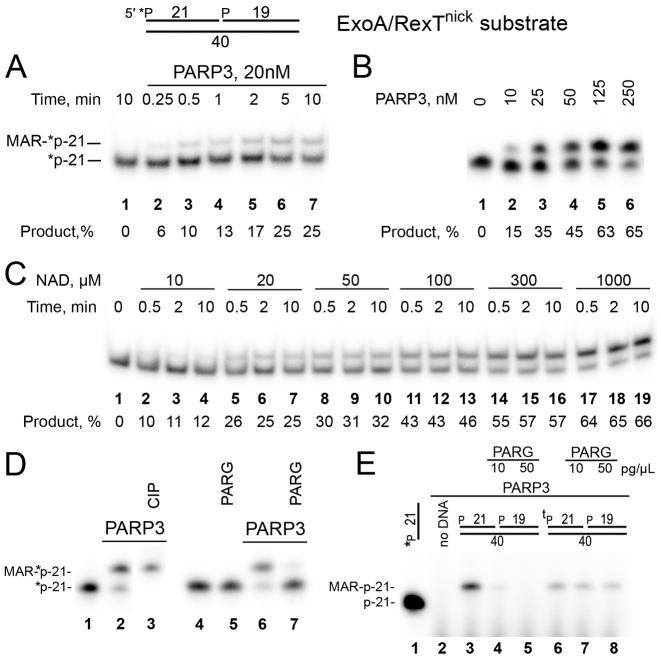
Figure 1.
PARP3-catalyzed formation of MAR–DNA adducts with nicked 5′-phosphorylated oligonucleotide duplexes. Denaturing PAGE analysis of PARP3 generated products of ADP-ribosylation of the 5′-[32P]labeled ExoA•RexTnick duplex. (A) Time dependence of PARP3-catalyzed MARylation of 50 nM DNA. (B) The dependence of DNA (20 nM) MARylation on PARP3 concentration. (C) PARP3 (100 nM) MARylation activity toward DNA (50 nM) in the presence of varying concentrations of NAD+. (D) PARG and CIP treatments of DNA MARylation products. After incubation with 500 nM PARP3, the DNA (40 nM) samples were heated for 20 min at 80°C and the resulting [32P]labeled DNA MARylation products were further incubated with 50 pg/μl PARG or 10 U of CIP for 30 min at 37°C. (E) One μM PARP3 was incubated with 1 μM cold ExoA•RexTnick-based oligonucleotide duplexes containing either a 5′-phosphate or a 5′-thiophosphate (tP) at the 5′ DSB termini in the presence of 0.5 μM [adenylate-32P]NAD+ for 30 min at 37°C. The reaction products were treated with PARG at 10 or 50 pg/μl for 30 min at 37°C.
PARP3-dependent DNA ADP-ribosylation reached a plateau already after 5 min of incubation, requiring relatively high protein concentrations (>100 nM) to obtain more than 60% of the modification 5′P-ExoA (Figure 1A and B). Contrary to PARP3, PARP1- and PARP2-dependent DNA ADP-ribosylation after the initial rapid phase (∼50% of the final product was formed in 5 min) was followed by a slow phase (another 50% of the product was formed in ∼1 h) [Supplementary Figure S2]. These differences in the kinetics of PARPs’ DNA ADP-ribosylation activity can be explained by gradual dissociation of auto-PARylated PARPs (PARP1 and PARP2)—but not of auto-MARylated PARP3—from DNA breaks; this dissociation limits the reaction turnover. Then, we examined NAD+ dependence of the PARP3-mediated catalysis of DNA ADP-ribosylation. Under the reaction conditions used, we observed formation of MAR–DNA adducts already at 10 μM NAD+ (with a 10–12% yield), and their steadily increased formation at higher concentrations of NAD+ (Figure 1C). As expected from our previous data on ADP-ribosylation of terminal phosphates by PARP1 and PARP2 (25), additional treatments of the PARP3 reaction products with calf-intestinal alkaline phosphatase (CIP) and PARG showed that the 5′[32P] label of the MARylated ExoA oligonucleotide was protected from CIP dephosphorylation (Figure 1D, lane 3), and the MAR adduct was completely removed by PARG (Figure 1D, lane 7). Moreover, in the experiments with the cold 5′P-ExoA•RexTnick duplex and [adenylate-32P]NAD+ (Figure 1E), replacement of 5′-phosphate with a 5′-thiophosphate (tP) group in oligonucleotide ExoA rendered PARP3-generated MAR–DNA adducts resistant to PARG treatment. This finding confirmed PARP-dependent modification of the terminal phosphate group and suggests that tP may serve for the stabilization of MAR–DNA adducts in vitro and in vivo. ADP-ribosylation of [32P] residues of 5′-[32P]labeled ExoA was in agreement with the analysis of products of BsmAI restriction endonuclease digestion after PARP3 or PARP2 ADP-ribosylation of [32P]labeled ExoA•RexTnick duplexes (Supplementary Figure S3). Thus, BsmAI digestion of the PARP3 MARylated 5′-[32P]labeled ExoA•RexTnick duplex resulted in formation of a MAR-[32P]labeled 3mer (Supplementary Figure S3A, lanes 6 and 7), suggestive of MARylation of the 5′-[32P]labeled terminus of 5′P-ExoA. In the case of PARP2-dependent PARylation of 3′-3′dAM32P-labeled 5′P-ExoA•RexTnick duplexes, BsmAI treatment led to disappearance of the [32P]labeled DNA–PAR complex (Supplementary Figure S3B, lane 8 versus 11), again pointing to PARylation of the unlabeled 5′ terminus of 5′P-ExoA. As expected, BsmAI digestion of the PARP2-PARylated 5′-[32P]labeled 5′P-ExoA•RexTnick duplex could not influence the mobility of the [32P]labeled PAR–DNA adduct owing to the high molecular weight of PAR (lane 14 versus 17). Similar results were obtained with additional PARG treatment after PARP2 treatments (lanes 9 and 15 versus 12 and 18, respectively).
It should be noted that the DNA–MAR adducts were substantially unstable during extensive heating (∼50% degradation after 1 h of incubation at 90°C and neutral or basic pH) and particularly at acidic pH (∼90% degradation after 10 min of incubation at 90°C and pH 3.3), resulting in a release of the initial [32P]labeled DNA substrate (Supplementary Figure S4). These results are consistent with the previously observed acid-labile phosphodiester linkage of ADP-ribose to a phosphoserine in histones (45).
Overall, these results strongly indicate that PARP3 uses the 5′ phosphate at the DSB end of the 5′P-ExoA•RexTnick duplex as an acceptor for transfer of the ADP-ribose group and that PARG can reverse ADP-ribosylation of phosphate residues.
Comparison of DNA substrate specificity of PARP2-catalyzed and PARP3-catalyzed DNA ADP-ribosylation
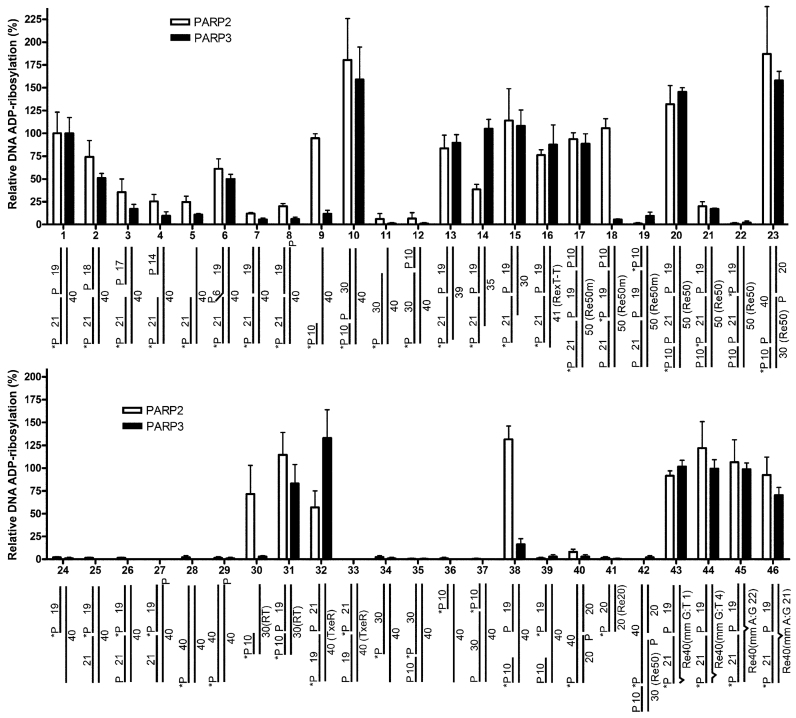
Next, we assessed in more detail the influence of the type of DNA breaks, the size, terminal phosphate positions and other structural characteristics of DNA duplexes (Supplementary Table S1) that mimic different types of DNA damage and intermediate products of the DNA repair pathways on PARP-catalyzed DNA ADP-ribosylation (Figure 2). Previously, we have already shown that PARP2 preferentially PARylates the 5′-phosphorylated 21mer oligonucleotide in the nicked or gapped ExoA•RexT duplex with a 5′-phosphate residue located at the double-strand termini (25). Thus, the extent of PARP2- or PARP3-catalyzed ADP-ribosylation of 5′P-ExoA in the 5′-[32P]labeled nicked duplex was assumed to be 100% (substrate N° 1), when compared with PARP activities toward other DNA structures.
Figure 2.
Effects of the type of DNA structure and termini on the PARP3- or PARP2-catalyzed formation of MAR– and PAR–DNA adducts, respectively. The bar graphs present the relative ratio of DNA ADP-ribosylation activities of PARP3 or PARP2 on different DNA structures as compared to the control ExoA•RexTnick duplex (N° 1). For oligonucleotide duplexes with non-standard RexT sequences, the names of sequences are shown in parentheses. The data on PARP-catalyzed formation of DNA ADP-ribosylation products are presented as mean ± SD from three independent experiments.
Each PARP efficiently ADP-ribosylated the 5′-phosphate residue located at the double-strand termini of DNA duplexes with a 5′-phosphorylated 6-nt flap (DNA substrate N° 6) and 5′-phosphorylated nick containing (i) a blunt DSB end (N° 1, 10, 17, 20, 31, 32); (ii) 1-, 5- and 10-nt 5′ overhangs (N° 13–15); (iii) 1-nt 3′ overhang (N° 16); (iv) single-nucleotide mismatches at different positions in 5′-[32P]labeled ExoA•RexTnick duplexes (N° 43–46). A significant increase in the efficiency of DNA ADP-ribosylation by PARP2 or PARP3 (∼1.8- and ∼1.6-fold, respectively) was observed when the 5′-phosphorylated nick was situated 10 nt downstream from the 5′P DSB terminus (substrate N° 10), and its strong inhibition was observed when the 5′-phosphorylated nick was 30 nt downstream from the 5′P acceptor terminus (substrate N° 12) in the same ExoA•RexT 40mer context. Efficiency of PARP2- and especially PARP3-catalyzed ADP-ribosylation progressively diminished with the increase of the gap size in the 5′-[32P]labeled ExoA•RexT gapped or recessed duplexes (Figure 2, DNA substrates N° 1–5). PARP3 retained only 10% or less of its activity toward duplexes containing a ≥5-nt gap and toward the recessed DNA duplex under study (substrates N° 4, 5, 9, 11, 30 and 38). No modification of the 5′P DSB terminus was observed when the 5′-phosphorylated nick was moved to the opposite strand (substrates N° 40 and 42). Given that PARP2 and PARP3 are selectively activated by 5′-phosphorylated DNA nicks (24), these results suggest that the discontinued strand in the duplex and the distance between the 5′P acceptor terminus and 5′P activator nick or gap site are important factors, in that the distance should not exceed ∼21 nt for efficient ADP-ribosylation of the acceptor terminus. These results may also reflect a decrease in PARP2 and particularly PARP3 activation by the 5′ phosphate in gapped structures (24). As expected from our previous results on PARP2 (25), no ADP-ribosylation was observed for 5′-phosphorylated oligonucleotides of the full/unnicked duplexes tested (substrates N° 28, 29 and 41) and for 5′-phosphorylated oligonucleotides downstream of a nick or gap in duplexes containing a single nick or gap, respectively (substrates N° 24–27, 33–37, 39 and 42) for both PARPs. These results are in agreement with structural data, where DNA-binding (activator) sites are distant from catalytic sites of DNA-dependent PARPs (41,46) and may be sterically protected from modification.
Moreover, when DNA duplexes contained two nicks on the same strand (substrates N° 17–22), we observed not only DNA ADP-ribosylation at 5′P DSB termini (substrates N° 17 and 20) but also PARP2-dependent oligo(ADP-ribosyl)ation at the site of the 5′ nick of DNA substrate N° 18 (Figure 2 and Supplementary Figure S5A), which was 21 nt downstream of the 5′P DSB terminus. Both PARPs showed negligible ADP-ribosylation of the 21mer oligonucleotide of DNA substrate N° 21 at the site of a nick 10 nt away from the 5′P DSB terminus (Figure 2). These data are suggestive of ADP-ribosylation of 5′-phosphorylated nicks by the PARP2 enzyme in case of multiple vicinal nicks. Efficiency and processivity of ADP-ribosylation at a particular SSB site possibly depend on competition between PARP’s binding to the SSB site and its ADP-ribosylation by another PARP molecule bound to and activated on an SSB nearby. We can hypothesize that in a specific configuration of multiple SSBs, PARP3 also catalyzes MARylation of 5′P at an SSB site, which has low affinity for PARP3 and therefore is less sterically protected by another PARP3 molecule bound to it. It should be noted that contrary to PARP3, PARP2 showed efficient DNA ADP-ribosylation of 5′P DSB termini of gapped (10-nt) and recessed DNA substrates, when the break in the modified strand started 10 nt downstream from the DSB terminus (substrates N° 9 and 38 versus 10; N° 30 versus 31). These results suggest that PARP2 may efficiently recognize and be activated on an SSB near a DSB in a 5′-phosphorylated nick/gap-independent manner.
Next, we further tested whether PARP3 can utilize 3′P at a nick or 1-nt gap as the acceptor site for MARylation of duplex DNA (Supplementary Figure S5B). No 3′P ADP-ribosylation at SSB sites was detected with the DNA constructs being analyzed (lanes 8, 9, 14 and 15). As expected from our previous work on PARP2 (25), DNA ADP-ribosylation activities of both PARPs at the 5′P DSB terminus were strongly inhibited by the presence of 3′P at SSB sites (lanes 5, 6, 11, 12, 17 and 18). Furthermore, the presence of a 3′-terminal phosphate residue at the 5′-phosphorylated nick site of oligonucleotide duplex ExoA•RexTnick also significantly inhibited auto-ADP-ribosylation of proteins PARP2 and PARP3 (Supplementary Figure S6). Taken together, these results suggested that both 3′OH and 5′P termini of a nick (competent for DNA ligation) participate in PARP2 and PARP3 DNA nick-dependent activation.
The results showed that PARP2- and PARP3-catalyzed DNA ADP-ribosylation activities on DNA substrates were not dependent on DNA sequence context or on the nature of the phosphorylated 5′-terminal nucleotide at a DNA acceptor site (Figure 2, DNA substrates N° 1 versus 32; N° 10 versus 20, 23 and 31). Furthermore, regarding the nature of the DNA base at the 3′OH end of the nick (Supplementary Figure S7), single-nucleotide mismatches at different positions in duplexes 5′-[32P]labeled ExoA•RexTnick (Figure 2, DNA substrates N° 43–46) and 5′ overhangs (N° 1 versus 13–15) did not exert a significant influence on the extent of DNA ADP-ribosylation by PARP2 and -3. Taken together, these data suggest that DNA ADP-ribosylation activities of PARP2 and PARP3 do not depend on a sequence context but rather on the presence of a phosphate and 3′-hydroxyl at DNA strand break termini and on the topological configuration of strand breaks relative to each other.
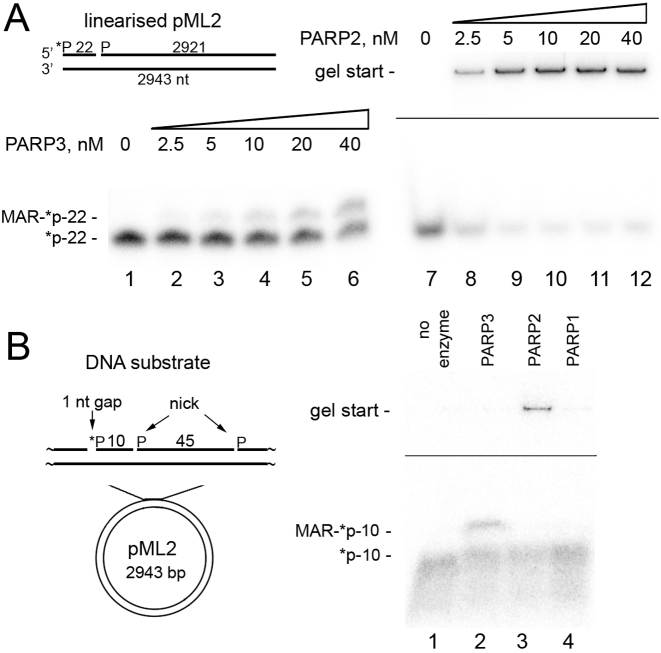
To test whether PARP2 or PARP3 can also ADP-ribosylate high-molecular-weight DNA substrates, at first we constructed a linear plasmid-based DNA substrate ∼3 kb in length containing a single nick 22 nt away from the 5′-[32P]-labeled blunt-ended DSB (Figure 3A). As shown in Figure 3A, PARP3 and especially PARP2 showed efficient ADP-ribosylation of the 5′-phosphorylated 22mer at the DSB terminus, suggesting that the DNA ADP-ribosylation activity of PARPs is not limited to short oligonucleotide duplexes and can be effective on breaks in high-molecular-weight DNA. To further substantiate the PARPs’ capacity for ADP-ribosylation of high-molecular-weight DNA at SSB sites, we used circular ∼3 kb plasmid DNA containing a 1 nt gap placed 10 nt upstream of a nick. Both PARP3 and PARP2 ADP-ribosylated 5′P in the gap, thereby generating mono- and poly(ADP-ribosyl)ated 5′-[32P]labeled 10mer oligonucleotides, respectively (Figure 3B and Supplementary Figure S8). In contrast to PARP2 and PARP3, PARP1 did not exert the DNA PARylation activity on the same plasmid DNA substrate (Figure 3B, lane 4). Further studies are needed to characterize the dependence of PARP1-catalyzed ADP-ribosylation of circular DNA on the distance between SSBs and on the size of a gap. These data revealed the ability of enzymes PARP2 and PARP3 to ADP-ribosylate plasmid DNA molecules containing multiple DNA strand breaks (SSB plus DSB or multiple SSBs).
Figure 3.
PARP3- and PARP2-catalyzed formation of MAR(PAR)–DNA adducts at the phosphorylated DSB and SSB termini of plasmid-based DNA substrates. (A) Denaturing PAGE analysis of PARP-generated products of ADP-ribosylation of 1 nM 5′-[32P]labeled linearized nicked pML2 plasmid under the standard reaction conditions. (B) ADP-ribosylation of a circular plasmid containing multiple SSBs. Fifty-nanomolar PARPs were incubated with 2 nM pML2 circular plasmid containing a 5′-[32P]labeled 1 nt gap placed upstream of two nicks under the standard reaction conditions.
Taken together, these data revealed that in general, both PARPs have broad DNA substrate specificity with similar DNA substrate requirements for efficient DNA ADP-ribosylation though only PARP2 shows significant DNA ADP-ribosylation at DSB sites of short recessed DNA structures.
PARP2 or PARP3 catalyzes ADP-ribosylation of terminal phosphates at the DSB end, depending on the nick location
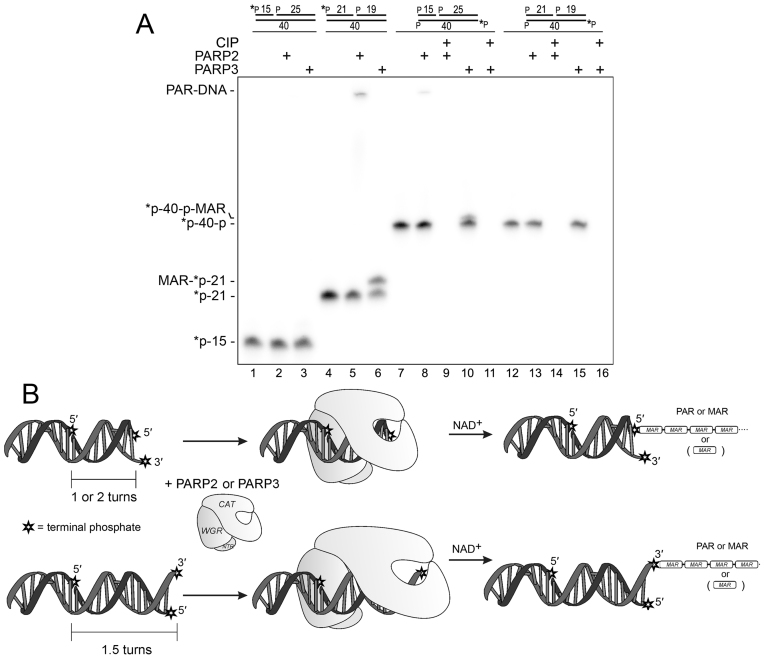
In the above experiments, the efficiency of DNA ADP-ribosylation was dependent on the distance between the 5′P DSB acceptor terminus and the nick harboring the 5′P activator, and stayed efficient at 10- and 21-nt distances. To further substantiate the mechanism of PARP’s action on duplex oligonucleotides, we characterized how the ADP-ribosylation of 5′ and 3′ acceptor phosphates at the DSB terminus depends on the distance from the 5′-phosphorylated nick. As shown in Figure 4A, neither PARP2 nor PARP3 could ADP-ribosylate a 5′[32P]-labeled DSB terminus when the nick was situated 15 nt away from the 5′ end of the nicked strand (lanes 2 and 3, respectively) contrary to the control 5′-[32P]labeled ExoA•RexTnick duplex (lanes 5 and 6, respectively). Taking into account that the 6-nt difference in the distance represents approximately a half of the turn of the DNA helix, we confirmed that in the first case, we could observe ADP-ribosylation at 3′P of the opposite strand at the same DSB terminus. Indeed, the 5′-[32P]labeled complementary 40mer (RexT) containing 3′P, was ADP-ribosylated by PARP2 and PARP3 (separately) when the nick was 15 nt away but not 21 nt away from the 5′ end of the nicked strand (lanes 8 and 10, respectively). Modification of 3′P and not 5′[32P] of the 40mer was confirmed by additional CIP treatment, which completely removed the unprotected 5′[32P] label (lanes 9 and 11). Taken together, these results suggest that PARP2 and PARP3 can produce PAR- and MAR–DNA adducts, respectively, not only on 5′- but also on 3′-terminal phosphate residues at DSB termini depending on the strand and position of the PARP-activating nick.
Figure 4.
PARP3 and PARP2 modify 5′- or 3′-terminal phosphates of the opposite strands at DSB termini depending on the distance from the nick. (A) Denaturing PAGE analysis of PARP-generated products of ADP-ribosylation of [32P]labeled nicked RexT sequence-based duplexes. ADP-ribosylation of 3′-unlabeled or 5′-terminally [32P]labeled phosphates of 40mer matrix strand was assessed by additional treatment with 10 U of CIP for 30 min at 37°C (lanes 9, 11, 14 and 16). (B) Schematic representation of the putative model of DNA modification by PARP2 or PARP3.
On the basis of the above results, we propose a putative model of PARP2 and PARP3 DNA ADP-ribosylation (Figure 4B), where the binding of PARP2 or PARP3 to a 5′-phosphorylated nick activates their catalytic domain, which in turn starts to ADP-ribosylate all sterically accessible acceptor groups on PARP itself as well as on the neighbouring DNA terminus. Thus, location of the nick within 1 and 2 turns of the DNA helix downstream of DSB termini renders its 5′ terminal phosphate accessible for the PARPs’ catalytic site. Similarly, a nick 1.5 turns downstream of a DSB promotes the modification of its 3′ terminal phosphate. Taking into account recent data showing that PARP2 preferentially binds to an SSB as a monomer (41), this model implies that the PARP molecule, which is involved in nick recognition, catalyzes modification of the DSB end of the same DNA–protein complex. Indeed, incubation of unlabeled ExoA•RexTnick-based DNA substrates competent for PARP activation with 5′-[32P]labeled full 40mer duplexes containing potential 5′P-terminal acceptor groups but not nicks, did not reveal their modification by activated PARPs bound to cold 5′P-ExoA•RexTnick (Supplementary Figure S9).
A DNA strand break's termini bearing terminal phosphates can be major acceptors of ADP-ribosylation by PARP2 or PARP3 as compared to auto-ADP-ribosylation
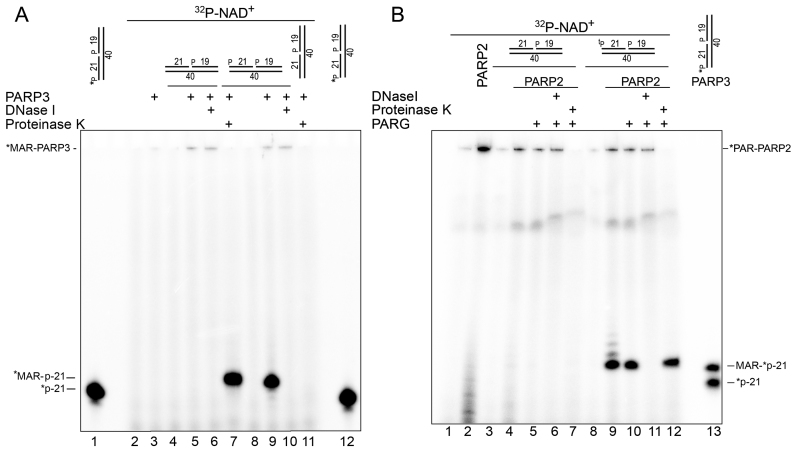
Previously, we already tried to assess the relative efficiency of PARP1 and PARP2 auto- and DNA-PARylation by fractionation of the biotinylated DNA substrates on streptavidin-coated magnetic beads, but formation of high-molecular-weight PAR, possible contamination with free PAR polymers at a high NAD+ concentration (1 mM) and the presence of biotinylated nucleotides did not allow us to obtain clear-cut results (25). Here, we used near-equimolar concentrations (0.5–1.0 μM) of unlabeled DNA substrates, PARPs and [adenylate-32P]NAD+, which enabled us to avoid the formation of long PAR polymers and to undoubtedly separate both types of ADP-ribosylation products by denaturing PAGE (Figure 5 and Supplementary Figure S10). The ADP-ribosylation action of PARP3 on the ExoA•RexTnick duplex containing only one 5′P group at the nick site (substrate N° 25) resulted in formation of a single [32P]labeled band at the top of the gel; this band was DNase I resistant but proteinase K sensitive, thus pointing to auto-MARylation of the PARP3 protein (Figure 5A, lanes 5, 6, 10 and 11). Addition of the 5′P group at the DSB terminus of the ExoA•RexTnick duplex (substrate N° 1) did not significantly change intensity of the auto-MARylated PARP3 bands at the top of the gel (Figure 5A, lanes 9 and 10) but led to the emergence of an additional ∼50-fold more intense band corresponding to MARylation of the 5′P-ExoA 21mer (lane 9), because it migrated like a 22mer (lanes 1 and 12) and was DNase I sensitive (lane 10) and proteinase K resistant (lane 7). These results suggest that PARP3 can preferentially ADP-ribosylate DNA rather than itself if the DNA substrate is prone to ADP-ribosylation.
Figure 5.
Comparison of efficiency of PARP3- or PARP2-catalyzed auto- and DNA ADP-ribosylation. One μM PARP3 (A) or PARP2 (B) was incubated with 1 μM cold oligonucleotide duplexes in the presence of 0.5 μM [adenylate-32P]NAD+ for 30 min at 37°C. The reaction products were analyzed by denaturing PAGE. tP: thiophosphate.
In similar experiments with the PARP2 protein, we used the ExoA•RexTnick duplex with ExoA containing or not containing a 5′-terminal tP group and additional PARG treatment (Figure 5B). Notably, replacement of 5′P with 5′tP at the nick site and at the DSB terminus had little (5−28% inhibition) or no effect on DNA ADP-ribosylation activity of enzymes PARP2 and PARP3 (Supplementary Figure S11). Incubation of PARP2 with a DNA duplex containing both activating and acceptor phosphates resulted in appearance of several radioactive bands migrating as mono-, di- and tri(ADP-ribosyl)ated P-ExoA adducts (Figure 5B, lane 9). We expected that PARG would be unable to remove the last MAR residue from the 5′tP group of ExoA, leaving MAR adducts not only on the PARP2 protein but also on DNA. Indeed, additional PARG treatment removed oligo(ADP-ribose)–DNA adducts and left only a slightly diminished mono-adduct band (lane 10), confirming that PARP2-generated MAR–DNA adducts on the terminal thiophosphate residue of DNA are also resistant to PARG treatment. Intensity of ADP-ribosylated 5′tP-ExoA bands was ∼5- and 4-fold higher than that of the bands of auto-ADP-ribosylated PARP2 before (lane 9) and after PARG treatment (lane 10), respectively.
Taken together, these results suggest that PARP2- or PARP3-catalyzed DNA ADP-ribosylation can be more efficient than their auto-ADP-ribosylation activities depending on the DNA substrates in question.
The presence of a PARP-dependent DNA PARylation activity in human cell-free extracts
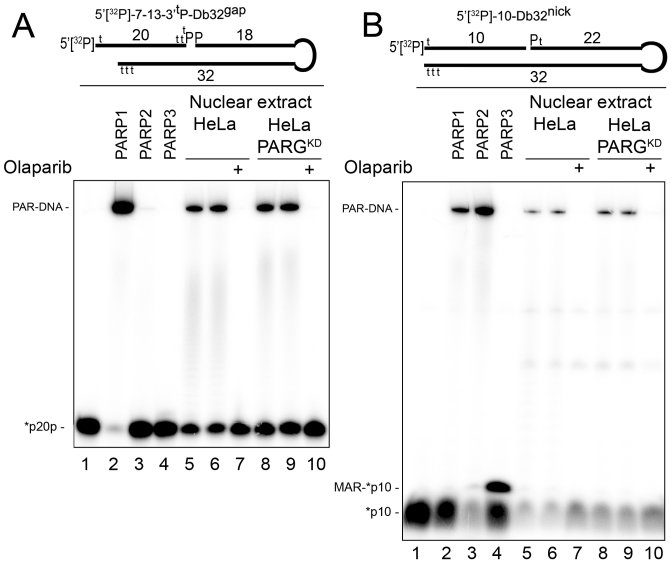
To measure PARP-mediated DNA ADP-ribosylation activity in the extracts of human cells, we used short DNA molecules mimicking a DSB and SSB (known as Dbait) structures (47). Dbait molecules were constructed by tethering two complementary oligonucleotides with a hexaethyleneglycol linker at one extremity of the duplex and protecting the other termini by adding several phosphorothioate modifications (Supplementary Figure S12). Dbait molecules are known to be resistant to nuclease degradation and to induce an efficient DNA damage response and activation of PAR synthesis after transfection into human cells (47). As shown in Figure 6, the 5′[32P]-7–13-3′tP-Db32gap molecule containing a 5′ overhang (7 nt), 5′[32P] at the DSB end and a 3′tP group at the gap site was specifically and efficiently ADP-ribosylated only by purified PARP1 but not by PARP2 or PARP3 protein (Figure 6A, lanes 2–4) obviously due to the presence of the 3′tP group at the gap site. Conversely, a 5′[32P]-10-Db32nick molecule containing the 3′OH group at the nick site was more efficiently ADP-ribosylated by PARP2 or PARP3 than by the PARP1 protein (Figure 6B, lanes 2–4). The incubation of 5′[32P]-7–13-3′tP-Db32gap with nuclear extracts from control HeLa BD650 and PARG-deficient HeLa PARGKD cells resulted in PARylation of most of the DNA substrate in both extracts (Figure 6A, lanes 5, 6, 8 and 9). As expected, addition of the PARP inhibitor Olaparib abrogated PARylation of this Dbait molecule (Figure 6A, lanes 7 and 10). We observed similar results with the 5′[32P]-10-Db32nick molecule, which is prone to PARP2 and PARP3 ADP-ribosylation, but the efficiency of its PARylation in the extracts was markedly lower and no formation of the corresponding MAR-[32P]-10mer product was detected (Figure 6B, lanes 5–10). These results are in agreement with other studies showing that PARP1 is the most abundant PARP family member in the nucleus of mammalian cells and accounts for most of the cellular PARylation activity after DNA damage (13–15). It should be noted that in a similar experiment with [adenylate-32P]NAD+ as described above for the PARP2 protein, despite efficient PARylation of 7–13-Db32gap-based Dbait molecules by PARP1, the level of its auto-PARylation was still 5- to 10-fold higher than PARylation of the 5′tP-7–13-Db32gap DNA substrate (Supplementary Figure S13, lanes 9 and 10) in agreement with the ratio obtained in our previous work on biotinylated DNA substrates (25).
Figure 6.
Formation of PAR–DNA adducts in nuclear extracts from HeLa cells. Twenty nanomolar 5′-[32P]labeled 7–13-3′tP-Db32gap (A) or 10-Db32nick (B) Dbait molecules were incubated with the indicated amount of HeLa extracts, 200 nM olaparib, 180 nM PARP1 and 40 nM PARP2 or 50 nM PARP3 under standard reaction conditions for a PARP-dependent DNA ADP-ribosylation assay. Incubation periods were 10 min for extracts, 30 min for PARP1 and PARP2, and 2 min for PARP3. Lanes 6 and 9 show repeats of the experiments in lanes 5 and 8, respectively. The reaction products were analyzed by denaturing PAGE. For details, see ‘Materials and Methods’ section and Supplementary Figure S12.
Altogether, these results suggest that PARP1 was the major enzyme responsible for the DNA PARylation activity in the HeLa cell-free extracts and that this activity can be efficient, depending on the structure of DNA breaks.
The search for PAR–DNA adducts in genomic DNA after a genotoxic treatment
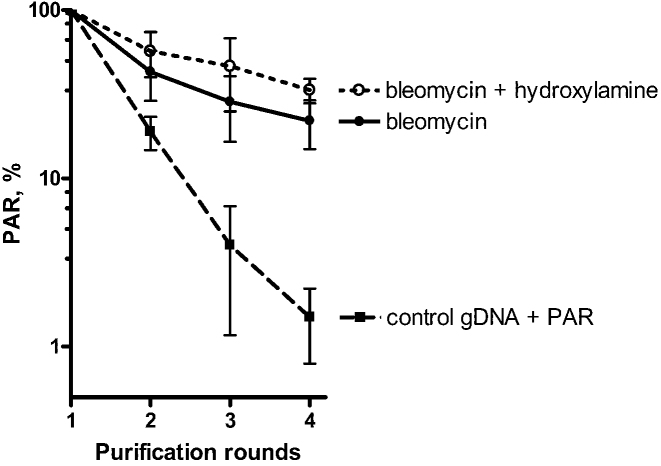
To find PARylated DNA adducts in vivo, HeLa PARGKD cells were treated with high doses of bleomycin to generate massive DSB, SSB and clustered/complex DNA damage and to induce PARP-dependent PARylation. Next, gDNA from treated HeLa PARGKD and untreated control HeLa BD650 cells was repeatedly purified, including extensive Proteinase K treatment, and was analyzed on each iteration for the presence of a PAR polymer by the dot blotting technique with an anti-PAR monoclonal antibody (Supplementary Figure S14). Taking into account the possibility of contamination of gDNA with PARylated proteins and to elucidate the chemical nature of the PAR linkage in gDNA samples, we used additional incubation of gDNA from bleomycin-treated cells with hydroxylamine after the first round of purification. It is known that the O-glycosidic linkage of ADP-ribose to acidic residues in proteins is sensitive to neutral hydroxylamine (48,49). In contrast, 2 h of incubation of a MARylated oligonucleotide with 1 M hydroxylamine (pH 8.0) did not cause detectable degradation of the (ADP-ribose)phosphate bond in the MAR–DNA adduct (Supplementary Figure S14D). Our dot blotting results revealed notable stabilization of the PAR signal in gDNA samples from bleomycin-treated cells with or without additional hydroxylamine treatment already after the second round of purification, contrary to the mix of control gDNA with free PAR which showed progressive loss of the PAR signal in every round of purification (Figure 7). These results are suggestive of the possible presence of PARylated DNA adducts in gDNA of human cells after genotoxic treatment. It should be noted that the in vitro auto-PARylated PARP1 protein was obtained by incubation of PARP1 with the unphosphorylated ExoA•RexT recessed duplex (substrate N° 5), which does not have acceptor sites for PAR. Nevertheless, this kind of approaches cannot completely rule out possible (i) contamination of the obtained gDNA samples with PARylated proteins or peptides that are tightly bound or cross-linked to DNA and highly resistant to proteinase and hydroxylamine treatments (due to PARylation of serine, lysine or other residues for example); (ii) non-specific recognition by anti-PAR antibodies of some DNA structures formed during bleomycin and hydroxylamine treatments; and (iii) presence of hypothetical noncovalent tangled complexes of PAR with gDNA.
Figure 7.
Differential loss of the PAR signal in dot blotting experiments during successive procedures of gDNA purification from HeLa PARGKD cells treated with bleomycin as compared to control gDNA mixed with free PAR. Graphical representation of the PAR level detected in 1200-ng dots of gDNA after each purification round. Control gDNA was purified from untreated HeLa BD650 cells. The PAR signal after the first purification round was set to 100%. The error bars represent SD (n = 3). For details, see ‘Materials and Methods’ section and Supplementary Figure S14.
Thus, these results provide additional albeit indirect evidence of DNA PARylation in vivo. Further studies including advanced mass spectrometry analysis are needed to detect the ADP-ribosylated DNA adducts in live cells beyond any doubt.
DISCUSSION
Although the phenomenon of NAD+-dependent PARylation of proteins was discovered more than 50 years ago, it is still unclear how this modification governs a multitude of cellular processes. Previously, using duplex DNA oligonucleotides, we have demonstrated that PARP1 and PARP2 can PARylate DNA strand break termini on 5′-terminal phosphates at DSB termini of nicked, gapped or recessed DNA duplexes (25). We suggested covalent attachment of C1′ of ADP-ribose to the terminal phosphates or the 2′-OH group of modified deoxy- and regular ribonucleotides in a DNA duplex by PARPs. In the present work, we show that PARP3 is a bona fide DNA mono(ADP-ribosyl)transferase which MARylates DNA strand break termini via covalent addition of a single ADP-ribose moiety to a 5′- or 3′-terminal phosphate residues in DNA. PARP3-catalyzed MARylation of DNA phosphates can be considered a new type of reversible post-replicative DNA modification. Previously, other examples of DNA MARylation have been demonstrated for different families of toxins: (i) guanine MARylation by pierisin 1 and CARP-1 from cabbage butterfly and shellfish, respectively (50); and (ii) thymine MARylation by DarT from bacterial toxin–antitoxin system DarTG (51). PARP2-catalyzed and PARP3-catalyzed modification of the terminal phosphate residues was confirmed by (i) their protection from CIP dephosphorylation after ADP-ribosylation (Figure 1D); (ii) resistance of MAR–DNA adducts at the tP residue to PARG digestion (Figure 5B); (iii) increased liability of MAR–DNA adducts at low pH (Supplementary Figure S4). Of note, acid-labile ADP-ribose modification of phosphoserine residues in histones from rat liver was observed 40 years ago (45), but the enzymes responsible for their formation are still unknown. Recently, serine in histones is proposed to be one of the major ADP-ribose acceptor sites for PARP signaling during the DNA damage response (52). We propose that enzymes PARP1–3 are involved in ADP-ribosylation of the phosphate groups present in DNA strand breaks and in proteins.
In the present study, we further substantiate our previous observations on PARylated DNA and demonstrate that PARP3-catalyzed DNA MARylation can be completely reversed by PARG (Figure 1D). While we revised the manuscript, Ahel’s laboratory in an independent study also showed that human PARP3 catalyzes MARylation of DNA oligonucleotide duplexes in vitro (53). In agreement with our data, they demonstrated that the mono-ADP-ribose moiety that is covalently attached to oligonucleotide termini can be removed by PARG. Of note, other cellular hydrolases such as MACROD2, TARG1 and ARH3 can remove the MAR adduct from DNA but with much lower efficacy as compared to PARG (53). In contrast to DNA, PARG does not completely remove ADP-ribose moieties in proteins and leaves the MAR adduct attached to an amino acid. This reversibility of DNA ADP-ribosylation suggests that in vivo PARP-mediated DNA strand break termini modifications serve rather as transient marks as compared to that of proteins. Nevertheless, we believe that the half-life of DNA–PAR or DNA–MAR adducts should be similar to that of PARylated proteins and may depend on the recruitment of PARG to the sites of ADP-ribosylation. In the absence of PARG being a major glycohydrolase in the nucleus, ADP-ribosylated DNA strand break termini may lead to persistent DNA damage and thus would be highly genotoxic as compared to PARylated PARPs and histones; indeed, a gene knockout of PARG causes early embryonic mortality (54). Therefore, combining DNA damage with the inhibition of PARG activity may generate cytotoxic DNA–PAR/MAR adducts and this event in turn could be exploited for the development of new powerful anticancer treatments.
Characterization of DNA substrate specificity of structurally similar enzymes PARP3 and PARP2 revealed that specificity of PARP3-catalyzed DNA MARylation is almost identical to that of PARP2. PARP3 and PARP2 efficiently ADP-ribosylate 5′-phosphorylated blunt, 5′- or 3′-overhang DSB termini of DNA oligonucleotide duplexes containing 5′-phosphorylated nicks, 1-nt gaps, mismatches or flaps (Figure 2). Contrary to PARP3, and in line with our previous results, PARP2 shows effective DNA ADP-ribosylation of 5′P DSB termini of recessed DNA substrates with a short (10 bp) double-stranded part, suggesting that PARP2 can effectively PARylate DNA in a 5′-phosphorylated nick- or gap-independent manner. The observed differences between the PARPs in DNA substrate specificity may be due to significant dissimilarities in the sequences and lengths of PARP3 and PARP2 NTRs, which are important for their affinity and specificity for an SSB in DNA (24,41). It was shown that PARP3 and PARP2 are selectively activated by 5′- but not 3′-phosphorylated DNA strand breaks (24). In line with these observations, PARP2 and PARP3 activation and DNA ADP-ribosylation activities are inhibited by 3′-terminal phosphate in the 5′-phosphorylated nicks or 1-nt gaps (Supplementary Figures S5B and S6). These results point to the involvement of the 3′OH in addition to the 5′P group in nick-dependent activation of PARPs, further supporting possible participation of these PARPs in the DNA ligation step. The types of DNA lesions used in this study can be generated either by direct action of reactive oxygen species, during DNA replication, or by different DNA repair pathways when they act on DNA damage. For example, nicks, gaps and flaps mimic intermediate products of the BER, NER and other DNA excision repair pathways. DNA duplexes containing DSB and proximal SSB can form in HR and NHEJ repair pathways. It has been reported that the MRN–CtIP complex generates an internal nick located ∼20 nt downstream of 5′-termini of a DSB (55). Moreover, PARP2- or PARP3-mediated DNA ADP-ribosylation is not limited to short oligonucleotide duplexes but is also efficient on high-molecular-weight DNA molecules (Figure 3). We demonstrated PARP2- and PARP3-catalyzed DNA ADP-ribosylation activities not only at DSB sites but also at the site of a 1 nt gap in plasmid DNA substrates containing downstream 5′-phosphorylated nicks nearby. These data clearly reveal the ability of enzymes PARP2 and PARP3 to ADP-ribosylate DNA containing multiple neighbouring DNA damage sites that could affect both DSB and SSB DNA repair pathways.
Previously, we have proposed that binding of a PARP to one DNA breakage site will activate the CAT domain which in turn could target and ADP-ribosylate an acceptor group at the second breakage site of the same DNA molecule (25). Recently, solution structures of PARP2 complexes with nicked DNA revealed that the main interface interacting with the nick is in the middle of the protein between the WGR and the CAT domains (41), suggesting that the DNA strand break, which is bound to the DNA-binding site of PARP2, is not accessible for the CAT domain of the same PARP2 molecule. The 3D structure of full-length PARP1 bound to dumbbell DNA with an SSB (39,46) also supports this notion. Our present study revealed that PARP2 and PARP3 (separately) modify DNA in a nick/gap-oriented and distance-dependent manner (Figure 4). We demonstrated that PARP3- and PARP2-catalyzed DNA ADP-ribosylation necessitates the presence of at least two DNA strand breaks either in the same strand (if these breaks are separated by a distance of one or two turns of the DNA helix) or in opposite strands, in which case, the breaks should be separated by a distance of 1.5 helix turns. These results are in line with the notion that both the activation of a PARP and the accessibility of the DNA acceptor groups for the activated CAT domain of a DNA-bound PARP are necessary for effective DNA ADP-ribosylation. Accordingly, for effective DNA ADP-ribosylation, PARPs should have high affinity for one of the strand breaks: for example, PARP1 binds to DNA in the following order of preference: blunt DSB end ≥ nicks > overhangs, whereas PARP2 and -3 have the preference order as follows: nicks > overhangs > blunt DSB end (23,56–58). Nevertheless, the DNA acceptor site should be free from the bound protein to be accessible for modification by the CAT domain of PARP proteins. In agreement with this model, PARP1 modifies protruding ends of DNA with greater efficacy as compared to nicks and blunt DSB ends. In contrast, PARP2 and PARP3 effectively modify both protruding DNA ends and blunt DSB ends and do not modify the single nicks. Overall, these data suggest that the DNA ADP-ribosylation activity strongly depends on the type of DNA damage and DNA repair intermediates that are generated during processing of the initial DNA damage by cellular enzymes. It should be noted that PARPs may also be activated on DNA hairpins, cruciforms, stably unpaired regions and other non-B-conformations of DNA (59). Thus, the presence of closely spaced clustered DNA strand breaks or breaks in proximity to some non-B DNA structures (and in particular configurations on a chromosome) may trigger DNA ADP-ribosylation by a PARP enzyme. Nonetheless, single, non-phosphorylated or distant DNA strand breaks will instead result in protein-targeted ADP-ribosylation only.
On DNA substrates prone to ADP-ribosylation and at a limiting concentration of NAD+ [which leads to mono- and oligo(ADP-ribosylation)], PARP3- or PARP2- but not PARP1-mediated DNA ADP-ribosylation turned out to be more efficient as compared to their simultaneous auto-ADP-ribosylation (Figure 5 and Supplementary Figure S13). These results suggest that termini of certain DNA–PARP3 and DNA–PARP2 complexes may be even more accessible for modification than their own acceptor amino acid residues. Nevertheless, our data revealed that PARP1 is the major enzyme responsible for the DNA PARylation activity in HeLa cell-free extracts, when we use the model Dbait molecules mimicking a DSB and SSB in DNA (Figure 6). This PARP1 dependence is not only due to the abundance of the latter but may also be due to the presence of other factors (in the nucleus of human cells) that can modulate PARP1 specificity for acceptors. For example, it has been demonstrated that histone PARylation factor 1 (HPF1) forms a robust protein complex with PARP1, thus limiting its auto-modification and promoting PARP1-dependent ADP-ribosylation of histones (52). DNA ADP-ribosylation activities that we observed using pure enzymes or cell-free extracts are indicative of the PARP-catalyzed ADP-ribosylation of DNA break termini in the cellular response to DNA damage. This notion is supported by the immunoblotting results that revealed the presence of a PAR signal in gDNA samples from bleomycin-treated PARG-depleted HeLa cells, and by its relative stabilization as compared to that in the mixture of control gDNA with a free PAR polymer (Figure 7). Evidently, new more sophisticated tools are needed in future studies to reliably distinguish ADP-ribosylated proteins and ADP-ribosylated DNA products in live cells and to assess the physiological relevance of PARP-dependent DNA ADP-ribosylation.
Finally, it is tempting to speculate that covalent modification of DNA strand break termini may enable creation of bulky and nucleotide size marks directly at DNA damage sites for (i) precise recruitment of the specific factors and for coordination of DNA repair pathways on DNA strand breaks; (ii) temporary blockade of processing of DNA strand break termini to protect them from non-specific degradation or aberrant error-prone end joining; iii) an apoptotic signal if not removed. We propose that the possibility of PARP-mediated DNA ADP-ribosylation during the DNA damage response should be taken into account for better understanding of the respective biological roles of DNA-dependent PARPs.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Murat Saparbaev for critical reading of the manuscript and thoughtful discussions. We are grateful to Valerie Schreiber and Françoise Dantzer for the vectors encoding proteins PARP2 and PARP3.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondation ARC (http://www.arc-cancer.net) [PJA20151203415 to A.A.I.]; ERA.Net RUS Plus (www.eranet-rus.eu) [DNA PARYLATION #306 to A.A.I., RFBR-16–54-76010 to O.I.L.]; Ministry of Education and Science of the Republic of Kazakhstan [programs 0115RK02473 and 0115RK03029 to B.T.M.]; NU ORAU (http://www.nu.edu.kz) (to B.T.M.); RSF Grant [14–24-00038 to O.I.L.]; French National Research Agency ‘Labex program’ [ARCANE project ANR-11-LABX-0003–01 to C.S.-P., D.G.]; Fondation ARC Postdoctoral Fellowship (http://www.arc-cancer.net) [PDF20110603195 to I.T.]; CIENCIACTIVA/CONCYTEC Doctoral Fellowship (www.cienciactiva.gob.pe) (to G.Z.). Funding for open access charge: National Laboratory Astana, Nazarbayev University, Astana, Republic of Kazakhstan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schreiber V., Dantzer F., Ame J.C., de Murcia G.. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006; 7:517–528. [DOI] [PubMed] [Google Scholar]
- 2. Kim M.Y., Zhang T., Kraus W.L.. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005; 19:1951–1967. [DOI] [PubMed] [Google Scholar]
- 3. Hottiger M.O., Hassa P.O., Luscher B., Schuler H., Koch-Nolte F.. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010; 35:208–219. [DOI] [PubMed] [Google Scholar]
- 4. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 5. Hottiger M.O. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 2015; 84:227–263. [DOI] [PubMed] [Google Scholar]
- 6. Posavec Marjanovic M., Crawford K., Ahel I.. PARP, transcription and chromatin modeling. Semin. Cell Dev. Biol. 2016; 63:102–113. [DOI] [PubMed] [Google Scholar]
- 7. Wei H., Yu X.. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics. 2016; 14:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin-Hernandez K., Rodriguez-Vargas J.M., Schreiber V., Dantzer F.. Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin. Cell Dev. Biol. 2016; 63:92–101. [DOI] [PubMed] [Google Scholar]
- 9. Pascal J.M., Ellenberger T.. The rise and fall of poly(ADP-ribose): an enzymatic perspective. DNA Repair. 2015; 32:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mortusewicz O., Ame J.C., Schreiber V., Leonhardt H.. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007; 35:7665–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F. et al. . Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J., Poirier G.G.. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008; 283:1197–1208. [DOI] [PubMed] [Google Scholar]
- 13. Shieh W.M., Ame J.C., Wilson M.V., Wang Z.Q., Koh D.W., Jacobson M.K., Jacobson E.L.. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998; 273:30069–30072. [DOI] [PubMed] [Google Scholar]
- 14. Ame J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Hoger T., Menissier-de Murcia J., de Murcia G.. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999; 274:17860–17868. [DOI] [PubMed] [Google Scholar]
- 15. Huber A., Bai P., de Murcia J.M., de Murcia G.. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair. 2004; 3:1103–1108. [DOI] [PubMed] [Google Scholar]
- 16. de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M. et al. . Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schreiber V., Ame J.C., Dolle P., Schultz I., Rinaldi B., Fraulob V., Menissier-de Murcia J., de Murcia G.. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002; 277:23028–23036. [DOI] [PubMed] [Google Scholar]
- 18. Menissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Ame J.C., Dierich A., LeMeur M. et al. . Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003; 22:2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupte R., Liu Z., Kraus W.L.. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017; 31:101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T.. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009; 28:2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanzlikova H., Gittens W., Krejcikova K., Zeng Z., Caldecott K.W.. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017; 45:2546–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yelamos J., Schreiber V., Dantzer F.. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol. Med. 2008; 14:169–178. [DOI] [PubMed] [Google Scholar]
- 23. Kutuzov M.M., Khodyreva S.N., Ame J.C., Ilina E.S., Sukhanova M.V., Schreiber V., Lavrik O.I.. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie. 2013; 95:1208–1215. [DOI] [PubMed] [Google Scholar]
- 24. Langelier M.F., Riccio A.A., Pascal J.M.. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014; 42:7762–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Talhaoui I., Lebedeva N.A., Zarkovic G., Saint-Pierre C., Kutuzov M.M., Sukhanova M.V., Matkarimov B.T., Gasparutto D., Saparbaev M.K., Lavrik O.I. et al. . Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 2016; 44:9279–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H., Schwede F., Yu Y., Kraus W.L.. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016; 353:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L.. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013; 52:272–285. [DOI] [PubMed] [Google Scholar]
- 28. Daniels C.M., Ong S.E., Leung A.K.. The promise of proteomics for the study of ADP-ribosylation. Mol. Cell. 2015; 58:911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghosh R., Roy S., Kamyab J., Dantzer F., Franco S.. Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways. DNA Repair. 2016; 45:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loseva O., Jemth A.S., Bryant H.E., Schuler H., Lehtio L., Karlberg T., Helleday T.. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010; 285:8054–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grundy G.J., Polo L.M., Zeng Z., Rulten S.L., Hoch N.C., Paomephan P., Xu Y., Sweet S.M., Thorne A.W., Oliver A.W. et al. . PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B(Glu2). Nat. Commun. 2016; 7:12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beck C., Boehler C., Guirouilh Barbat J., Bonnet M.E., Illuzzi G., Ronde P., Gauthier L.R., Magroun N., Rajendran A., Lopez B.S. et al. . PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 2014; 42:5616–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rouleau M., McDonald D., Gagne P., Ouellet M.E., Droit A., Hunter J.M., Dutertre S., Prigent C., Hendzel M.J., Poirier G.G.. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 2007; 100:385–401. [DOI] [PubMed] [Google Scholar]
- 34. Rulten S.L., Fisher A.E., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W.. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011; 41:33–45. [DOI] [PubMed] [Google Scholar]
- 35. Rouleau M., El-Alfy M., Levesque M.H., Poirier G.G.. Assessment of PARP-3 distribution in tissues of cynomolgous monkeys. J. Histochem. Cytochem. 2009; 57:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Wang J., Ding M., Yu Y.. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013; 10:981–984. [DOI] [PubMed] [Google Scholar]
- 37. Leutert M., Pedrioli D.M., Hottiger M.O.. Identification of PARP-specific ADP-ribosylation targets reveals a regulatory function for ADP-ribosylation in transcription elongation. Mol. Cell. 2016; 63:181–183. [DOI] [PubMed] [Google Scholar]
- 38. Oliver A.W., Ame J.C., Roe S.M., Good V., de Murcia G., Pearl L.H.. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004; 32:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dawicki-McKenna J.M., Langelier M.F., DeNizio J.E., Riccio A.A., Cao C.D., Karch K.R., McCauley M., Steffen J.D., Black B.E., Pascal J.M.. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell. 2015; 60:755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Obaji E., Haikarainen T., Lehtio L.. Characterization of the DNA dependent activation of human ARTD2/PARP2. Sci. Rep. 2016; 6:34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weeks S.D., Drinker M., Loll P.J.. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr. Purif. 2007; 53:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bryksin A., Matsumura I.. Overlap extension PCR cloning. Methods Mol. Biol. 2013; 1073:31–42. [DOI] [PubMed] [Google Scholar]
- 44. Ame J.C., Fouquerel E., Gauthier L.R., Biard D., Boussin F.D., Dantzer F., de Murcia G., Schreiber V.. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 2009; 122:1990–2002. [DOI] [PubMed] [Google Scholar]
- 45. Ord M.G., Stocken L.A.. Adenosine diphosphate ribosylated histones. Biochem. J. 1977; 161:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D.. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell. 2015; 60:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Croset A., Cordelieres F.P., Berthault N., Buhler C., Sun J.S., Quanz M., Dutreix M.. Inhibition of DNA damage repair by artificial activation of PARP with siDNA. Nucleic Acids Res. 2013; 41:7344–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burzio L.O., Riquelme P.T., Koide S.S.. ADP ribosylation of rat liver nucleosomal core histones. J. Biol. Chem. 1979; 254:3029–3037. [PubMed] [Google Scholar]
- 49. Gagne J.P., Ethier C., Defoy D., Bourassa S., Langelier M.F., Riccio A.A., Pascal J.M., Moon K.M., Foster L.J., Ning Z. et al. . Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair. 2015; 30:68–79. [DOI] [PubMed] [Google Scholar]
- 50. Nakano T., Takahashi-Nakaguchi A., Yamamoto M., Watanabe M.. Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. Curr. Top. Microbiol. Immunol. 2015; 384:127–149. [DOI] [PubMed] [Google Scholar]
- 51. Jankevicius G., Ariza A., Ahel M., Ahel I.. The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol. Cell. 2016; 64:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bonfiglio J.J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., Matic I.. Serine ADP-ribosylation depends on HPF1. Mol. Cell. 2017; 65:932–940.e936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Munnur D., Ahel I.. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017; 284:4002–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koh D.W., Lawler A.M., Poitras M.F., Sasaki M., Wattler S., Nehls M.C., Stoger T., Poirier G.G., Dawson V.L., Dawson T.M.. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17699–17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 56. D'Silva I., Pelletier J.D., Lagueux J., D’Amours D., Chaudhry M.A., Weinfeld M., Lees-Miller S.P., Poirier G.G.. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim. Biophys. Acta. 1999; 1430:119–126. [DOI] [PubMed] [Google Scholar]
- 57. Sukhanova M.V., Abrakhi S., Joshi V., Pastre D., Kutuzov M.M., Anarbaev R.O., Curmi P.A., Hamon L., Lavrik O.I.. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2016; 44:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Riccio A.A., Cingolani G., Pascal J.M.. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 2016; 44:1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lonskaya I., Potaman V.N., Shlyakhtenko L.S., Oussatcheva E.A., Lyubchenko Y.L., Soldatenkov V.A.. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 2005; 280:17076–17083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.