Abstract
The exponentially increasing volumes of DNA sequence data highlight the need for new DNA cloning methods to explore the new information. Here, we describe ‘ExoCET’ (Exonuclease Combined with RecET recombination) to directly clone any chosen region from bacterial and mammalian genomes with nucleotide precision into operational plasmids. ExoCET combines in vitro exonuclease and annealing with the remarkable capacity of full length RecET homologous recombination (HR) to retrieve specified regions from genomic DNA preparations. Using T4 polymerase (T4pol) as the in vitro exonuclease for ExoCET, we directly cloned large regions (>50 kb) from bacterial and mammalian genomes, including DNA isolated from blood. Employing RecET HR or Cas9 cleavage in vitro, the directly cloned region can be chosen with nucleotide precision to position, for example, a gene into an expression vector without the need for further subcloning. In addition to its utility for bioprospecting in bacterial genomes, ExoCET presents straightforward access to mammalian genomes for various applications such as region-specific DNA sequencing that retains haplotype phasing, the rapid construction of optimal, haplotypic, isogenic targeting constructs or a new way to genotype that presents advantages over Southern blotting or polymerase chain reaction. The direct cloning capacities of ExoCET present new freedoms in recombinant DNA technology.
INTRODUCTION
Recombinant DNA cloning, mutagenesis and engineering are central to molecular biology and biotechnology. Although relatively short DNA segments can be obtained by polymerase chain reaction (PCR) or de novo synthesis, DNA cloning of regions longer than about 10 kb has traditionally relied on the construction of DNA libraries followed by screening protocols to find the desired cloned sequences. This classical approach to DNA cloning has been powerful but is laborious and the target DNA segment once obtained usually must be recloned, reduced or reassembled from the piece(s) identified by the library screen(s) for functional studies. Recently we established a new path for direct cloning from genomic DNA samples that bypasses DNA library construction, screening and subcloning (1). This direct DNA cloning breakthrough was based on the discovery that the full-length Rac prophage protein RecE, with its partner RecT, mediates highly efficient homologous recombination (HR) between two linear DNA substrates. To promote bioprospecting, we applied full length RecE/RecT to directly clone secondary metabolite gene clusters up to 50-kb long from prokaryotic genomes into expression vectors (1–6).
However the application of full length RecET direct cloning to mammalian genomes proved to be more challenging. Bacterial and mammalian genomes differ by approximately three orders of magnitude (∼5 × 106 versus ∼3 × 109 bp) and the efficiency of RecET direct cloning is obviously constrained by the chance that the linear cloning vector and the target genomic fragment simultaneously enter into an Escherichia coli host cell upon electroporation. The work described here began with the idea that direct cloning efficiencies could be improved by annealing the linear vector and target genomic fragment together in vitro prior to transformation into E. coli for HR in vivo by full length RecE/RecT. We first evaluated a variety of protocols and reagents for the in vitro assembly step. Several exonucleases were found to be suitable and we settled on the 3′ exonuclease activity of T4 polymerase (T4pol) as the best for direct cloning from genomic DNA preparations. Having established an efficient T4pol protocol, we explored mechanistic aspects of the in vitro annealing and in vivo HR combination before pursuing various challenging applications including direct cloning from mammalian genomes.
MATERIALS AND METHODS
Bacteria strains and pSC101 expression plasmids
Escherichia coli GB2005 was derived from DH10B by deleting fhuA, ybcC and recET (7). GB05-dir was derived from GB2005 by integrating the PBAD-ETgA operon (full length recE, recT, redγ and recA under the arabinose-inducible PBAD promoter) at the ybcC locus (1). GB08-red was derived from GB2005 by integrating the PBAD-gbaA operon (redγ, redβ, redα and recA under the arabinose-inducible PBAD promoter) at the ybcC locus (8).
pSC101-BAD-ETgA-tet (1) conveys tetracycline resistance and carries the PBAD- full length ETgA operon and a temperature sensitive pSC101 replication origin which replicates at 30°C but not at 37°C so it can be easily eliminated from the host by temperature shift in the absence of selection (9).
Genomic DNA isolation and digestion
Gram-negative Photobacterium phosphoreum ANT-2200 and Photorhabdus luminescens DSM15139 were cultured overnight in 50 ml medium. After centrifugation, the cells were resuspended thoroughly in 8 ml of 10 mM Tris–Cl (pH 8.0). Five hundred microliters of 20 mg ml−1 proteinase K and 1 ml of 10% sodium dodecyl sulphate (SDS) were added and incubated at 50°C for 2 h until the solution became clear. Genomic DNA was recovered from the lysate by phenol-chloroform-isoamyl alcohol (25:24:1, pH 8.0) extraction and ethanol precipitation. The DNA was dissolved in 10 mM Tris–Cl (pH 8.0) and digested with BamHI + KpnI for cloning of the 14-kb lux gene cluster.
Gram-positive Streptomyces albus DSM41398 was cultured in 50 ml of tryptic soy broth at 30°C for 2 days. The genomic DNA was isolated according to the method described in ref. (10) with slight modification. After centrifugation, the cells were resuspended thoroughly in 8 ml of SET buffer (75 mM NaCl, 25 mM ethylenediaminetetraacetic acid (EDTA), 20 mM Tris, pH 8.0) and 10 mg lysozyme was added. After incubation at 37°C for 1 h, 500 μl of 20 mg ml−1 proteinase K and 1 ml of 10% SDS were added and incubated at 50°C for 2 h until the solution became clear. Three and a half milliliters of 5 M NaCl was added into the lysate. Genomic DNA was recovered from the lysate by phenol-chloroform-isoamyl alcohol (25:24:1, pH 8.0) extraction and ethanol precipitation. The DNA was dissolved in 10 mM Tris–Cl (pH 8.0).
Genomic DNA was purified from mouse melanoma B16 cells, human embryonic kidney 293T cells and human blood using Qiagen Blood & Cell Culture DNA Kits according to the manufacturer’s instructions, except DNA was recovered from the Proteinase K treated lysate by phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) extraction and ethanol precipitation. The DNA was dissolved in 10 mM Tris–Cl (pH 8.0). Restriction digested genomic DNA was extracted with phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) and precipitated with ethanol. The DNA was dissolved in 10 mM Tris–Cl (pH 8.0). End cut pipette tips were used to avoid shearing genomic DNA.
The genomic DNA of P. luminescens DSM15139 was digested with XbaI for plu3535-plu3532 cloning, and XbaI + XmaI for plu2670 cloning. The genomic DNA of S. albus was digested with EcoRV or Cas9–gRNA complexes for cloning of the salinomycin gene cluster. The mouse genomic DNA was digested with HpaI for Prkar1a cloning, BamHI + KpnI for Dpy30 cloning, and SwaI for Wnt4 or Lmbr1l-Tuba1a cloning. The human genomic DNA was digested with SpeI for DPY30 cloning, NdeI + BstZ17I for IGFLR1-LIN37 cloning, BstZ17I for IGFLR1-ARHGAP33 cloning and NdeI for ZBTB32-LIN37 cloning. Digested genomic DNA was extracted with phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) and precipitated with ethanol. The DNA was dissolved in ddH2O and concentrated to 1 μg μl−1. End cut pipette tips were used to avoid shearing genomic DNA. Ten micrograms of digested genomic DNA was used for ExoCET cloning.
Cas9 digestion of S. albus genomic DNA
The S. pyogenes Cas9 protein was provided by David N. Drechsel (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). To obtain the best Cas9 in vitro cleavage efficiency, four CRISPR guide sequences were selected for both regions flanking the salinomycin gene cluster (Figure 3A). The eight CRISPR guide sequences were incorporated into the oligos used to amplify the CRISPR minigenes (Supplementary Table S1). The CRISPR minigenes were amplified with PCR using BstZ17I-linearized pBR322-U6-ccdB-cm-tracrRNA (11) as a template and the proof reading PrimeSTAR Max DNA polymerase (Takara). The eight CRISPR minigenes were extracted from agarose gels after electrophoresis and purified using the QIAquick gel extraction kit (Qiagen). The HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, cat. no. # E2040S) was used for in vitro transcription of CRISPR gRNAs and 500 ng minigenes were used as the template. gRNAs were purified with a RNA clean Kit (Tiangen, Beijing China; cat. no. DP412) and eluted into DNase/RNase-free ddH2O.
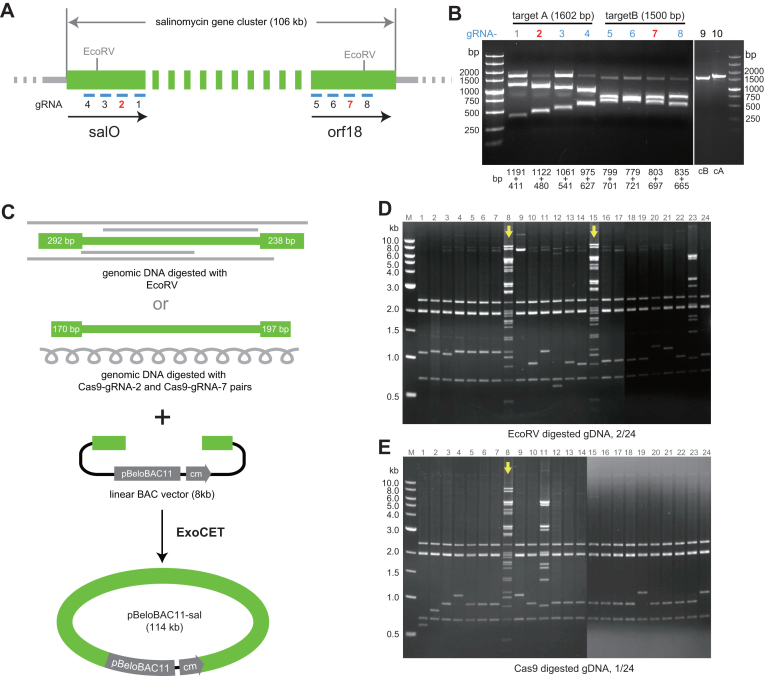
Figure 3.
Direct cloning of the 106-kb salinomycin gene cluster from EcoRV or Cas9 digested genomic DNA of Streptomyces albus. (A) Positions of EcoRV sites and eight Cas9 guide sequences on the salinomycin gene cluster. (B) In vitro cleavage with the eight Cas9–gRNAs to evaluate gRNA efficiency on PCR products amplified from the cleavage sites; gRNAs 2 and 7 were selected. cB (lane 9) and cA (lane 10) are negative controls with Cas9 and without gRNA. (C) The salinomycin gene cluster released from genomic DNA with EcoRV or Cas9–gRNA2/Cas9–gRNA7 was cloned into the pBeloBAC11 vector using ExoCET. Homology arms (blue) had been inserted into the BAC as previously described (6) and then cleaved with BamH1 to generate the illustrated direct cloning vector. The amount of sequence overlap between the ends of the genomic DNA and vector is indicated at the ends of the genomic DNA. (D and E) PvuII restriction analysis of the recombinant DNA obtained with ExoCET cloning. Correct clones are indicated with arrows.
The cleavage efficiency of Cas9–gRNA complexes were tested with the PCR amplified DNA fragment containing the Cas9 cutting sites (Figure 3B). The primers used to amplify the target DNA fragments, A (1602 bp) and B (1500 bp), are listed in Supplementary Table S1. The cleavage efficiency of Cas9–gRNAs was evaluated by digesting 100 ng of target DNA fragments in a 10-μl reaction containing 150 ng of Cas9, 400 ng of gRNA and 1 μl of Cas9 nuclease reaction buffer (NEB). After incubating at 37°C for 2 h, 4 μg RNase A (Thermo Scientific) was added and incubated at 37°C for 15 min. After addition of 1 μl stop solution (30% glycerol, 1.2% SDS, 250 mM EDTA pH 8.0) and incubation at 37°C for 15 min, an aliquot was analyzed on an agarose gel.
Cas9 digestion of S. albus genomic DNA was carried out in a 800 μl reaction containing 80 μl of 10 × Cas9 nuclease reaction buffer (NEB), 80 μg of genomic DNA, 40 μg of gRNA-2, 40 μg of gRNA-7 and 20 μg of Cas9. The genomic DNA used for Cas9 digestion was extracted three times with phenol–chloroform–isoamyl alcohol (25:24:1, pH8.0) and dissolved in DNase/RNase-free ddH2O. After incubating at 37°C for 6 h, 100 μg of RNase A (Thermo Scientific) was added and incubated at 37°C for 1 h. Then 100 μg of proteinase K (Roche) was added and incubated at 50°C for 1 h. Then, the digested genomic DNA was extracted with phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) and precipitated with ethanol. The genomic DNA was dissolved in ddH2O and concentrated to 1 μg μl−1. End cut pipette tips were used to avoid shearing genomic DNA. Ten micrograms of digested genomic DNA was used for ExoCET cloning.
Preparation of linear cloning vectors
The p15A-cm vector was amplified with PCR (Supplementary Figure S3A) using the PrimeSTAR Max DNA Polymerase (Takara) according to the manufacturer's instructions. Oligonucleotides containing the 80 nt homology arms and ∼20 nt standard PCR primers at the 3′ end (Supplementary Tables S2 and 3) were polyacrylamide gel electrophoresis (PAGE) purified. The PCR products were extracted from agarose gels after electrophoresis and purified using the QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions, except that DNA was eluted from the column with ddH2O and concentrated to 200 ng μl−1. Two hundred nanograms of p15A-cm vectors were used for ExoCET cloning unless otherwise stated.
The pBeloBAC11 vector used to clone the salinomycin gene cluster and the pBAC2015 vector used to clone plu3535-3532 were previously described (6,12). Bacterial artificial chromosome (BAC) vectors were linearized with BamHI to expose both homology arms, and extracted with phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0) and precipitated with isopropanol. The DNA was dissolved in ddH2O and concentrated to 1 μg μl−1. One microgram of linear BAC vectors were used for ExoCET cloning.
Preparation of the mVenus-PGK-neo cassette for recombineering
The mVenus-PGK-neo cassette was amplified from pR6K-2Ty1-2PreS-mVenus-Biotin-PGK-em7-neo (13) with PCR using the proof reading PrimeSTAR Max DNA Polymerase (Takara) according to the manufacturer’s instructions. The primers are listed in Supplementary Table S4. The PCR products were purified with QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions, except that DNA was eluted from the column with ddH2O and concentrated to 100 ng μl−1. Two hundred nanograms of the cassette was used for recombineering.
In vitro assembly
Ten micrograms of genomic DNA and 200 ng of 2.2-kb p15A-cm linear vector (1 μg of 8-kb linear BAC vector) were assembled in 20 μl reactions consisting of 2 μl of 10 × NEBuffer 2.1 and 0.13 μl of 3 U μl−1 T4pol (NEB, cat. no. M0203). Assembly reactions were prepared in 0.2 ml PCR tubes and cycled in a thermocycler as follows: 25°C for 1 h, 75°C for 20 min, 50°C for 30 min, then held at 4°C.
Assembly reactions with other exonucleases were cycled as follows: T5 exonuclease (T5exo; NEB, cat. no. M0363): 50°C for 30 min, then held at 4°C; T7 exonuclease (T7exo; NEB, cat. no. M0263): 25°C for 20 min, 50°C for 30 min, then held at 4°C; DNA polymerase I Klenow fragment (Kle; NEB, cat. no. M0210), T7 DNA polymerase (T7pol; NEB, cat. no. M0274) and λ exonuclease (λexo; NEB, cat. no. M0262): 25°C for 20 min, 75°C for 20 min, 50°C for 30 min, then held at 4°C; Exonuclease III (ExoIII; NEB, cat. no. M0206): 37°C for 20 min, 75°C for 20 min, 50°C for 30 min, then held at 4°C; Phusion DNA polymerase (Phu; NEB, cat. no. M0530): 37°C for 20 min, 50°C for 30 min, then held at 4°C.
Gibson assembly was performed at 50°C for 1 h with Gibson Assembly Master Mix (NEB, cat. no. E2611).
For reactions performed in triplicates, we used single genomic DNA preparations with same vector preparation and the same batch of electrocompetent cells. Three in vitro reactions, subsequent electroporations, plating and counting were performed in parallel. The in vitro assembly products were desalted at room temperature for 30 min by drop dialysis against ddH2O using Millipore Membrane Filters (Merck-Millipore, cat. no. VSWP01300), then 5 μl of each reaction product was electroporated into E. coli cells.
Electroporation
Forty microliters of E. coli overnight culture (OD600 = 3∼4) was inoculated into 1.4 ml of LB medium supplemented with appropriate antibiotics and incubated at 30°C for 2 h with shaking at 950 r.p.m. in an Eppendorf thermomixer (OD600 = 0.35∼0.4). Thirty five microliters of 10% L-arabinose (Sigma-Aldrich, cat. no. A3256; w/v, in ddH2O) was added to induce the expression of ETgA or gbaA. The cells were grown at 37°C for another 40 min (OD600 = 0.7∼0.8), then centrifuged at 9400 × g for 30 s, 2°C. The supernatant was discarded and the cell pellet was resuspended in 1 ml of ice-cold ddH2O. The cell suspension was centrifuged again to repeat the washing once more. Cells were resuspended in 20 μl of ice-cold ddH2O.
Five microliters of desalted in vitro assembly products were added into cell suspensions in ExoCET cloning experiments. In recombineering exercises, 200 ng plasmid and 200 ng PCR products were used. Electroporation was performed using chilled 1-mm cuvettes (Bio-Rad, cat. no. 1652089) and an Eppendorf electroporator 2510 at 1350 V, 10 μF, 600Ω. After electroporation, 1 ml of LB medium was added into the cuvette to suspend the cells and the cell suspension was transferred into a 1.5-ml Eppendorf tube with a puncture in its cap for aeration. The culture was incubated at 37°C for 1 h with shaking at 950 r.p.m. in an Eppendorf thermomixer. Appropriate volumes of the cell suspension were taken from the culture and spread on LB plates with antibiotics (chloramphenicol, 15 μg ml−1; kanamycin, 15 μg ml−1). The number of colonies was counted after overnight incubation and the colony number per ml (c.f.u./ml) was calculated.
RESULTS
Concerted action of in vitro assembly and full length RecE/RecT improves the efficiency of direct cloning
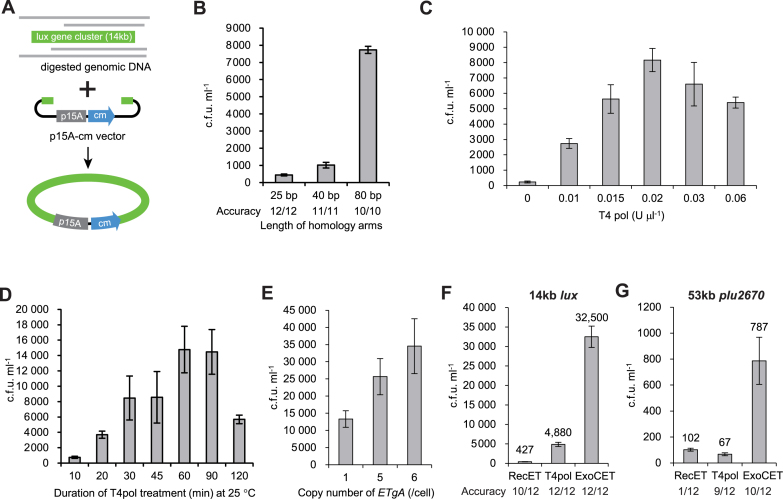
The efficiency of RecET direct cloning is limited by the need for the linear vector and the target genomic segment to simultaneously enter one E. coli cell before productive HR can take place. To associate the two DNA molecules in vitro before electroporation into E. coli, we evaluated a variety of exonucleases and annealing protocols using a model experiment based on direct cloning of a 14kb fragment encoding the lux gene cluster from the Gram-negative luminous piezophile marine bacterium P. phosphoreum ANT-2200 (14) (Figure 1A). The basic protocol involved mixing restriction digested genomic DNA (10 μg) with a linear direct cloning vector (2.2 kb p15A-cm; 200 ng) that was flanked by short sequence regions (homology arms) identical to the ends of the 14 kb lux restriction fragment. For reaction and process optimization, the cloning efficiency was compared using the total colony number on selection plates from 1 ml culture (c.f.u. ml−1) and the fidelity was assessed by restriction analysis (Figures 1 and 2). Various exonucleases were tested and several were satisfactory (Supplementary Figure S1) before we settled on the 3′ exonuclease activity of T4 polymerase (T4pol) because it consistently gave the highest efficiency and fidelity. For annealing after exonuclease digestion, results were largely indifferent to the cooling rate so we opted for the convenience of the default rate of the Eppendorf MC nexus gradient thermocycler (2°C s−1) (Supplementary Figure S2). We next evaluated the contribution of homology arm length to the outcome of ExoCET direct cloning. Not unexpectedly, the longest homology arm length tested (80 bp) was the most efficient (Figure 1B). Because synthesis of 100mer oligonucleotides is convenient and reliable, all further experiments employed 100mers (i.e. 5′ 80 nucleotide (nt) homology arm plus 3′ 20 nt PCR primer to the p15A plasmid; Supplementary Figure S3A). We also evaluated the impact of T4pol input (Figure 1C), time of exonuclease digestion (Figure 1D) and RecE/RecT expression level (Figure 1E) on ExoCET. For the latter experiment, we expressed full length RecE/RecT from either one integrated copy of the arabinose-inducible BADrecE/recT/redγ/recA (ETgA) operon in GB05-dir or a pSC101 plasmid borne version of the same operon (approximately five copies per cell) or both together (approximately six copies per cell). Expression from pSC101 delivered better direct cloning efficiency than from the chromosomal single copy in GB05-dir, which was further increased when both the plasmid and chromosomal operons were employed (Figure 1E), thereby indicating that the results were reflective of ETgA expression levels and also that the toxicity associated with overexpressed RecT had not been provoked. However, ETgA expression from a pR6K-pir plasmid (∼15 copies per cell) appeared to provoke toxicity (data not shown). Regarding the other genes in the ETgA operon: Redγ inhibits the major E. coli exonuclease RecBCD (15) so linear DNA molecules persist for much longer and hence their ability to promote recombination is greater; and RecA promotes transformation efficiency (7).
Figure 1.
Concerted action of in vitro assembly and full length RecE/RecT improves the efficiency of direct cloning. (A) A schematic diagram illustrating direct cloning of the 14-kb lux gene cluster from Photobacterium phosphoreum ANT-2200. The linear p15A-cm vector and target genomic segment have identical sequences at both ends. (B) Longer homology arms increase the cloning efficiency of ExoCET. The linear vector flanked by 25-, 40- or 80-bp homology arms was mixed with genomic DNA and treated with 0.02 U μl−1 T4pol at 25°C for 20 min before annealing and electroporation into arabinose induced Escherichia coli GB05-dir. Error bars, s.d.; n = 3. (C) Titration of T4pol amount for ExoCET. The linear vector with 80-bp homology arms and genomic DNA were treated as in (B) except the amount of T4pol was altered as indicated. (D) Incubation time of T4pol on cloning efficiency. As for (C) using 0.02 U μl−1 T4pol except the incubation time was altered as indicated. (E) Higher copy number of ETgA increases ExoCET cloning efficiency. As for (D) using 1 h and electroporation into arabinose induced E. coli GB05-dir (one copy of ETgA on the chromosome), GB2005 harboring pSC101-BAD-ETgA-tet (approximately five copies of ETgA on pSC101 plasmids) or GB05-dir harboring pSC101-BAD-ETgA-tet (approximately six copies of ETgA) as indicated. (F) ExoCET increases direct cloning efficiency. As for (E) using E. coli GB05-dir harboring pSC101-BAD-ETgA-tet (ExoCET) or omission of T4pol from the in vitro assembly (ETgA) or omission of the arabinose induction of pSC101-BAD-ETgA-tet (T4pol). (G) As for (F) except the 53 kb plu2670 gene cluster was directly cloned. Accuracy denotes the success of direct cloning as evaluated by restriction digestions (Supplementary Figure S4). Each experiment was performed in triplicate (n = 3) and error bars show standard deviation (s.d).
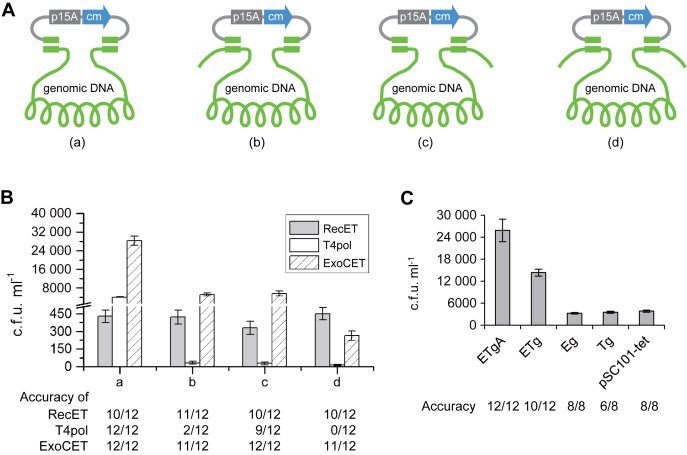
Figure 2.
ExoCET mechanism. (A) Juxtaposition of the 80-bp homology arms between the p15A-cm (chloramphenicol) vector and the 14-kb lux genomic segment is illustrated: (a) both homology arms were located at the termini; (b and c) one homology arm was located at a terminus and the other 1 kb from the other end; (d) both homology arms were 1 kb from each end. (B) Number of colonies obtained from ETgA, T4pol or ExoCET using the homology arm combinations (a–d) as indicated. Reaction conditions were the same as for Figure 1F. (C) Protein combinations as indicated expressed from pSC101 plasmids in GB2005 were tested for direct cloning of the 14-kb lux gene cluster using terminal homology arms and ExoCET conditions except for the omission of RecA (ETg); RecA and RecT (Eg), RecA and RecE (Tg) and all (pSC101-tet). Error bars, s.d.; n = 3. Corresponding DNA analyses are shown in Supplementary Figure S5.
Having established optimal reaction conditions (Supplementary Figure S3B), we compared direct cloning frequencies using full length RecE/RecT alone, the T4pol in vitro annealing protocol alone or both together (Figure 1F). As expected from our previous work, direct cloning of the 14kb lux gene cluster by RecE/RecT alone was successful at high fidelity (Supplementary Figure S4). However, T4pol in vitro assembly followed by electroporation into a standard E. coli recombinant host was much better (4880 versus 427). This success indicates (i) the endogenous E. coli machinery is highly adept at completing and sealing plasmid scaffolds; and (ii) our T4pol in vitro assembly protocol is efficient. However the ExoCET combination of T4pol in vitro assembly and full length RecE/RecT in vivo HR was significantly better than either process alone. To further validate our conclusions, we compared RecET, T4pol alone and ExoCET for direct cloning of a 53 kb gene cluster, plu2670. As expected for a much larger target region, overall yield was reduced. However ExoCET was clearly superior (Figure 1G).
Mechanistic aspects of ExoCET
To analyze the synergism between combining in vitro annealing with full length RecET HR, we designed the experiment illustrated in Figure 2A. The experiments in Figure 1 employed a direct cloning vector with 80nt homology arms to the termini of the 14 kb lux gene cluster. Three variations were generated by moving either 80 nt homology arm, or both, to 80 nt regions located 1 kb from the termini. ExoCET efficiencies were again compared to T4pol alone or RecET alone (Figure 2B). The highest ExoCET and T4pol alone efficiencies were achieved when both homology arms were terminal. When one homology arm (or both) was internal, T4pol alone was ineffective indicating that direct cloning using annealing after T4pol exonuclease action depends on terminal complementarities and that annealing at only one end is insufficient to promote direct cloning without subsequent RecET HR. In contrast, RecET HR efficiently utilized the internal homology arms and did not require terminal homologies. Together, the combination of T4pol annealing in vitro at one end with RecET HR at the other promoted direct cloning yields about 12× over RecET alone. These data indicate that the major contribution of T4pol to ExoCET is due to the in vitro annealing of an end, which greatly increases the co-transformation efficiency. RecET HR then promotes recombination at the other end in vivo. Notably, when both homology arms were positioned at the very end of the target genomic fragment, ExoCET was about six to eight times more efficient than T4pol alone (Figures 1F and 2B). This indicates that most (>85%; Supplementary Note S1) of the in vitro assembly products with two terminal homology arms were associated at only one end.
Because RecT is a single strand annealing protein (SSAP), we also considered the possibility that RecT annealing of the single stranded regions exposed by T4pol could contribute to ExoCET. To test this possibility, T4pol annealed products were electroporated into E. coli cells induced for expression of RecT without RecE (from pSC101-Tg). No co-operation between T4pol and RecT was observed (Figure 2C). RecE/RecT is a 5′-3′ exonuclease/SSAP syn/exo pair (16) and a specific protein–protein interaction is required for HR using double stranded DNAs (17). No recombination was observed when RecT was omitted (pSC101-Eg). Therefore ExoCET requires both RecE and RecT. Omission of RecA had only a modest impact (Figure 2C), consistent with the previous finding that RecA increased transformation efficiency rather than recombination (7).
Validation of ExoCET and combination with CRISPR/Cas9 cleavage in vitro
To validate ExoCET, we applied it to several tasks that had previously proven challenging with RecET alone. First, direct cloning of the 38kb plu3535-3532 gene cluster from Photohabdus luminescens into an expression plasmid where previously we achieved efficiencies of 2/12 (1). With ExoCET we achieved 12/12 (Table 1). Similarly our previous attempts to directly clone the 106 kb salinomycin biosynthesis gene cluster from S. albus in one step had been fruitless and to succeed we had to break the task into three pieces (6). To apply ExoCET to this task, we introduced PCR generated homology arms from each end of the 106 kb EcoRV restriction fragment that encompasses the gene cluster into a BAC vector. After linearization of the BAC between the homology arms and mixing with EcoRV digested genomic DNA, 2/24 clones examined were correct (Figure 3 and Table 1). This exercise benefitted from a fortuitous disposition of EcoRV sites. Potentially, the use of a programmable nuclease, specifically the RNA guided endonuclease, Cas9 (18,19), can obviate the reliance on similar good fortune for other exercises. To evaluate this idea, we repeated the 106kb salinomycin direct cloning exercise with the same BAC vector and genomic DNA digested in vitro using Cas9 programmed with guide RNAs to deliver cleavages very close to the EcoRV sites. A similar success was achieved (Figure 3 and Table 1). Compared to our previous experiences with RecET, ExoCET delivered significantly improved performance for direct cloning of large regions and this advantage can also be coupled with use of Cas9 as a programmable endonuclease. As with RecET alone, most of the wrong ExoCET products arose from self-circularization of the empty cloning vector. Microrepeats larger than 6 bp shared between the homology arms promote self-circularization and there were five pairs of 8∼10bp direct repeats between the two homology arms used to clone the 106-kb region. Potentially Cas9 cleavages can be chosen to minimize or avoid the presence of microrepeats and hence reduce unwanted background. For optimal efficiency, the homology arms should be adjacent to the restriction sites used to liberate the target genomic DNA fragment. Restriction analysis showed several clones that were neither empty vector, which is the usual source of unwanted product, nor the correct product (lane 9 and 23 in Figure 3D and lane 11 in Figure 3E). These clones contained some of the intended DNA region rearranged by intramolecular recombination among repetitive sequences in the salinomycin gene cluster to delete segments.
Table 1. Large genomic segments directly cloned from bacteria, mammalian cells and human blood with ExoCET.
| Target | Source | Genome (Mb) | Digestion enzymes | Fragment (kb) | Vector | c.f.u. (/ml) | Correct/checked |
|---|---|---|---|---|---|---|---|
| plu3535-3532 | P. luminescens DSM15139 | 5.69 | XbaI | 38 | pBAC2015 | 1815±132 | 12/12 |
| plu2670 | P. luminescens DSM15139 | 5.69 | XbaI+XmaI | 53 | p15A | 787±194 | 10/12 |
| salinomycin cluster | S. albus DSM41398 | 8.38 | EcoRV | 106 | pBeloBAC11 | 425±91 | 2/24 |
| salinomycin cluster | S. albus DSM41398 | 8.38 | Cas9 | 106 | pBeloBAC11 | 260±14 | 1/24 |
| Wnt4 | Mouse melanoma B16 cell | 2800.06 | SwaI | 45 | p15A | 76±16 | 8/25 |
| Lmbr1l-Tuba1a | Mouse melanoma B16 cell | 2800.06 | SwaI | 53 | p15A | 52±6 | 1/12 |
| Prkar1a | Mouse melanoma B16 cell | 2800.06 | HpaI | 8 | p15A | 205±17 | 10/12 |
| IGFLR1-ARHGAP33 | Human blood | 3221.49 | BstZ17I | 41 | p15A | 275±76 | 5/48 |
| ZBTB32-LIN37 | Human blood | 3221.49 | NdeI | 45 | p15A | 115±35 | 2/48 |
| Dpy30 | Mouse melanoma B16 cell | 2800.06 | BamHI+KpnI | 8.7 | p15A | 273±18 | 9/12 |
| DPY30 | HEK 293T cell | 3221.49 | SpeI | 9.1 | p15A | 40±10 | 17/24 |
| DPY30 | Human blood | 3221.49 | SpeI | 9.1 | p15A | 45±2 | 5/24 |
| Oct4-Venus | Mouse R1 ES cells | 2800.06 | EcoRV+PacI | 9.6 | p15A | 34±1 | 9/36 |
| Nanog-Cherry | Mouse R1 ES cells | 2800.06 | NdeI | 13 | p15A | 49±12 | 17/54 |
| Gata2-Venus | Mouse GM8 ES cells | 2800.06 | BstZ17l | 16.8 | p15A | 212±27 | 5/45 |
| Mll4 (1) | Mouse R1 ES cells | 2800.06 | SspI+SpeI | 17.1 | p15A | 127±38 | 7+3/24 |
| Mll4 (2) | 323±65 | 2+2/36 | |||||
| Mll4 (3) | 142±27 | 6+9/72 | |||||
| Mll4 (4) | 483±91 | 3+5/36 |
All experiments were done in triplicate; c.f.u. includes standard deviation and fidelity was monitored by restriction analysis of the indicated number of colonies. For the Mll4 experiments, fidelity shows the targeted allele + wt allele/colonies examined. DNA analyses are shown in Supplementary Figure S6.
Direct cloning from mammalian genomes using ExoCET
We next evaluated whether the added efficiencies of ExoCET would deliver the additional reach required for direct cloning from mammalian genomic DNA. A 45-kb restriction fragment containing the mouse Wnt4 gene was selected as a suitable challenge. Of 25 colonies checked, 8 were correct (Table 1). Illumina sequencing revealed the single-nucleotide polymorphism (SNP) haplotype linkage patterns of both alleles of this 45-kb region (Supplementary Table S5), thereby indicating that application of ExoCET to SNP analysis has the potential to bypass haplotype scrambling inherent in PCR-based approaches. Given the Wnt4 success, we explored the potential for SNP analysis by directly cloning another large segment (53 kb) including the Lmbr1l-Tuba1a genes followed by, using purified genomic DNA from human blood, a 41-kb region including the IGFLR1-ARHGAP33 genes. Next, a 45-kb region including the ZBTB32-LIN37 genes was directly cloned (Table 1). These results indicate that ExoCET offers an unrivalled capacity for haplotype phasing in SNP analyses.
ExoCET mammalian applications: haplotypic isogenic targeting constructs
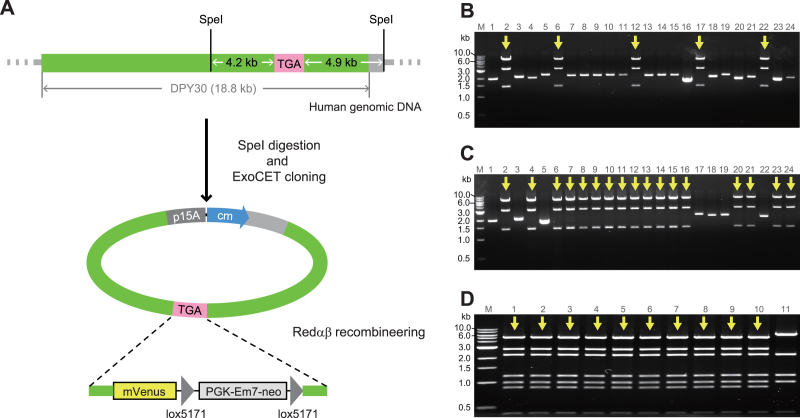
To exploit ExoCET access to mammalian genomes, we developed two further applications. First, the generation of isogenic targeting constructs for mammalian genome engineering. Drawing upon experience gained in mouse embryonic stem (ES) cells (20), we aimed to directly clone 8–10 kb sections to provide the optimal length for gene targeting employing isogenic homology arms. Notably with ExoCET, these sections are not only isogenic but also maintain haplotypic linkages of polymorphisms. Hence we call them ‘HIT’ (haplotypic isogenic targeting) constructs. Approximately 9-kb HIT segments for two genes, Dpy30/DPY30 and Prkar1a, were directly cloned from mouse or human genomic DNA, either isolated from a cultured cell line or from blood (Table 1). Thereafter they were modified by Red recombineering to insert a selectable gene cassette for targeting in mammalian cell lines (8) (Figure 4). The cassette insertion site was selected to destroy a Cas9 guide RNA recognition site so that the HIT construct can be used with Cas9-assisted targeting (11).
Figure 4.
Generation of HIT constructs from mammalian genomic DNA applied to DPY30. (A) Scheme illustrating the DPY30 stop codon region cloned from human genomic DNA after SpeI digestion. Once directly cloned, the C-terminus of DPY30 was tagged with a mVenus cassette using Redαβ recombineering and standard cassettes (8,13). (B) EcoRI restriction analysis of the recombinant clones obtained by ExoCET using genomic DNA isolated from human blood. (C) EcoRI restriction analysis of the recombinant clones obtained by ExoCET using genomic DNA isolated from human embryonic kidney 293T cells. (D) PvuII restriction analysis after mVenus cassette insertion using Redαβ recombineering. As expected, every clone was correct. Lane 11 is the original plasmid without mVenus cassette insertion. Correct clones are indicated with arrows.
ExoCET mammalian applications: an alternative to southern analysis with the option to sequence
In a second application, we aimed to develop an alternative to Southern blotting for genotyping. To pilot the exercise, we used three mouse ES cell lines carrying previously validated targeted genes, namely Oct4, Nanog and Gata2. All three targeted alleles included a neomycin resistance gene with a bacterial promoter that conveys kanamycin resistance in E. coli (8). After appropriate restriction digestion of genomic DNA preparations and ExoCET, chloramphenicol resistant candidate colonies were evaluated for co-resistance to kanamycin and by restriction digestion. The success rate, that is the number of correctly retrieved targeted alleles compared to the total number of chloramphenicol resistant colonies, ranged from 11 to 31% (Table 1). We then applied ExoCET to genotyping candidate ES cell clones from a Cas9 assisted RAC tag targeting exercise (11) to insert a tag at the N-terminus of Mll4. Of four successfully targeted ES clones, both targeted and untargeted Mll4 alleles were retrieved by ExoCET at about the same frequency (Table 1) and the remaining ExoCET candidates contained empty vectors and so were readily discounted (Supplementary Figure S6). Except for Gata2 when 1.5 μg genomic DNA was used, the above exercises all used 10 μg. ExoCET was also successful using 0.5 and 1.0 μg genomic DNA inputs (Supplementary Table S6) indicating that higher throughput applications can be developed.
ExoCET metagenomic applications
Environmental samples usually contain more than 104 species (21–23). To test if ExoCET can retrieve a gene cluster from metagenomic samples, we diluted 10, 5, 2 and 1 ng of P. phosphoreum genomic DNA into 10 μg of Bacillus subtilis genomic DNA. We managed to directly clone the 14 kb lux gene cluster with a reasonable efficiency even from the 10−4 diluted genomic samples (Table 2).
Table 2. Direct cloning of the 14 kb lux gene cluster from diluted P. phosphoreum genomic DNA with ExoCET.
| P. phosphoreum (ng) (BamHI + KpnI) | B. subtilis (μg) (BamHI) | Vector | c.f.u. (/ml) | Correct/checked |
|---|---|---|---|---|
| 10 | 10 | p15A-cm | 200 ± 2 | 7/12 |
| 5 | 10 | p15A-cm | 142 ± 22 | 5/12 |
| 2 | 10 | p15A-cm | 102 ± 8 | 2/12 |
| 1 | 10 | p15A-cm | 104 ± 18 | 2/24 |
DISCUSSION
Functional analysis of the exponentially increasing volumes of genome sequencing data will benefit from faster and simpler ways to build expression constructs and targeting vectors. Here we extend the reach of direct DNA cloning by adding exonuclease digestion and annealing in vitro to full length RecE/RecT recombination in E. coli. Now regions up to 50kb and beyond can be readily retrieved from genomes with complexities up to at least 3.0 × 109 bp. Furthermore, the ability to retrieve DNA segments from a metagenomic mimic DNA preparation (Table 2) points towards the application of ExoCET for direct cloning in metagenomics.
The promise that direct cloning can bypass library construction and screening has driven various exercises in the past, including our recent description of the properties of full length RecE/RecT. Larionov et al. developed the yeast-based transformation-associated recombination (TAR) cloning approach (24). Ongoing improvements to TAR include a recent combination with Cas9 cleavage. Although only one example was shown, the combination with Cas9 enhances the possibility that TAR will be less reliant on skills and could become more widely applicable (25,26). Recently a method called ‘CATCH’ (Cas9-Assisted Targeting of CHromosome Segments) used in vitro Cas9 cleavage and Gibson assembly (27) to clone large regions into BACs (28). This method, which has only been applied to prokaryotic genomes, appears to rely on a substantial primary PCR screen to identify candidate colonies for closer examination and is considerably more work than ExoCET. We also tested whether Gibson assembly would be an effective alternative first step for ExoCET using direct cloning of the mouse Wnt4 as the assay and found that Gibson assembly promoted a huge amount of empty vector background with or without RecET (Supplementary Table S7). Several other direct cloning applications have been described (Supplementary Table S8) but they also appear to be inefficient or restricted to specialist applications and none have been widely employed. Consequently, there is a need for a direct cloning method that is easy to use and broadly applicable across a wide range of fragment sizes and genome complexities. By improving upon RecET direct cloning, we suggest that ExoCET will fulfill this need because it is a simple and efficient method generically applicable to a broad range of direct cloning challenges from size (up to 106 kb so far) and genome complexities (to at least 3 × 109 bp). Furthermore, ExoCET presents advantages over PCR for amplification of DNA because it has a much higher fidelity, is not limited in size and does not scramble haplotypes.
Homology arm position plays an important role in the design of ExoCET direct cloning exercises. To obtain the highest efficiency, both homology arms should be positioned at the very end of the target fragment. Indeed, to benefit from the in vitro T4pol exonuclease and subsequent annealing step, one homology arm must be terminal. However, the other homology arm can be internal because RecE/RecT can locate it for HR. This information can be employed when using direct cloning to establish an expression construct. One homology arm can be directed to the very 3′ end of the target fragment whereas the 5′ end of the target gene can be precisely positioned under a promoter and ribosome recognition sequence by the choice of an internal homology arm. Alternatively, in the absence of usefully positioned restriction sites, it is possible to use Cas9 as an in vitro endonuclease to optimize by design the release of the target fragment for direct cloning by ExoCET into an expression construct.
Among other applications, direct cloning from mammalian genomes, including DNA isolated from blood or patient-specific cell lines, will facilitate haplotype phasing of SNPs (29) as well as rapid cloning of HIT constructs for nuclease assisted targeting in human stem cells. The increasing relevance of human stem cells for biomedical research, isolated either from patients, cord blood or by somatic cell reprogramming has focused attention on methods to accurately engineer stem cell genomes. Engineering human genomes presents greater challenges than engineering laboratory mouse genomes because humans are genetically diverse. For HR, isogenicity, that is sequence identity, is obviously a critical issue as recognized many years ago for gene targeting in mouse ES cells (30). However, the impact of sequence mismatches in regions involved in HR is still not well understood. How much does a single mismatch (e.g. an SNP or an indel) debilitate HR? And how does the position of the mismatch close to or far from the intended recombination site affect efficiency? How do multiple mismatches affect HR efficiencies? These questions, and more, remain largely unanswered. Nevertheless it is clearly advisable to use exactly identical sequences for gene targeting. ExoCET presents a rapid way to obtain the ideal targeting construct both in terms of isogenicity and the optimal lengths of homology arms. As opposed to the application of PCR to amplify homology arms from genomic DNA, ExoCET is not severely limited in size or prone to mutagenesis and furthermore will also maintain haplotypic linkages. The ends of the homology arms can be selected according to both targeting efficiency and the genotyping strategy, either by Southern blotting, junction PCR or now ExoCET. Hence ExoCET offers several advantages for personal genome surgery especially when combined with CRISPR/Cas9 (11).
ExoCET is also potentially the most reliable method for genotyping. Whereas, both Southern blotting and junction PCR may produce false positive signals, this is highly unlikely with ExoCET. Furthermore Southern blotting in mammalian genomes is often complicated by the laborious search for a good hybridization probe and junction PCR is restricted to short amplification products that do not encompass high GC or secondary structural contents. ExoCET bypasses these problems.
Through the ability to selectively acquire large DNA segments from complex genomic preparations including blood, ExoCET also presents options for diagnostics and pathology tests such as directed sequence acquisition for personal medicine or the isolation of DNA viruses from patient materials. ExoCET will have broad applications in functional and comparative genomics, as well as bioprospecting with prokaryotic biosynthetic pathways.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David N. Drechsel (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) for providing the Cas9 protein, Longfei Wu (Laboratoire de Chimie Bactérienne, Aix-Marseille Université, Marseille, France) for providing Photobacterium phosphoreum ANT-2200, Junying Miao (College of Life Science, Shandong University, Jinan, People’s Republic of China) for providing the mouse melanoma B16 cell line and human embryonic kidney 293T cell line, Jinan Blood Center for providing human blood from multiple anonymous donors.
Author contributions: H.W., L.X., Y.Y., R.M., J.F., A.F.S. and Y.Z. designed the experiments. H.W., Z.L., R.J., J.Y. and A.L. performed the experiments. H.W., J.F., Y.Z. and A.F.S. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International S&T Cooperation Program of China [ISTCP 2015DFE32850 to J.F.]; National Natural Science Foundation of China [31670097 to Y.Z.]; Shandong Innovation and Transformation of Achievements Grant [2014ZZCX02601 to Y.Z.]; Major Project of Science and Technology of Shandong Province [2015ZDJS04001 to Y.Z.]; 111 Project [B16030 to Y.Z.]; Recruitment Program of Global Experts in Shandong University (to Y.Z.); China Postdoctoral Science Foundation [2015T80710 to H.W.]; Postdoctoral Innovation Program of Shandong Province [201303110 to H.W.]; Key Research and Development Program of Shandong Province [2015GSF12101 to A.L.]; TUD Elite University Support the Best program (to A.F.S.); Deutches Krebshilfe [110560 to A.F.S.]. Funding for open access charge: International S&T Cooperation Program of China [ISTCP 2015DFE32850].
Conflict of interest statement. R.M., A.F.S. and Y.Z. are shareholders in Gene Bridges GmbH, which holds exclusive commercial rights to recombineering.
REFERENCES
- 1. Fu J., Bian X., Hu S., Wang H., Huang F., Seibert P.M., Plaza A., Xia L., Müller R., Stewart A.F. et al. . Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 2012; 30:440–446. [DOI] [PubMed] [Google Scholar]
- 2. Bian X., Huang F., Stewart F.A., Xia L., Zhang Y., Müller R.. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. Chembiochem. 2012; 13:1946–1952. [DOI] [PubMed] [Google Scholar]
- 3. Bian X., Plaza A., Zhang Y., Müller R.. Luminmycins A-C, cryptic natural products from Photorhabdus luminescens identified by heterologous expression in Escherichia coli. J. Nat. Prod. 2012; 75:1652–1655. [DOI] [PubMed] [Google Scholar]
- 4. Bian X., Huang F., Wang H., Klefisch T., Müller R., Zhang Y.. Heterologous production of glidobactins/luminmycins in Escherichia coli Nissle containing the glidobactin biosynthetic gene cluster from Burkholderia DSM7029. Chembiochem. 2014; 15:2221–2224. [DOI] [PubMed] [Google Scholar]
- 5. Tang Y., Frewert S., Harmrolfs K., Herrmann J., Karmann L., Kazmaier U., Xia L., Zhang Y., Müller R.. Heterologous expression of an orphan NRPS gene cluster from Paenibacillus larvae in Escherichia coli revealed production of sevadicin. J. Biotechnol. 2015; 194:112–114. [DOI] [PubMed] [Google Scholar]
- 6. Yin J., Hoffmann M., Bian X., Tu Q., Yan F., Xia L., Ding X., Stewart A.F., Müller R., Fu J. et al. . Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM41398 in Streptomyces coelicolor A3(2). Sci. Rep. 2015; 5:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J., Sarov M., Rientjes J., Fu J., Hollak H., Kranz H., Xie W., Stewart A.F., Zhang Y.. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 2006; 32:43–53. [DOI] [PubMed] [Google Scholar]
- 8. Fu J., Teucher M., Anastassiadis K., Skarnes W., Stewart A.F.. A recombineering pipeline to make conditional targeting constructs. Methods Enzymol. 2010; 477:125–144. [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto-Gotoh T., Sekiguchi M.. Mutations of temperature sensitivity in R plasmid pSC101. J. Bacteriol. 1977; 131:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pospiech A., Neumann B.. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995; 11:217–218. [DOI] [PubMed] [Google Scholar]
- 11. Baker O., Gupta A., Obst M., Zhang Y., Anastassiadis K., Fu J., Stewart A.F.. RAC-tagging: recombineering and Cas9-assisted targeting for protein tagging and conditional analyses. Sci. Rep. 2016; 6:25529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H., Li Z., Jia R., Hou Y., Yin J., Bian X., Li A., Müller R., Stewart A.F., Fu J. et al. . RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 2016; 11:1175–1190. [DOI] [PubMed] [Google Scholar]
- 13. Hofemeister H., Ciotta G., Fu J., Seibert P.M., Schulz A., Maresca M., Sarov M., Anastassiadis K., Stewart A.F.. Recombineering, transfection, Western, IP and ChIP methods for protein tagging via gene targeting or BAC transgenesis. Methods. 2011; 53:437–452. [DOI] [PubMed] [Google Scholar]
- 14. Zhang S., Barbe V., Garel M., Zhang W., Chen H., Santini C.L., Murat D., Jing H., Zhao Y., Lajus A. et al. . Genome sequence of luminous piezophile Photobacterium phosphoreum ANT-2200. Genome Announc. 2014; 2:e0009614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy K.C. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 1991; 173:5808–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weller S.K., Sawitzke J.A.. Recombination promoted by DNA viruses: phage lambda to herpes simplex virus. Annu. Rev. Microbiol. 2014; 68:237–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muyrers J.P., Zhang Y., Buchholz F., Stewart A.F.. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000; 14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 18. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. et al. . A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011; 474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torsvik V., Goksoyr J., Daae F.L.. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990; 56:782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rappe M.S., Giovannoni S.J.. The uncultured microbial majority. Annu. Rev. Microbiol. 2003; 57:369–394. [DOI] [PubMed] [Google Scholar]
- 23. Charlop-Powers Z., Milshteyn A., Brady S.F.. Metagenomic small molecule discovery methods. Curr. Opin. Microbiol. 2014; 19:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larionov V., Kouprina N., Graves J., Chen X.N., Korenberg J.R., Resnick M.A.. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee N.C., Larionov V., Kouprina N.. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015; 43:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kouprina N., Larionov V.. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma. 2016; 125:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 28. Jiang W., Zhao X., Gabrieli T., Lou C., Ebenstein Y., Zhu T.F.. Cas9-assisted targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 2015; 6:8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nedelkova M., Maresca M., Fu J., Rostovskaya M., Chenna R., Thiede C., Anastassiadis K., Sarov M., Stewart A.F.. Targeted isolation of cloned genomic regions by recombineering for haplotype phasing and isogenic targeting. Nucleic Acids Res. 2011; 39:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riele H.T., Maandag E.R., Berns A.. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:5128–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.