Abstract
Acetylation of histone H3 at lysine-56 by the histone acetyltransferase Rtt109 in lower eukaryotes is important for maintaining genomic integrity and is required for C. albicans pathogenicity. Rtt109 is activated by association with two different histone chaperones, Vps75 and Asf1, through an unknown mechanism. Here, we reveal that the Rtt109 C-terminus interacts directly with Asf1 and elucidate the structural basis of this interaction. In addition, we find that the H3 N-terminus can interact via the same interface on Asf1, leading to a competition between the two interaction partners. This, together with the recruitment and position of the substrate, provides an explanation of the role of the Rtt109 C-terminus in Asf1-dependent Rtt109 activation.
INTRODUCTION
Eukaryotic DNA is packaged into a protein–DNA complex called chromatin. Nucleosomes, consisting of two copies each of histones H2A, H2B, H3 and H4, are the basic packing unit of chromatin, binding ∼145 bp of DNA (1). Histones can undergo post-translational modifications (PTMs), which play an important role in a multitude of processes including transcriptional regulation and DNA repair (2). Incorporation of PTMs begins immediately after synthesis (3), when histones are bound to both general and histone-specific chaperones (3,4).
Asf1 is a histone-specific chaperone necessary for both histone processing and deposition (5–8), as well as transcriptional regulation (9) and DNA repair (5,10). It binds the H3:H4 hetero-dimer, thereby preventing formation of the (H3:H4)2 tetramer (11,12).
In lower eukaryotes, newly synthesized histone H3 is acetylated on lysine-56 (K56) by Rtt109 and on lysine-9 (K9) by Rtt109 and Gcn5 (8,13–14). The role of K9 acetylation is largely unknown, while K56 acetylation has been shown to be important for genomic stability (15) and is required for recognition of the (H3:H4)2 tetramer by the downstream chaperone Rtt106 (16).
Rtt109 is a fungal histone acetyltransferase with only remote homology to human p300 and is required for Candida albicans pathogenesis (17). The lack of close homologues in higher eukaryotes makes it an attractive target for antifungal therapy, although recent high throughput screens have failed to identify suitable lead candidates (18–20). Alone, Rtt109 has very low acetyltransferase activity, but is strongly activated by the two histone chaperones Asf1 and Vps75 (13–14,21–22). Depending on chaperone association and substrate, different levels of activation and specificity have been observed in vitro (23,24). In addition, Rtt109 has been reported to regulate transcription together with Asf1 via an acetylation-dependent pathway that does not target H3:H4 (25).
Previously, it was reported that the Rtt109 C-terminus is required for Asf1-dependent activation of Rtt109-mediated acetylation of K56 in vitro (26). In vivo, the C-terminal tail of Rtt109 is necessary for K9 acetylation, but K56 acetylation is impaired only by the concurrent absence of both the Rtt109 C-terminus and Vps75 (26). This data prompted us to study the functional role of the Rtt109 C-terminus in the activation of acetylation. While Rtt109 forms a tight complex with Vps75 (27,28), its interaction with Asf1 has remained elusive. Here, we show that in Saccharomyces cerevisiae the Rtt109 C-terminus directly interacts with Asf1 via a surface shared by multiple Asf1 interaction partners (29,30). Interestingly, the same surface can also interact with the H3 tail in the Asf1−H3:H4 complex, indicating a role for Asf1 in chaperoning the H3 tail. Using NMR methods designed for the study of high-molecular-weight complexes, we show that the interaction between the Rtt109 C-terminus and Asf1 is retained when Rtt109 is bound to the Vps75 homodimer (Vps752) and when Asf1 forms a hetero-trimer with H3:H4. These findings suggest that the interaction of Asf1 with the Rtt109 C-terminus contributes to Rtt109 activation both in vitro and in vivo, where Rtt109 is in complex with Vps75.
MATERIALS AND METHODS
Protein expression and purification
Plasmids for Xenopus laevis histone H3/H4 (pET3d/a), Saccharomyces cerevisiae Vps75 (pET28a) and Saccharomyces cerevisiae Asf1 (2–169, pGEX6P-2) were a kind gift from K. Luger (Colorado State).
Asf12–169 was sub-cloned into pETM11, containing an N-terminal hexa-histidine tag, followed by a TEV protease cleavage site. The plasmid was transformed into Escherichia coli BL21(DE3) and expression was induced by addition of 0.5 mM IPTG at OD600 = 0.6–0.8 and continued overnight at 16°C. Cells were lysed by sonication in lysis buffer (1× phosphate-buffered saline (PBS) + 500 mM NaCl, 10 mM imidazole, 5 mM BME (β-mercapto-ethanol), 1× protease inhibitor (Roche; 11697498001)). The protein was purified by immobilized-metal-ion affinity-chromatography (IMAC, GE Healthcare Histrap HP 5 ml). The column was equilibrated and washed with wash buffer (25 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM BME, 10 mM imidazole) and eluted with a gradient to 75% elution buffer (25 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM BME, 500 mM imidazole) over 15 CV (column volumes). The fractions containing the tagged protein were pooled and the his-tag removed by overnight incubation with TEV protease. To remove the tag and the TEV protein, the solution was passed over a Histrap HP column equilibrated in wash buffer and the flow-through collected. Cleaved Asf1 was further purified by ion-exchange (GE Healthcare Hitrap Q, 25 mM Tris–HCl pH 7.5, 100–1000 mM NaCl, 5 mM BME) and size-exclusion chromatography (Superdex S200 16/60; 25 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM BME). Full-length human Asf1b was purified identically to S. cerevisiae Asf1. Vps75 was purified identically to Asf1 except that different buffers were used during the first Histrap step (wash buffer: 25 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM BME, 20 mM imidazole; elution buffer: 25 mM Tris–HCl pH 7.5, 500 mM NaCl, 5 mM BME, 400 mM imidazole).
15N- and 15N,13C-labeled proteins were expressed in M9 minimal medium containing 15NH4Cl or 15NH4Cl and 13C glucose and purified as described above. Perdeuterated and 1H,13C methyl isoleucine-, leucine- and valine-labeled Asf1 (Val-[13CH3]γ, Leu-[13CH3]δ, Ile-[13CH3]δ1) was obtained by expression in E. coli BL21(DE3) grown in M9 minimal medium in 100% D2O. α-ketobutyric acid sodium salt (60 mg/l, Sigma, 589276) and α-ketoisovaleric acid sodium salt (120 mg/ml, Sigma, 691887) were added 45 min before induction at OD600 = 0.6–0.8.
Codon-optimized full-length Saccharomyces cerevisiae Rtt109 was cloned in a pETM-11 vector containing a N-terminal hexa-histidine tag, followed by a TEV protease cleavage site. Rtt109 was expressed in E. coli BL21(DE3) and purified as previously described (31). Xenopus laevis histones were expressed and purified as reported by Luger et al. (32). Labeled histones were expressed in M9 minimal medium containing 15NH4Cl and purified identically to the unlabeled proteins.
Peptides were synthesized by the Peptide Synthesis Unit of the Helmholtz Center for Infection Research (Braunschweig) under the supervision of Dr Werner Tegge.
Microscale thermophoresis measurement
Asf1 was labeled with NT-647 via an NHS coupling using a kit from Nanotemper (MO-L0001) following the manufacturer's instructions. Thermophoresis temperature-jump titration series were performed in standard capillaries (Nanotemper, MO-K002) using 100 nM labeled Asf1 and concentrations of 0.058–475 μM of Rtt109 (2× serial dilutions) and 0.228–590 μM of Rtt109−Vps752 (2× serial dilutions). Experiments were performed at 25°C, in 50 mM citrate buffer at pH 6.5, 150 mM NaCl, 5 mM BME, with an excitation power of 20% and an MST power of 10–20%. Measurements were performed in triplicates, KD values were calculated independently and used to obtain the average and error values. Higher MST powers resulted in poor-quality curves at higher Rtt109 concentrations, probably due to Rtt109 aggregation. This effect was not observed for the Rtt109−Vps752 complex. Data was analyzed using MO.Affinity Analysis (Nanotemper, v2.1.2333).
Isothermal titration calorimetry (ITC)
ITC measurements were performed on a NANO ITC (TA Instruments) in PBS pH 7.4 with 5 mM BME at 20°C using an equilibration time of 3000 s and a stir-rate of 200 rpm. The cell concentration was 185 μM Asf1 (in a volume of 220–230 μl) and the peptide concentration in the syringe was 2.78 mM for Rtt109419–436 and 2.76 mM for Rtt109419–428. For the measurement of the Rtt109419–433 peptides, the cell concentration was 160 μM Asf1 (in a volume of 240–250 μl) and the peptide concentration in the syringe was 1.95 mM for Rtt109419–433, 2.75 mM for Rtt109419–433 RK427DD and 1.45 mM for Rtt109419–433 KK428DD. The first injection of 0.48 μl was not considered for the analysis. Twenty to twenty-two injections of 1.98 μl of peptide followed, with 300 s between injections. Data was analyzed using NanoAnalyze (TA Instruments, v3.6.0). Data points that suffered from baseline distortions were not included in the analysis.
Nuclear magnetic resonance spectroscopy
NMR data were acquired on Bruker Avance III HD 850 MHz and 600 MHz spectrometers equipped with He-cooled (TCI) and N2-cooled (CPP) cryogenic inverse HCN probe-heads, respectively. All experiments were performed at 298 K.
Asf1 data collection and assignment
2D 1H–15N and 1H–13C (ILV-methyl) correlation spectra were acquired using 15N-HSQC (33–35) and 13C-HMQC (methyl-TROSY) (36–39) pulse sequences. All assignment experiments were performed in PBS pH 7.4 with 5 mM BME for free Asf1 and PBS pH 6.2 with 5 mM BME for peptide-bound Asf1. Free and peptide-bound Asf1 backbone resonances were assigned using standard TROSY-HNCA, TROSY-HNCACB, TROSY-HN(CO)CACB and TROSY-HNCO experiments (40,41) with a deuterated and 13C,15N-labeled Asf1 sample at 500 μM protein and 3 mM Rtt109 peptide. All other experiments for structure determination were recorded using 13C,15N-labeled Asf1. Side-chain resonances for the peptide-bound Asf1 were assigned using an HC(C)H-TOCSY experiment (42,43) and assignment of the aromatic resonances was aided by an (HB)CB(CGCD)HD spectrum (44). ILV methyl resonances in the Asf1–H3:H4 complex (Asf1 perdeuterated and 1H,13C methyl-ILV-labeled; H3:H4 unlabeled) were assigned using a 4D 13C-HMQC–NOESY–13C-HMQC experiment (45,46) recorded with non-uniform-sampling. The sampling schedule was generated using the ist@HMS online schedule generator (http://gwagner.med.harvard.edu/intranet/istHMSv2/index.html). The 4D spectrum was reconstructed with iterative-soft-thresholding as implemented in the istHMS software (47–49). Distances for structure calculation of the Asf1–Rtt109419–433 complex were derived from 3D NOESY–15N-HSQC and NOESY–13C-HSQC spectra (120 ms NOESY mixing time) (50–53). Intermolecular Asf1–Rtt109419–433 NOEs in the NOESY–13C-HSQC spectrum were identified with the assistance of a 3D 13C,15N-double-filtered-NOESY–13C-HSQC experiment (54). Intramolecular Rtt109419–433 NOEs were measured from a 2D 1H-NOESY (150 ms NOESY mixing time) spectrum (55–57) recorded on a sample containing 150 μM Rtt109419–433 and 500 μM perdeuterated and 13C,15N-labeled Asf1. Because the Rtt109 peptide has only moderate affinity for Asf1 and is in fast exchange, the intermolecular Asf1-Rtt109419–433 resonances observed in the 3D 13C,15N-double-filtered-NOESY–13C-HSQC spectrum of Asf1 in the presence of excess Rtt109 peptide correspond closely to the free form of the peptide. To assign these resonances, we recorded 2D 1H-NOESY (150 ms NOESY mixing time) and 1H-TOCSY (70 ms DIPSI-2 mixing) experiments (57–59) with 3 mM Rtt109419–433 peptide in the presence of 500 μM perdeuterated and 13C,15N-labeled Asf1.
Non-exchangeable protons were identified by rapidly buffer-exchanging 15N-labeled Asf1 into 100% D2O buffer (15 mM NaH2PO4 pH 6.2, 150 mM NaCl, 5 mM BME) using a HiTrap desalting column (GE Healthcare). Backbone amide 1H–15N RDCs were determined by measuring the 1H–15N splitting in isotropic and anisotropic samples from the difference in the 15N frequencies of the amide peaks in the 15N-HSQC spectrum and a 15N-TROSY-HSQC spectrum recorded with Hα/Hβ band-selective decoupling (60,61). The anisotropic sample was prepared by addition of Pf1 bacteriophage (ASLA biotech) (62) to a concentration of 8.7 mg/ml (giving a residual 2H quadrupolar splitting of the HOD signal of 7.3 Hz).
Titrations were performed with 50–100 μM Asf1 with the indicated equivalents of interaction partners.
Data processing and analysis
NMR data was processed with Topspin (Bruker), istHMS and nmrPipe (63) and analysed using CcpNmr Analysis (64). The magnitude (Da) and rhombicity (R) of the alignment tensor were derived by fitting the experimental RDCs to PDB 1ROC using singular-value decomposition as implemented in PALES (65). CSPs were calculated as 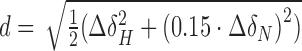 or
or 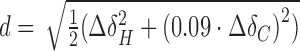 .
.
Structure calculation and analysis
Structure calculations were performed using ARIA v2.3/CNS v1.21 (66,67) via the standard ARIA v2 iterative-assignment protocol. Distance restraints were generated from NOESY peak heights with application of spin-diffusion correction for the calibration factors. For the 15N-edited NOESY spectrum, peak heights were scaled according to the intensities of the corresponding amide peaks in the 15N HSQC spectrum prior to generation of distance restraints. Amide-to-carbonyl hydrogen-bond restraints were applied for non-exchangeable amide protons. RDC restraints for backbone amide bonds of residues in canonical secondary structure elements were applied using the SANI potential of CNS (Da = 7.2 Hz; R = 0.52). Dihedral-angle restraints for backbone ϕ and ψ angles were generated from backbone chemical shifts using TALOS-N (68). Restraints were applied for residues where the TALOS-N classifications was ‘strong’ or ‘generous’, using respective error ranges of twice and three times the uncertainty reported by TALOS-N. Initial structure calculations indicated a conflict between the hydrogen-bond and dihedral restraints for residue F106. A calculation where both types of restraints were omitted for this residue revealed a well-defined conformation in good agreement with the predicted hydrogen bond (and differing from the TALOS-predicted dihedral restraints). Therefore the dihedral restraints for F106 were removed for the final structure calculation. The numbers of steps for the hot, cool1, cool2 and refine stages of the molecular-dynamics simulated-annealing protocol were 40 000, 20 000, 16 000 and 16 000, respectively. All distance restraints were used in every iteration. One hundred structures were calculated in the final iteration, of which the 10 lowest-energy structures were refined in explicit water to generate the final structural ensemble.
Structural quality reports and statistics were generated using the PSVS web-server (69). Buried surface area was determined using the Intersurf tool (prune distance: 6.0, molecule bias: 1.0) and the measure volume and area tools in Chimera (70). Figures were prepared using gnuplot, CcpNmr Analysis (71) and Pymol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Acetylation assays
Asf1−H3:H4, Vps752−Rtt109 and Vps752−Rtt1091–424 complexes were reconstituted by size-exclusion chromatography (Superdex S200 10/300; 10 mM HEPES pH 8.0, 100 mM NaCl, 5 mM BME for complexes with Rtt109, 10 mM HEPES pH 8.0, 500 mM NaCl, 5 mM BME for Asf1−H3:H4). Acetylation reactions were performed using 0.3 μM Rtt109–Vps752 or Rtt109, 0.3 μM Asf1−H3:H4 and 3 μM Ac-CoA in 10 mM HEPES pH 8.0, 100 mM NaCl, 5 mM BME. Reactions were stopped with 0.5 M sodium acetate, pH 5.5; 1.6 ul of the reaction mixtures were spotted on a nitrocellulose membrane. Membranes were air-dried for ca. 30 min, blocked for 1 h in 5% BSA–TBST buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% Tween-20), and subsequently incubated for 1 h with a 1:2000 dilution of rabbit anti-histone H3 (acetyl K9) primary antibody (ab4441 Abcam, lot no. GR304381–1) in TBST with 0.1% BSA. After washing three times with TBST buffer, the membranes were incubated for 1 h with the secondary antibody, an anti-rabbit IgG (whole molecule)–alkaline phosphatase (A3687 Sigma, lot no. SLBK3154V) solution diluted 1:10 000 with 0.1% BSA and TBST. After three washes with TBST buffer, the blots were developed with BCIP®/NBT Liquid Substrate System (B1911 Sigma, lot no. SLBQ3317V).
The density of the dots was quantified using the ImageJ software (71). After background subtraction, the measured intensities were normalized to the negative control (Asf1−H3:H4 alone). The results are the average of three experiments with the standard deviation representing the error.
RESULTS
Asf1 interacts with the C-terminus of Rtt109
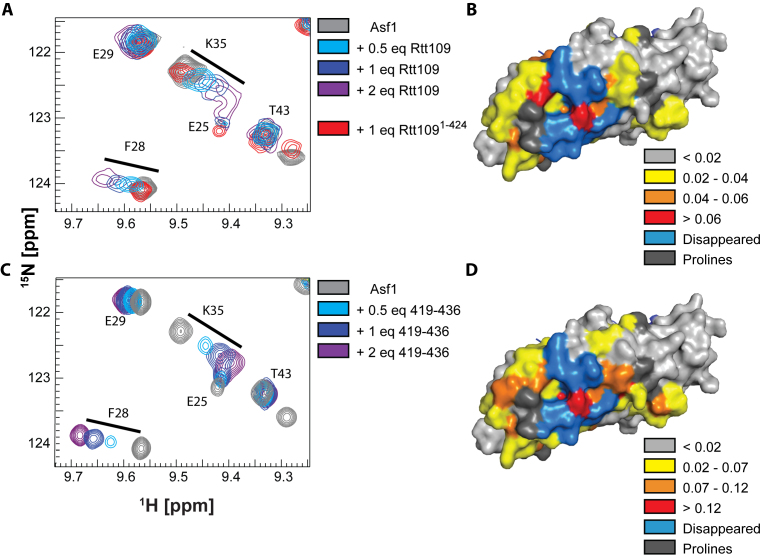
We characterized the interaction of Rtt109 and Asf1 by NMR spectroscopy (Figures 1 and 2, Supplementary Figures S1–S3). Rtt109 is prone to aggregation and precipitation in commonly used buffers. In previous work, we used a thermal-shift assay to optimize Rtt109 stability and found optimal conditions in a buffer containing citric acid at low pH (5.5–6.5) (31). Using this buffer, we titrated 15N-labeled Asf1 with increasing amounts of unlabeled full-length Rtt109 and observed significant changes in the Asf1 15N-HSQC spectrum (33–35) (Figure 1A and Supplementary Figure S2A). Mapping these chemical shift perturbations (CSPs) on the structure of Asf1 revealed that they cluster on the surface opposite to the histone interaction surface (Figure 1B), suggesting that the interaction of Asf1 with Rtt109 is compatible with histone binding. Next we performed an identical titration with a truncated Rtt109 construct (1–424) lacking the C-terminal 12 amino acids (26). In contrast to full-length Rtt109, only small changes in the Asf1 spectrum were observed upon addition of Rtt1091–424 (Figure 1A), indicating that the interaction between Asf1 and Rtt109 is dependent on the C-terminus of Rtt109. Similarly, when we added a peptide corresponding to the C-terminal 18 residues of Rtt109 (Rtt109419–436) to labeled Asf1 (Figure 1C and Supplementary Figure S2B), we observed large CSPs, which mapped onto the same Asf1 surface as those from adding full-length Rtt109 (Figure 1D).
Figure 1.
Rtt109 interacts with Asf1 via its C-terminus. (A) Overlay of 15N-HSQC spectra of Asf1 (100 μM) in the presence of Rtt109 and Rtt1091–424. The target concentrations of full-length Rtt109 corresponded to 0.5, 1 and 2 equivalents of Rtt109, but the actual achieved concentrations (estimated from 1D 1H spectra) were lower due to partial precipitation of Rtt109 in the NMR tube. The spectra were acquired in 50 mM sodium citrate pH 6.5, 150 mM NaCl, 5 mM BME at 600 MHz and 298 K. (B) Mapping of the CSPs observed upon addition of 1 equivalent of full-length Rtt109 onto the Asf1 structure in complex with the H3:H4 dimer (PDB ID 2HUE). (C) Overlay of 15N-HSQC spectra of Asf1 (100 μM) in the presence of Rtt109419–436. The spectra were acquired in 50 mM sodium citrate pH 6.5, 150 mM NaCl, 5 mM BME at 850 MHz and 298 K. (D) Mapping of the CSPs observed upon addition of 1 equivalent of Rtt109419–436 onto the Asf1 structure as in (B). The chemical shifts perturbations observed upon addition of Rtt109419–436 were larger than those observed upon addition of full-length Rtt109, which can be recapitulated by the on-going precipitation of full-length Rtt109 during the NMR experiment. Asf1 is in surface representation.
Figure 2.
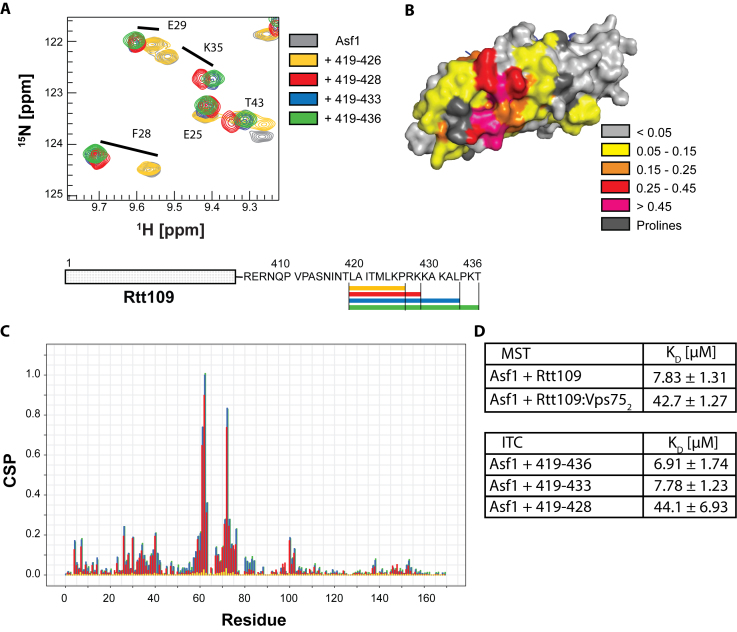
Rtt109 interacts with Asf1 via its C-terminus. (A) Overlay of 15N-HSQC spectra of Asf1 (100 μM) in the presence of Rtt109-derived peptides (all at 3 mM) of various lengths. The spectra were acquired in PBS pH 6.2 with 5mM BME at 850 MHz and 298 K. (B) Mapping of the CSPs observed upon addition of 3 mM Rtt109419–436 onto the Asf1 structure in complex with the H3:H4 dimer (from PDB ID 2HUE). (C) CSP analysis of the spectra in (A). (D) Results of ITC and MST titrations of Asf1 with different peptides and proteins. Experiments were performed in triplicate.
To better define the residues of Rtt109 responsible for the interaction with Asf1, we performed NMR titrations with successively shorter peptides derived from the Rtt109 C-terminus. Rtt109419–433 interacted with Asf1 in an identical manner to Rtt109419–436 (Figure 2A and C). Rtt109419–428 showed a somewhat changed pattern of CSPs (Figure 2A and C), while deletion of the 10 C-terminal amino acids (Rtt109419–426) completely abolished the interaction with Asf1 (Figure 2A and C).
Next, we used isothermal titration calorimetry (ITC) (72) and microscale thermophoresis (MST) (73) to quantify the interactions of Asf1 with the Rtt109 C-terminal peptides and full-length Rtt109 (Figure 2D, Supplementary Figures S4 and S5). The ITC titrations revealed comparable KD values for Rtt109419–436 and Rtt109419–433 (6.91 ± 1.74 μM and 7.78 ± 1.23 μM, respectively), which increased to 44.1 ± 6.93 μM for the shorter peptide Rtt109419–428. MST titrations with full-length Rtt109 gave a KD of 7.63 ± 1.31 μM, in agreement with the hypothesis that the C-terminal residues of Rtt109 are solely responsible for the interaction with Asf1. In the presence of Vps752, the affinity of full-length Rtt109 for Asf1 was slightly weaker (KD = 42.7 ± 1.27 μM).
Structural characterization of the Asf1–Rtt109 interaction
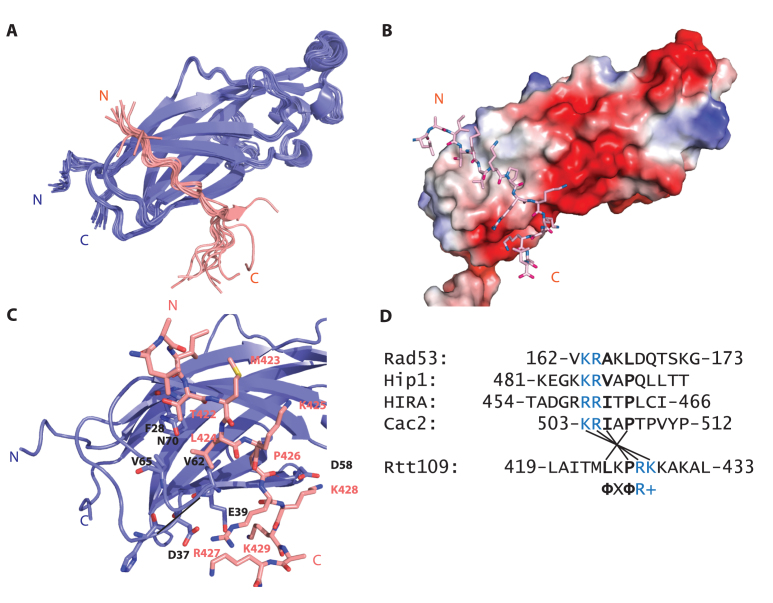
We used NMR to solve the structure of Asf1 in complex with the Rtt109 C-terminus (419LAITMLKPRKKAKAL433) (Figure 3 and Table 1). The Rtt109 C-terminal peptide structure was defined to a precision of 0.69 Å (backbone root-mean-square deviation, RMSD, calculated over residues 421–429 for the 10 lowest-energy structures) by 231 (199 unambiguous) intra-peptide NOEs (derived from a 2D 1H NOESY spectrum (55–57) of the peptide in complex with perdeuterated Asf1) and 159 (67 unambiguous) intermolecular Asf1–peptide NOEs (assigned using a 3D 13C,15N-filtered-NOESY–13C-HSQC spectrum (54) of the peptide in complex with 13C,15N-labeled Asf1). A further 6732 (4117 unambiguous) Asf1 intramolecular NOEs derived from standard 3D NOESY–15N-HSQC and NOESY–13C-HSQC spectra (50–53) defined the complex to a precision of 0.47 Å (backbone RMSD calculated over residues 2–47 and 54–156 of Asf1 and residues 421–429 of Rtt109419–433 for the 10 lowest-energy structures). Comparison with a crystallographic structure of Asf1 (PDB ID: 1ROC) demonstrates that the Asf1 conformation is not significantly perturbed by Rtt109 binding (backbone RMSD of 0.80 Å for residues 1–155).
Figure 3.
Structure of the Asf1–Rtt109419–433 complex.( A) The water-refined ensemble of the 10 lowest-energy structures obtained for the Asf1–Rtt109419–433 complex (Asf1 in blue; peptide in pink). Only residues 1–155 of Asf1 are shown for clarity; the stretch 156–169 is disordered. (B) Charged surface representation of Asf11–155 in complex with the Rtt109419–433 peptide (in sticks). The main interaction surface is hydrophobic with additional ionic contacts at the C-terminus of the peptide. (C) Detailed view of the Asf1–Rtt109 interface. Interacting residues of Asf1 are shown as sticks and labeled in black text. (D) Sequence alignment of known Asf1 interaction partners, highlighting conserved hydrophobic and positively charged residues.
Table 1. Assignment and restraint statistics and structural quality parameters for the structure of the Asf12–169−Rtt109419–433 complex. For the structural quality parameters, the first column (headed 〈SA〉) contains average values for the ensemble (standard deviations in parentheses), while the second column (headed 〈SA〉c) contains the corresponding values for the structure closest to the mean. Structural RMSDs were calculated over the well-ordered residues, defined as residues 2–47 and 54–156 of Asf12–169 and residues 421–429 of Rtt109419–433.
| Resonance assignments | ||
| All protons | 98.9% | |
| Asf12–169 | 98.8% | |
| Rtt419–433 | 100.0% | |
| Asf12–169 backbone atoms (N, Cα, C’, HN, Hα, Cβ) | 94.9% | |
| NMR restraints | ||
| NOE distance restraints | 7121 | |
| Unambiguous | 4383 | |
| Classified by sequence-separation: | ||
| Intra-residue | 1584 | |
| Sequential | 1051 | |
| Short-range | 410 | |
| Medium-range | 73 | |
| Long-range (incl. inter-molecular) | 1265 | |
| Classified by intra-/inter-molecular: | ||
| Inter-molecular | 67 | |
| Intra-Asf12–169 | 4117 | |
| Intra-Rtt109419–433 | 199 | |
| Ambiguous | 2738 | |
| Classified by sequence-separation: | ||
| Intra-residue | 375.7 | |
| Sequential | 562.9 | |
| Short-range | 368.5 | |
| Medium-range | 98.3 | |
| Long-range (incl. inter-molecular) | 1332.6 | |
| Classified by intra-/inter-molecular: | ||
| Inter-molecular | 91.9 | |
| Intra-Asf12–169 | 2614.6 | |
| Intra-Rtt109419–433 | 31.5 | |
| Hydrogen-bond restraints | 81 | |
| Dihedral-angle restraints | 270 | |
| ϕ angle | 135 | |
| ψ angle | 135 | |
| RDC restraints (N–H) | 61 | |
| Structural quality | 〈SA〉 | 〈SA〉c |
| RMSDs from experimental restraints | ||
| NOE distances (Å) | 0.0216 (0.0005) | 0.0217 |
| H-bond distances (Å) | 0.0134 (0.0029) | 0.0130 |
| Dihedral angles (°) | 0.817 (0.056) | 0.825 |
| Violations | ||
| NOE distances > 0.5 Å | 0 (0) | 0 |
| NOE distances > 0.3 Å | 2.3 (0.9) | 3 |
| Dihedral angles > 10° | 0 (0) | 0 |
| Dihedral angles > 5° | 0.8 (0.9) | 0 |
| RMSDs from ideal geometry | ||
| Bond lengths (Å) | 0.00420 (0.00011) | 0.0044 |
| Bond angles (°) | 0.5472 (0.0087) | 0.5421 |
| Improper angles (°) | 1.592 (0.078) | 1.613 |
| Final energy (kcal mol–1) | –6914 (82) | –7006 |
| Structural RMSDs to mean structure | ||
| Backbone atoms (Å) | 0.47 (0.07) | 0.38 |
| Heavy atoms (Å) | 0.80 (0.11) | 0.70 |
| Ramachandran plot summary (PROCHECK) | ||
| Most favoured regions | 88.2% | |
| Additionally allowed regions | 9.8% | |
| Generously allowed regions | 0.5% | |
| Disallowed regions | 1.6% | |
| Global quality scores (–Raw/Z-score) | ||
| Verify3D | 0.35/–1.77 | |
| ProsaII | 0.30/–1.45 | |
| Procheck | –0.54/–3.19 | |
| Molprobity Clashscore | 34.34/–4.37 | |
The peptide interacts with a hydrophobic patch composed of residues 60–72 (60ILVGPVPVGVNKF72) on the side of Asf1 opposite to the histone interaction surface, resulting in a buried surface area of 739 Å2. This interaction interface is identical to the one identified by CSP analysis of Asf1 titrated with either full-length Rtt109 or Rtt109419–436 (Figures 1 and 2). Residues T422, L424 and P426 of the Rtt109 peptide are largely responsible for the hydrophobic interactions (Figure 3C). In addition, the basic residues R427, K428 and K429 participate in ionic interactions with the negatively charged surface adjacent to the hydrophobic patch, contacting residues D37, D58 and E39, respectively. The interaction is further stabilized by a hydrogen bond between the carbonyl group of peptide residue R427 and the amide group of Asf1 residue L61 (Figure 3C).
To validate this structure we generated two Rtt109419–433 mutants with two aspartate substitutions at either R427 and K428 or K428 and K429, referred to as Rtt109419–433 RK427DD and Rtt109419–433 KK428DD respectively. ITC measurements showed a complete loss of binding affinity for both mutant peptides, confirming that the ionic interactions mediated by these residues are necessary for Asf1 recognition (Supplementary Figure S4).
Next, we compared the Asf1–Rtt109419–433 structure with those available for Asf1 in complex with other interaction partners that engage the same surface (HIRA, Cac2, Rad53 and Hip1). We identified a common binding motif consisting of ΦXΦR+, in which Φ and + represent a small hydrophobic amino acid and a positively charged residue, respectively (Figure 3D). Interestingly, the Rtt109 peptide is reversed with respect to the other Asf1 interaction partners (Supplementary Figure S6A). The salt-bridge observed between D37 and R427 in our structure is conserved in all other Asf1–peptide complexes (Supplementary Figure S6B).
The Asf1–Rtt109 interaction in the presence of histone H3:H4 dimer
Rtt109 probably interacts with Asf1 while it is in complex with the Rtt109 substrate, the histone H3:H4 dimer. On the other hand, Rtt109 also forms a tight complex with Vps752. To determine whether the interaction between the Rtt109 C-terminus and Asf1 is retained in higher-order complexes, we produced Asf1 with 1H,13C-methyl-labeled Ile, Leu and Val residues in a perdeuterated background (74). This labeling scheme yields high-quality methyl group spectra for high-molecular-weight complexes that would otherwise be inaccessible by solution NMR methods (36,75). The Asf1–H3:H4 complex is prone to aggregation at high concentration in low-salt conditions, but by using a citrate buffer (pH 6.5), which also increases Rtt109 stability, we obtained a soluble complex at physiological ionic strength. 60% of the Asf1 methyl resonances in the Asf1–H3:H4 complex could be assigned by analysis of a 4D 13C-HMQC–NOESY–13C-HMQC spectrum (45,46) in conjunction with the Asf1 structure.
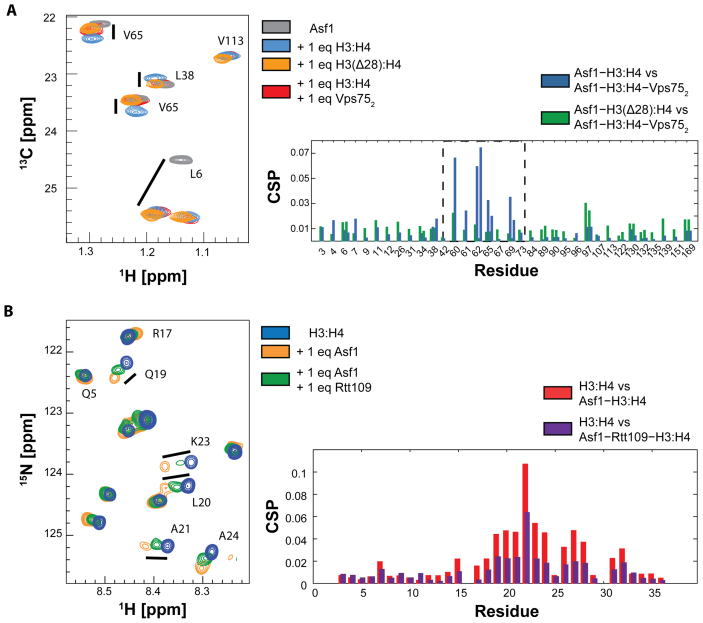
To investigate the Rtt109–Asf1 interaction in the presence of both Vps75 and histones, we first titrated the Asf1–H3:H4 complex with unlabeled Vps752. Surprisingly, we observed noticeable CSPs for several residues, which clustered on the same surface engaged by the Rtt109 C-terminal peptide (Figure 4A and Supplementary Figure S7). In contrast, residues belonging to the Asf1 surface in contact with the core of the H3-H4 dimer did not display any CSPs upon addition of Vps752 (e.g. L6 in Figure 4A). Interestingly, adding unlabeled Vps752 to Asf1 alone did not give any CSPs, suggesting that Vps75 does not interact directly with Asf1 (Supplementary Figure S8). To explain these observations, we reasoned that Vps75 modulates the interaction between histones and Asf1, leading to CSPs of Asf1 resonances in the Asf1–H3:H4 complex upon addition of Vps752. Because the region of Asf1 showing these CSPs is located far from the canonical histone interaction interface, we hypothesized that the long unstructured histone H3 tail folds back onto Asf1 in the Asf1–H3:H4 complex. To test this hypothesis, we reconstituted the H3:H4 complex using truncated H3 that lacks the N-terminal 28 aa. The chemical shift perturbations observed for the Asf1–H3:H4–Vps752 and Asf1–H3(Δ28):H4 complexes relative to Asf1–H3:H4 were very similar (Figure 4A), providing strong evidence that Vps75 binding results in release of the H3 tail from Asf1. In addition, we titrated the H3:H4 complex containing 15N-labeled H3 with unlabeled Asf1. As expected, we observed CSPs (Figure 4B) for H3 residues 19–26, confirming the interaction between the H3 tail and Asf1. When Rtt109 is added to this mixture, the CSPs of the H3 tail relative to free H3:H4 are reduced, showing that Rtt109 can out-compete the H3 tail for the same interaction surface on Asf1 (Figure 4B). Having discovered a previously unknown H3−Asf1 interaction, we wondered whether the same interaction mode is also preserved in higher eukaryotes. To address this question, we recorded spectra of the H3:H4 complex containing 15N-labeled H3 in the presence of human Asf1b. Interestingly, similar chemical shifts were observed for H3 in both the Asf1 and hAsf1b complexes (Supplementary Figure S9), suggesting conservation across species of the interaction of Asf1 with the H3 tail.
Figure 4.
The H3 tail and Rtt109 share the same interaction surface on Asf1. (A) 13C-HMQC spectra and CSP analysis of ILV-labeled Asf1 in the presence of the H3:H4 dimer (blue), H3:H4−Vps752 (red) or the H3(Δ28):H4 dimer (orange). CSPs are calculated between Asf1−H3:H4 and Asf1−H3:H4−Vps752 (blue), and between Asf1−H3(Δ28):H4 and Asf1−H3:H4−Vps752 (green). The spectra were acquired in 50 mM sodium citrate pH 6.5, 150 mM NaCl, 5 mM BME at 850 MHz and 298 K. (B) 15N-HSQC spectra and CSP analysis of 15N-labeled histone H3 in the H3:H4 complex (blue) and in the Asf1−H3:H4 complex with (green) or without (orange) Rtt109. CSPs of Asf1−H3:H4 and Asf1−H3:H4 in the presence of Rtt109 are calculated with respect to the H3:H4 complex. The spectra were acquired in 50 mM sodium citrate pH 6.2, 150 mM NaCl, 5 mM BME at 600 MHz and 298 K.
The Asf1–Rtt109 interaction is retained in a complex containing Vps75 and histone H3:H4
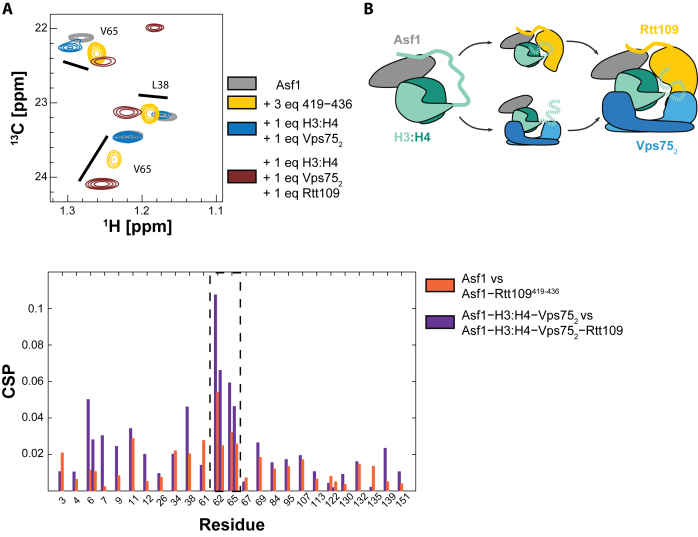
Next, we probed the binding of the Asf1–H3:H4 substrate to the Rtt109–Vps752 complex by monitoring CSPs of the Ile, Leu and Val methyl resonances of Asf1. In the presence of Vps75, we observe large Asf1 CSPs upon addition of Rtt109 (Figure 5A and Supplementary Figure S10). When comparing the Asf1 CSPs from the Rtt109419–433-to-Asf1 and Rtt109-to-Asf1–H3:H4–Vps752 titrations, we observe similar shifts on the same Asf1 interface described before, confirming that the interaction between Asf1 and the Rtt109 C-terminus is retained in the Asf1–H3:H4–Rtt109–Vps752 complex. Differences in CSPs were observed only for residues 6–9, which we presume is due to this region of Asf1 experiencing a different chemical environment in the Asf1–H3:H4–Rtt109–Vps752 complex compared to the Asf1–Rtt109 complex.
Figure 5.
The Rtt109–Asf1 interaction is retained in a larger enzymatic complex. (A) Overlay of 13C-HMQC spectra and CSP analysis of ILV-labeled Asf1 (grey) in complex with Rtt109419–436 (yellow), H3:H4−Vps752 (blue) or H3:H4−Vps752−Rtt109 (brown). The full spectrum is shown in Supplementary Figure S10. CSPs are calculated between Asf1 and Asf1–Rtt109419–436 (orange), and between Asf1–H3:H4–Vps752 and Asf1–H3:H4–Vps752–Rtt109 (purple). The spectra were acquired in 50 mM sodium citrate pH 6.5, 150 mM NaCl, 5 mM BME at 850 MHz and 298 K. (B) Schematic summarizing the described interactions. The H3 tail interacts with Asf1 in the Asf1−H3:H4 complex. This interaction can be released by addition of Vps75 or Rtt109.
The presence of Vps75 relieves the need for the Rtt109 C-terminus for efficient K9 acetylation
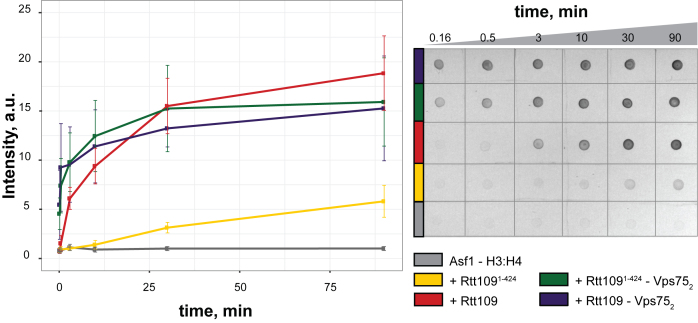
Lastly, we performed dot-blot assays to quantify the influence of the Rtt109 C-terminus on the efficiency of K9 acetylation (K9Ac). We found that in the absence of Vps75, truncation of the Rtt109 C-terminus abolishes K9 acetylation almost completely (Figure 6), suggesting that the interaction between Asf1 and the Rtt109 C-terminus is indispensable for K9 acetylation when Vps75 is absent. In the presence of Vps75, K9 acetylation is faster and is impacted only minimally by the absence of the Rtt109 C-terminus. This is in agreement with published data, which previously indicated that K9 acetylation in vitro and in the presence of Vps75 does not require the Rtt109 C-terminus (26). Asf1-dependent K9 acetylation (in the absence of Vps75) was not detected by western blotting in Fillingham et al. (26), but has been previously observed by mass spectrometry, albeit to a lower extent than K56 acetylation (76). We observe that the absence of Vps75 slows but does not abolish K9 acetylation and that the Rtt109 C-terminus is required for K9 acetylation only in the absence of Vps75.
Figure 6.
K9 acetylation in the absence of Vps75 depends on the Rtt109 C-terminus. Time-course of K9 acetylation (left panel) by full-length Rtt109 (red), Rtt1091–424 (yellow), Vps752−Rtt109 (blue) and Vps752− Rtt1091–424 (green), as measured in dot-blot assays (right panel). The control curve without Rtt109 is in grey.
DISCUSSION
We have shown that the Rtt109 C-terminus directly interacts with Asf1 via both hydrophobic and charge-mediated contacts. The interaction mode is similar to other reported Asf1–peptide complexes, with the exception that the Rtt109 peptide is inversely oriented relative to the peptide in all other complexes. Using NMR techniques designed for studying high-molecular-weight complexes, we showed that the interaction between Asf1 and the C-terminus of Rtt109 is retained in a complex containing histone H3:H4 dimer and Vps752. Interestingly, the histone H3 tail interacts with the same surface of Asf1 as the Rtt109 C-terminus in the Asf1−H3:H4 complex; the Rtt109 C-terminus can out-compete this interaction (Figure 5B), releasing the H3 tail from Asf1. Similarly, Vps75 binding also causes Asf1 to release the H3 tail (Figure 5B).
In acetylation assays, we observe that in vitro deletion of the Rtt109 C-terminus impedes Asf1-dependent H3-K9Ac, but does not affect acetylation in the presence of both Asf1 and Vps75. Similar results have been reported in the literature for H3-K56Ac both in vitro and in vivo (26). Our structural findings provide a rationale for this data. The Rtt109–Asf1 interaction can out-compete binding of the H3 tail to Asf1, releasing it for modification. In the absence of both the Rtt109 C-terminus and Vps75, the H3 tail remains bound to Asf1 and protected from the enzyme. In agreement with this scenario, the Rtt109 C-terminus has been demonstrated to be redundant for Vps75-dependent H3-K9Ac in vitro in the absence of Asf1 (77). In addition, the Asf1−Rtt109 interaction may help in recruiting the Asf1-bound histone substrate to Rtt109, possibly limiting the orientations of H3:H4 with respect to the catalytic site and thereby promoting productive binding of the substrate. In agreement with the observation that Vps75 can rescue the effect of Rtt109 C-terminus deletion in vitro, we find that both Vps752 and the Rtt109 C-terminus modulate the Asf1–H3 tail interaction (Figure 5B). In vivo, however, both Vps75 and the Rtt109 C-terminus seem to be required for K9 (but not for K56) acetylation (26). This discrepancy suggests that there are additional factors contributing to or competing with H3-K9Ac by Rtt109 in vivo.
The chaperoning of the H3 tail by Asf1 might have implications beyond the Rtt109 enzymatic activation in the other Asf1-dependent regulatory pathways. The interaction of the H3 tail with Asf1 is conserved in humans (Supplementary Figure S9), suggesting a general role of Asf1 in modulating the conformation and accessibility of the H3 tail. Binding to Asf1 may protect the H3 tail from either interaction or modification and represent a mechanism to regulate access to the tail with respect to other cellular factors. In support of this hypothesis, the presence of Asf1 can inhibit H3 and H4 acetylation by the SAS complex (78).
AVAILABILITY
The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID: 6F0Y); the NMR data has been deposited in the Biological Magnetic Resonance Bank (BMRB ID: 34201).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Andrea Scrima for help with MST experiments, Dr Christian Kleusch for the introduction to MST and Dr Luca Codutti for the ITC experiments. For the peptide synthesis, we would like to thank Dr Werner Tegge from the Peptide- and Chemical Synthesis Team of the Helmholtz Centre for Infection Research, Braunschweig.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
L.L. acknowledges the receipt of an EMBO Long-term Fellowship [ALTF 1474-2014, Marie Curie Actions, LTFCOFUND2013, GA-2103-609409]. Funding for open access charge: University Budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campos E.I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W.-H.W., Seepany H., Gao Z., Day L.A., Greenblatt J.F. et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010; 17:1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campos E.I., Smits A.H., Kang Y.-H., Landry S., Escobar T.M., Nayak S., Ueberheide B.M., Durocher D., Vermeulen M., Hurwitz J. et al. Analysis of the histone H3.1 interactome: a suitable chaperone for the right event. Mol. Cell. 2015; 60:697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tyler J.K., Adams C.R., Chen S.-R., Kobayashi R., Kamakaka R.T., Kadonaga J.T.. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999; 402:555–560. [DOI] [PubMed] [Google Scholar]
- 6. Groth A., Ray-Gallet D., Quivy J.-P., Lukas J., Bartek J., Almouzni G.. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 2005; 17:301–311. [DOI] [PubMed] [Google Scholar]
- 7. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y.. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004; 116:51–61. [DOI] [PubMed] [Google Scholar]
- 8. Adkins M.W., Carson J.J., English C.M., Ramey C.J., Tyler J.K.. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 2007; 282:1334–1340. [DOI] [PubMed] [Google Scholar]
- 9. Minard L.V., Williams J.S., Walker A.C., Schultz M.C.. Transcriptional regulation by Asf1: new mechanistic insights from studies of the DNA damage response to replication stress. J. Biol. Chem. 2011; 286:7082–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emili A., Schieltz D.M., Yates Iii J.R., Hartwell L.H.. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001; 7:13–20. [DOI] [PubMed] [Google Scholar]
- 11. English C.M., Maluf N.K., Tripet B., Churchill M.E.A., Tyler J.K.. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3−H4 heterotetramer on DNA. Biochemistry. 2005; 44:13673–13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. English C.M., Adkins M.W., Carson J.J., Churchill M.E.A., Tyler J.K.. Structural basis for the histone chaperone activity of Asf1. Cell. 2006; 127:495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsubota T., Berndsen C.E., Erkmann J.A., Smith C.L., Yang L., Freitas M.A., Denu J.M., Kaufman P.D.. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007; 25:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han J., Zhou H., Li Z., Xu R.-M., Zhang Z.. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 2007; 282:28587–28596. [DOI] [PubMed] [Google Scholar]
- 15. Masumoto H., Hawke D., Kobayashi R., Verreault A.. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005; 436:294–298. [DOI] [PubMed] [Google Scholar]
- 16. Su D., Hu Q., Li Q., Thompson J.R., Cui G., Fazly A., Davies B.A., Botuyan M.V., Zhang Z., Mer G.. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature. 2012; 483:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopes da Rosa J., Boyartchuk V.L., Zhu L.J., Kaufman P.D.. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopes da Rosa J., Bajaj V., Spoonamore J., Kaufman P.D.. A small molecule inhibitor of fungal histone acetyltransferase Rtt109. Bioorg. Med. Chem. Lett. 2013; 23:2853–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dahlin J.L., Nissink J.W.M., Strasser J.M., Francis S., Higgins L., Zhou H., Zhang Z., Walters M.A.. PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 2015; 58:2091–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dahlin J.L., Sinville R., Solberg J., Zhou H., Han J., Francis S., Strasser J.M., John K., Hook D.J., Walters M.A. et al. A cell-free fluorometric high-throughput screen for inhibitors of Rtt109-catalyzed histone acetylation. PLoS ONE. 2013; 8:e78877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berndsen C.E., Tsubota T., Lindner S.E., Lee S., Holton J.M., Kaufman P.D., Keck J.L., Denu J.M.. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat. Struct. Mol. Biol. 2008; 15:948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kolonko E.M., Albaugh B.N., Lindner S.E., Chen Y., Satyshur K.A., Arnold K.M., Kaufman P.D., Keck J.L., Denu J.M.. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20275–20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuo Y.-M., Henry R.A., Huang L., Chen X., Stargell L.A., Andrews A.J.. Utilizing targeted mass spectrometry to demonstrate Asf1-dependent increases in residue specificity for Rtt109-Vps75 mediated histone acetylation. PLoS ONE. 2015; 10:e0118516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abshiru N., Ippersiel K., Tang Y., Yuan H., Marmorstein R., Verreault A., Thibault P.. Chaperone-mediated acetylation of histones by Rtt109 identified by quantitative proteomics. J. Proteomics. 2013; 81:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin L.-j., Schultz M.C.. Promoter regulation by distinct mechanisms of functional interplay between lysine acetylase Rtt109 and histone chaperone Asf1. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:19599–19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Radovani E., Cadorin M., Shams T., El-Rass S., Karsou A.R., Kim H.-S., Kurat C.F., Keogh M.-C., Greenblatt J.F., Fillingham J.S.. The carboxyl terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryot. Cell. 2013; 12:654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang Y., Holbert M.A., Delgoshaie N., Wurtele H., Guillemette B., Meeth K., Yuan H., Drogaris P., Lee E.-H., Durette C. et al. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011; 19:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su D., Hu Q., Zhou H., Thompson J.R., Xu R.-M., Zhang Z., Mer G.. Structure and histone binding properties of the Vps75-Rtt109 chaperone-lysine acetyltransferase complex. J. Biol. Chem. 2011; 286:15625–15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiao Y., Seeger K., Lautrette A., Gaubert A., Mousson F., Guerois R., Mann C., Ochsenbein F.. Surprising complexity of the Asf1 histone chaperone-Rad53 kinase interaction. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang Y., Poustovoitov M.V., Zhao K., Garfinkel M., Canutescu A., Dunbrack R., Adams P.D., Marmorstein R.. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 2006; 13:921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kozak S., Lercher L., Karanth M.N., Meijers R., Carlomagno T., Boivin S.. Optimization of protein samples for NMR using thermal shift assays. J. Biomol. NMR. 2016; 64:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luger K., Rechsteiner T.J., Richmond T.J.. Becker PB. Chromatin Protocols. 1999; 119:Humana Press; 1–16. [Google Scholar]
- 33. Bodenhausen G., Ruben D.J.. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980; 69:185–189. [Google Scholar]
- 34. Piotto M., Saudek V., Sklenář V.. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992; 2:661–665. [DOI] [PubMed] [Google Scholar]
- 35. Sklenar V., Piotto M., Leppik R., Saudek V.. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. A. 1993; 102:241–245. [Google Scholar]
- 36. Tugarinov V., Hwang P.M., Ollerenshaw J.E., Kay L.E.. Cross-correlated relaxation enhanced 1H−13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 2003; 125:10420–10428. [DOI] [PubMed] [Google Scholar]
- 37. Mueller L. Sensitivity enhanced detection of weak nuclei using heteronuclear multiple quantum coherence. J. Am. Chem. Soc. 1979; 101:4481–4484. [Google Scholar]
- 38. Bax A., Griffey R.H., Hawkins B.L.. Correlation of proton and nitrogen-15 chemical shifts by multiple quantum NMR. J. Magn. Reson. 1983; 55:301–315. [Google Scholar]
- 39. Robin Bendall M., Pegg D.T., Doddrell D.M.. Pulse sequences utilizing the correlated motion of coupled heteronuclei in the transverse plane of the doubly rotating frame. J. Magn. Reson. 1983; 52:81–117. [Google Scholar]
- 40. Salzmann M., Pervushin K., Wider G., Senn H., Wüthrich K.. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eletsky A., Kienhöfer A., Pervushin K.. TROSY NMR with partially deuterated proteins. J. Biomol. NMR. 2001; 20:177–180. [DOI] [PubMed] [Google Scholar]
- 42. Bax A., Clore G.M., Gronenborn A.M.. 1H⋅1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 1990; 88:425–431. [Google Scholar]
- 43. Kay L.E., Xu G.Y., Singer A.U., Muhandiram D.R., Formankay J.D.. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B. 1993; 101:333–337. [Google Scholar]
- 44. Yamazaki T., Forman-Kay J.D., Kay L.E.. Two-dimensional NMR experiments for correlating carbon-13.beta. and proton.delta./.epsilon. chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993; 115:11054–11055. [Google Scholar]
- 45. Clore G.M., Kay L.E., Bax A., Gronenborn A.M.. Four-dimensional carbon-13/carbon-13-edited nuclear Overhauser enhancement spectroscopy of a protein in solution: application to interleukin 1.beta. Biochemistry. 1991; 30:12–18. [DOI] [PubMed] [Google Scholar]
- 46. Vuister G.W., Clore G.M., Gronenborn A.M., Powers R., Garrett D.S., Tschudin R., Bax A.. Increased resolution and improved spectral quality in four-dimensional 13C/13C-separated HMQC-NOESY-HMQC spectra using pulsed field gradients. J. Magn. Reson. B. 1993; 101:210–213. [Google Scholar]
- 47. Hyberts S.G., Milbradt A.G., Wagner A.B., Arthanari H., Wagner G.. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012; 52:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hyberts S.G., Arthanari H., Robson S.A., Wagner G.. Perspectives in magnetic resonance: NMR in the post-FFT era. J. Magn. Reson. 2014; 241:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tange O. GNU parallel: the command-line power tool. The USENIX Magazine. 2011; 42–47.Feb. 2011. [Google Scholar]
- 50. Fesik S.W., Zuiderweg E.R.P.. Heteronuclear three-dimensional nmr spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson. 1988; 78:588–593. [Google Scholar]
- 51. Marion D., Kay L.E., Sparks S.W., Torchia D.A., Bax A.. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J. Am. Chem. Soc. 1989; 111:1515–1517. [Google Scholar]
- 52. Ikura M., Kay L.E., Tschudin R., Bax A.. Three-dimensional NOESY-HMQC spectroscopy of a 13C-labeled protein. J. Magn. Reson. 1990; 86:204–209. [Google Scholar]
- 53. Muhandiram D.R., Farrow N.A., Xu G.Y., Smallcombe S.H., Kay L.E.. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O. J. Magn. Reson. B. 1993; 102:317–321. [Google Scholar]
- 54. Zwahlen C., Legault P., Vincent S.J.F., Greenblatt J., Konrat R., Kay L.E.. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997; 119:6711–6721. [Google Scholar]
- 55. Macura S., Ernst R.R.. Elucidation of cross relaxation in liquids by two-dimensional N.M.R. spectroscopy. Mol. Phys. 1980; 41:95–117. [Google Scholar]
- 56. Kumar A., Ernst R.R., Wüthrich K.. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980; 95:1–6. [DOI] [PubMed] [Google Scholar]
- 57. Thrippleton M.J., Keeler J.. Elimination of zero-quantum interference in two-dimensional NMR spectra. Angew. Chem. Int. Ed. 2003; 42:3938–3941. [DOI] [PubMed] [Google Scholar]
- 58. Braunschweiler L., Ernst R.R.. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983; 53:521–528. [Google Scholar]
- 59. Shaka A.J., Lee C.J., Pines A.. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Reson. 1988; 77:274–293. [Google Scholar]
- 60. Pervushin K., Riek R., Wider G., Wüthrich K.. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yao L., Ying J., Bax A.. Improved accuracy of 15N–1H scalar and residual dipolar couplings from gradient-enhanced IPAP-HSQC experiments on protonated proteins. J. Biomol. NMR. 2009; 43:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hansen M.R., Mueller L., Pardi A.. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Mol. Biol. 1998; 5:1065–1074. [DOI] [PubMed] [Google Scholar]
- 63. Delaglio F., Grzesiek S., Vuister G., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 64. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct. Funct. Bioinform. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 65. Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 2008; 3:679. [DOI] [PubMed] [Google Scholar]
- 66. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T.E., Nilges M.. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007; 23:381–382. [DOI] [PubMed] [Google Scholar]
- 67. Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 68. Shen Y., Bax A.. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013; 56:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bhattacharya A., Tejero R., Montelione G.T.. Evaluating protein structures determined by structural genomics consortia. Proteins: Struct. Funct. Bioinform. 2007; 66:778–795. [DOI] [PubMed] [Google Scholar]
- 70. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 71. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pierce M.M., Raman C.S., Nall B.T.. Isothermal titration calorimetry of protein–protein interactions. Methods. 1999; 19:213–221. [DOI] [PubMed] [Google Scholar]
- 73. Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S.. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010; 1:100. [DOI] [PubMed] [Google Scholar]
- 74. Tugarinov V., Kay L.E.. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003; 125:13868–13878. [DOI] [PubMed] [Google Scholar]
- 75. Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T.. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013; 502:519–523. [DOI] [PubMed] [Google Scholar]
- 76. Abshiru N., Ippersiel K., Tang Y., Yuan H., Marmorstein R., Verreault A., Thibault P.. Chaperone-mediated acetylation of histones by Rtt109 identified by quantitative proteomics. J. Proteomics. 2013; 81:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fillingham J., Recht J., Silva A.C., Suter B., Emili A., Stagljar I., Krogan N.J., Allis C.D., Keogh M.-C., Greenblatt J.F.. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 2008; 28:4342–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sutton A., Shia W.-J., Band D., Kaufman P.D., Osada S., Workman J.L., Sternglanz R.. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 2003; 278:16887–16892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.