Abstract
Protein-mediated DNA looping is ubiquitous in chromatin organization and gene regulation, but to what extent supercoiling or nucleoid associated proteins promote looping is poorly understood. Using the lac repressor (LacI), a paradigmatic loop-mediating protein, we measured LacI-induced looping as a function of either supercoiling or the concentration of the HU protein, an abundant nucleoid protein in Escherichia coli. Negative supercoiling to physiological levels with magnetic tweezers easily drove the looping probability from 0 to 100% in single DNA molecules under slight tension that likely exists in vivo. In contrast, even saturating (micromolar) concentrations of HU could not raise the looping probability above 30% in similarly stretched DNA or 80% in DNA without tension. Negative supercoiling is required to induce significant looping of DNA under any appreciable tension.
INTRODUCTION
Protein-mediated DNA looping is a ubiquitous feature of gene regulation and other DNA transactions (1–4). The paradigmatic lac repressor (LacI)-mediated loop inhibits the expression of the lac operon in Escherichia coli (5), and the looping probability is set by LacI tetramer concentration, binding site affinities, and the loop size (6,7). Indeed, loops secured by a single LacI tetramer bridging two binding sites (operators) are suppressed when separate LacI tetramers occupy both operators, or when either the stiffness of short, or entropy of long, intervening DNA overwhelms the free energy of protein binding (8,9). Thus, ∼500 bp-long DNA segments most easily form LacI-mediated loops (9) at intermediate LacI concentrations (7,10).
In vivo, bacterial DNA is extensively decorated and configured by nucleoid associated proteins (NAPs), including the abundant histone-like U (HU) protein (11), which compacts DNA into a flexible HU-DNA filament in 150–200 mM salt (12). HU potently changes the bacterial transcriptome (11), and binding correlates with negative supercoiling in stationary phase E. coli (13). 500 nM or more HU also drives LacI-mediated looping of ∼140 bp DNA to maxima without altering the binding of LacI (14).
Supercoiling has been also shown to dynamically coordinate the transcription of bacterial (15,16) as well as eukaryotic genes (17,18), and the overall level of supercoiling is an indicator of cell health (19,20). Supercoiling is widely and inhomogeneously distributed in the bacterial chromosome, being generated by the binding of nucleoid-associated proteins, such as HU (21,22), and by various DNA transactions (13,23). Negative supercoiling stabilizes regulatory DNA loops, for example, those that repress the lytic genes of bacteriophage λ (24,25) or the gal or lac operons (26,27). Such loops act as barriers to supercoiling diffusion and confine supercoiling density (25,28) as part of biochemical interplay in which supercoiling promotes looping and loops constrain stabilizing superhelical density.
Small and large loops have different energy barriers to formation, but supercoiling and nucleoid-associated proteins may enhance looping in both cases. They can be distinguished by comparing the loop segment to the persistence length of DNA, ∼150 bp (∼50 nm). For small loops on the order of a persistence length or less, energy is required to kink or bend the short, stiff segment of DNA and juxtapose DNA segments for looping, as well as to twist the intervening DNA segment to orient both DNA binding sites toward the protein that joins them. Supercoiling can modify the twist of DNA to minimize the difference in rotational orientations of two binding sites about the helical axis (29). In this way a small, bidentate protein can simultaneously bind two sites on the same face of the helix to produce a loop. Several reports in the literature have shown that the probability of forming loops oscillates as a function of the rotational offset determined by the contour length between the binding sites (30–32). This may be important for loops between the O1 and O3 operators of the lac operon in E. coli, which are separated by a torsionally rigid 92 bp segment, and may be relevant to observations of the supercoiling-driven modulation of in vivo repression of a reporter gene by protein-mediated DNA loops 70–85 bp-long (33). As shown previously, supercoiling-induced and HU-stabilized kinks likely bent the segment and optimized the orientation of the binding sites enhanced the formation of the 113 bp loop mediated by the gal repressor (27). Thus, there is clear evidence that HU and supercoiling may enhance the formation of short loops, whether or not the mechanism involves plectonemes.
For long, flexible loops, the foremost barrier is the separation between the binding sites (34). This is especially critical in DNA under tension which extends the molecule and favors twist over writhe, which opposes juxtaposition of the protein binding sites for looping. Proteins like RNA polymerase intermittently translocate DNA, creating as much as 20 pN of tension before stalling (35). This intermittent activity will modulate the twist/writhe equilibrium and thereby the opportunity for proteins like the lac repressor to secure loops. As will be shown below, supercoiling-induced plectonemes draw binding sites together to promote looping. The O2 and O1 operators of the lac operon in E. coli are separated by a 401 bp. The intrinsic flexibility of such a long span relieves flexural and torsional strain and randomizes the orientations of juxtaposed binding sites. Nevertheless, in vitro experiments from thirty years ago on similarly long segments showed that supercoiling enhanced looping (36). Those experiments which did not include protein factors, like HU, that might modify looping in vivo, did not lead to a mechanical view of how supercoiling enhances the looping of long segments and whether accessory proteins can produce similar effects.
More recently, in vivo levels of LacI-mediated looping were found to be higher than those measured in vitro for a range of large loop sizes in identical DNA templates (7). To determine whether NAPs and/or supercoiling were likely to have caused the differences, the probabilities of LacI-mediated DNA loops in single DNA molecules have been measured as a function of either the superhelicity of the tether, or the concentration of HU for DNA molecules under little or no tension. Both parameters progressively decreased the fractional extension of DNA under 0.25 pN of tension and enhanced looping, but only supercoiling drove the formation of stable loops in DNA under higher tension.
MATERIALS AND METHODS
Preparation of DNA constructs
All DNA fragments for TPM experiments were amplicons of PCR reactions with plasmid DNA pYY_I1_400_BstEII (37) or pO1O2 (26)) as templates with dNTPs (Fermentas-Thermo Fisher Scientific Inc., Pittsburgh, PA, USA), digoxigenin- and biotin-labeled primers (Integrated DNA Technologies, Coralville, IA, or Invitrogen, Life Technologies, Grand Island, NY, USA) (Supplementary Table S1) and Taq Polymerase (New England BioLabs, Ipswich, MA, USA). The total lengths of the DNA constructs Os-400-O1 and O1-400-O2 were 909, and 831 bp respectively with centrally located, 400 bp, LacI-inducible loops as diagramed in Supplementary Figure S1.
DNA tethers for magnetic-tweezers experiments were generated by ligating a 2115 bp main construct containing the O1-400-O2 sequence with a centrally positioned, 400 bp, LacI-inducible loop (Supplementary Figure S1), to ∼1 kb digoxigenin- and biotin- labeled DNA fragments at opposite ends with T4 DNA ligase (New England BioLabs, Ipswich, MA, USA). This procedure has been used previously to produce similar, torsionally restrained DNA tethers (38). The main construct was a PCR amplicon produced with an equimolar dNTP mix and primers including ApaI or XmaI restriction digest sites. The ∼1 kb biotin- or digoxigenin-labeled DNA fragments for anchorage were generated by ApaI or XmaI restriction digests of ∼2 kb PCR amplicons produced using dATP, dCTP, dGTP, dTTP (Fermentas-Thermo Fisher Scientific Inc., Pittsburgh, PA, USA) and digoxigenin-11-dUTP (Roche Life Science, Indianapolis, IN, USA) or biotin-11-dUTP (Invitrogen, Life Technologies, Grand Island, NY, USA) in a molar ratio of 1:1:1:0.9:0.1. After digestion and purification with silica-membrane-based kits (Qiagen, Germantown, MD, USA), the main and the anchorage fragments were ligated with T4 ligase. Random, 10% incorporation of the biotin- and digoxigenin-dUTP labels in the sequences bordering the ApaI and XmaI restriction sites in the pBluescriptKS(+) template used here, will place the first label from 6 to 71 bp away from the ligation junctions to the anchorage fragments in 95% of the molecules (Supplementary Figure S2). Therefore, the total length variation in the DNA tether due to labeling will not exceed 14–127 bp (38 nm). Multiple labels along the anchorage segments is necessary to torsionally restrain the DNA tethers.
Microchamber preparation
Microchambers were prepared similarly to what has been previously described (25,39). In brief, a microchamber with ∼30 μl volume was created between two glass coverslips (Fisherbrand, Thermo Fisher Scientific, Waltham, MA, USA) separated by a parafilm gasket with a narrow inlet and outlet to reduce evaporation of the reaction buffer, and a wide, central observation area (40). DNA tethers were attached through a digoxigenin at one end to the chamber bottom passivated with anti-digoxigenin (Roche Life Science, Indianapolis, IN, USA) and at the other end to either a 320-nm-diameter streptavidin-coated polystyrene bead (Spherotech, Lake Forest, IL, USA) for TPM experiments or a 1.0-μm–diameter streptavidin-coated paramagnetic bead (Dynabead MyOne Streptavidin T1, Invitrogen, Life Technologies, Grand Island, NY, USA) for magnetic tweezing. The chamber was filled with a 10 mM Tris–HCl (pH 7.4), 200 mM KCl, 0.5 mg/ml α-casein buffer and stored in a sealed box to maintain high humidity at 4°C. Before use the chamber was flushed with 200 μl of λ buffer (10 mM Tris–HCl (pH 7.4), 200 mM KCl, 5% DMSO, 0.1 mM EDTA, and 0.1 mg/ml α-casein).
Tethered particle motion (TPM) experiments
All LacI and HU titration TPM experiments were conducted in λ buffer at room temperature. For HU titration experiments, the concentrations of LacI were fixed at 5 or 2.5 nM for Os-400-O1 and O1-400-O2 DNA respectively, to give ∼25% looping probabilities without HU (Supplementary Figure S3).
A Leica DM LB-100 microscope (Leica Microsystems, Wetzlar, Germany) with an oil-immersion objective (63×, NA 0.6–1.4) and differential interference contrast was used to observe tethered beads through a CV-A60 video camera (JAI, Copenhagen, Denmark). The beads were tracked at 50 Hz using custom Labview (National Instruments, Austin, TX, USA) software, to record a time series of absolute XY positions of each bead. Vibrational or mechanical drift in the position of each tethered bead was removed by subtracting the position of one, or the average position of multiple, stuck bead(s) within the same field of view (41,42). The DNA excursion for each was calculated as 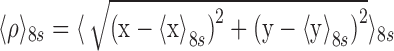 with (x, y) representing the momentary position of a given bead and
with (x, y) representing the momentary position of a given bead and 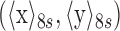 representing an eight second moving average, effectively the anchor point of the bead. Changes in the extent of the motion, ‘excursion’, reflect conformational transformations of the tethered molecule (39,42,43).
representing an eight second moving average, effectively the anchor point of the bead. Changes in the extent of the motion, ‘excursion’, reflect conformational transformations of the tethered molecule (39,42,43).
Then, beads that exhibited clouds of (x, y) positions for which the ratio of the major and minor axes of an elliptical fitting was greater than 1.07, were discarded, since they were likely to have had multiple DNA tethers. Furthermore, beads with anomalously low excursion values were discarded to exclude tethers which became frequently stuck on chamber surface or did not exhibit the entire free range of motion. Seventy-four percent of the beads recorded exhibited symmetrical motion of the proper magnitude and were included in looping analyses (Supplementary Table S2).
The time-series data for selected beads from the same experimental condition were pooled to generate histograms of the observed excursions (Supplementary Figure S4). Each histogram was fit with three Gaussian distributions, representing the excursion values of one unlooped and two looped states mediated by LacI. Looping probabilities for different conditions were calculated by dividing the area under Gaussian distribution of looped states by the total area under all three Gaussians.
Magnetic tweezer experiments
A magnetic tweezer (MT) was used to supercoil DNA and to assay the dynamics of LacI-mediated loop formation. The MT consists of permanent magnets that can be translated along and rotated about the optical axis of the microscope to vary the strength of a laterally oriented magnetic field in the microchamber (44,45). Single DNA tethers were identified through extension vs. twist measurements. Under a high tension of 2 pN, overwound DNA adopts a plectonemic form that smoothly reduces the DNA extension. In contrast, nicked DNA swivels about single bonds to relax any applied torsion, fails to form plectonemes, and the extension does not decrease. Stretching DNA tethers under high tension (∼2 pN) extends a 2115 bp DNA template to ∼0.7 μm.
The x, y, z and t coordinates of tethered bead and at least one non-specifically stuck bead were recorded using a custom MatLab routine at 10 Hz. Three-minute recordings under three levels of tension (0.25, 0.45 or 0.75 pN) and a series of twist settings were measured to determine the DNA extension in each condition before adding LacI protein. After adding 1 nM LacI protein, x, y, z and t data were recorded for 20 min at selected tension and twist settings. The DNA extension versus time data were then analyzed to identify probable looping events. A custom MatLab ‘change point’ algorithm followed by an expectation-maximization routine (46,47) was applied to determine the duration of looped (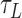 ) and unlooped (
) and unlooped (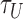 ) states, and the looping probability was calculated as:
) states, and the looping probability was calculated as: 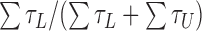 . The free energy change was calculated using
. The free energy change was calculated using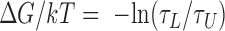 .
.
Estimation of the HU concentration in an E. coli cell
Escherichia coli is a rod-shaped bacterium measuring ∼1 μm in diameter and 2 μm in length. The volume of a cylindrical E. coli cell is therefore
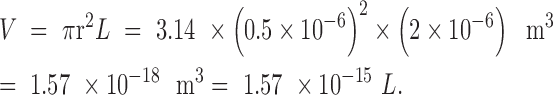 |
The number of HU proteins in an E. coli cell (in logarithmic phase) has been estimated to range between 30 000 and 55 000 copies (48,49). The number of moles of HU protein in an E. coli cell with 50 ∼000 copies equals
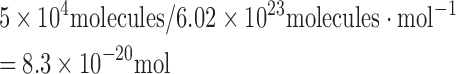 |
and the concentration of HU protein 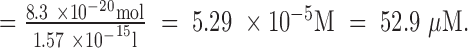 Thus, the concentration range of HU protein is ∼30–60 μM.
Thus, the concentration range of HU protein is ∼30–60 μM.
RESULTS
Looping probability depends on operator affinity
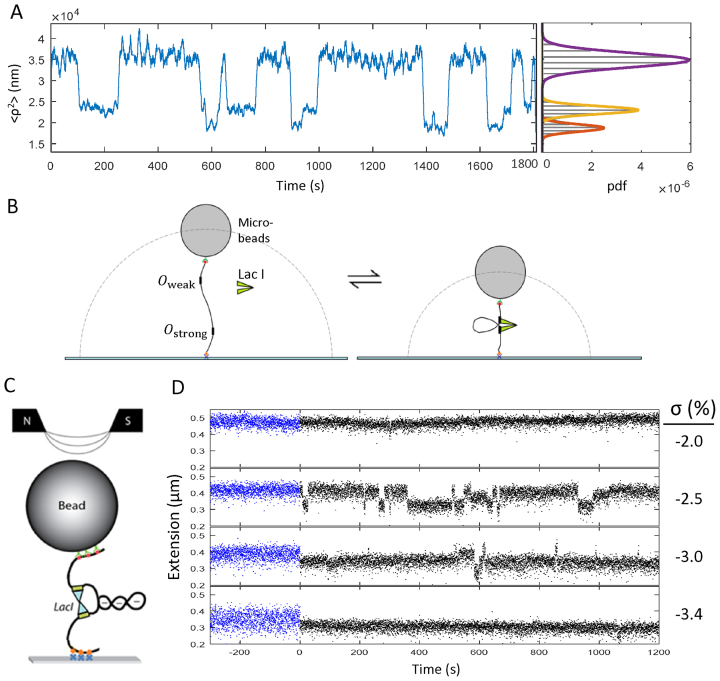
In tethered particle microscopy (TPM), the amplitude of the Brownian motion of a bead tethered by a single DNA molecule to the glass surface of a microscope flow-chamber is monitored over time. A typical experimental recording from TPM assays for two different constructs, Os-400-O1 and O1-400-O2 (Supplementary Figure S1) exhibited intermittent switching of the excursions of beads between one high and two low levels (Figure 1A), as reported for previous measurements of LacI-mediated looping (Figure 1B) (7,50,51). Multiple recordings from each condition were analyzed as described in the methods to determine the fractions of time spent in the looped and unlooped states.
Figure 1.
Experimental schematics and representative data. (A) A representative recording of mean squared excursion versus time shows intermittent changes in the average excursion of the bead as LacI-induced looping changes the overall tether length in a 909 bp Os-400-O1 DNA tether. At right, a histogram of the excursion values for the recordings exhibits three peaks which represent the two looped states and the unlooped state fitted with Gaussian curves, shown in red, yellow, and purple respectively. Similar data were recorded for 831-bp-long O1-400-O2 tethers (not shown). (B) A schematic representation of tether length changes due to LacI-induced DNA looping during a TPM experiment (not to scale). (C) A schematic representation of writhe created using a magnetic tweezer and trapped within a LacI-mediated loop (not to scale). Red/green dots indicate biotin/streptavidin linkages of DNA to beads. Orange diamonds/blue crosses indicate digoxigenin/antidigoxigenin linkages of DNA to glass. O1-400-O2 DNA looped by LacI is shown to trap three negative (-) supercoils. The north and south poles of magnets above the microscope stage create a magnetic field to attract and rotate the bead to allow stretching and twisting of the DNA. (D) Recordings of the extension of a 2115 bp-long O1–400-O2 DNA tether under 0.45 pN of tension at different levels of negative supercoiling in the absence (blue) and in the presence (black) of LacI. Extended (unlooped) states dominate at low negative linking number values but become intermittent and finally disappear altogether as negative supercoiling increases and favors the looped state.
At a LacI concentration of 0.1 nM, the looping probability in Os-400–O1 tethers reached a maximum of almost 90% versus only 45% for O1-400-O2 tethers (Supplementary Figure S3), indicating that the operator affinity affected the maximum looping probability as observed previously for smaller loops (32). Higher concentrations of LacI decrease the looping probability as both of the operator sites become occupied by separate LacI tetramers. We chose concentrations that produced 25% looping probabilities, so that changes in looping by increased HU concentration, or supercoiling, would have a measurable effect.
HU decreased tether lengths and promoted LacI-mediated looping
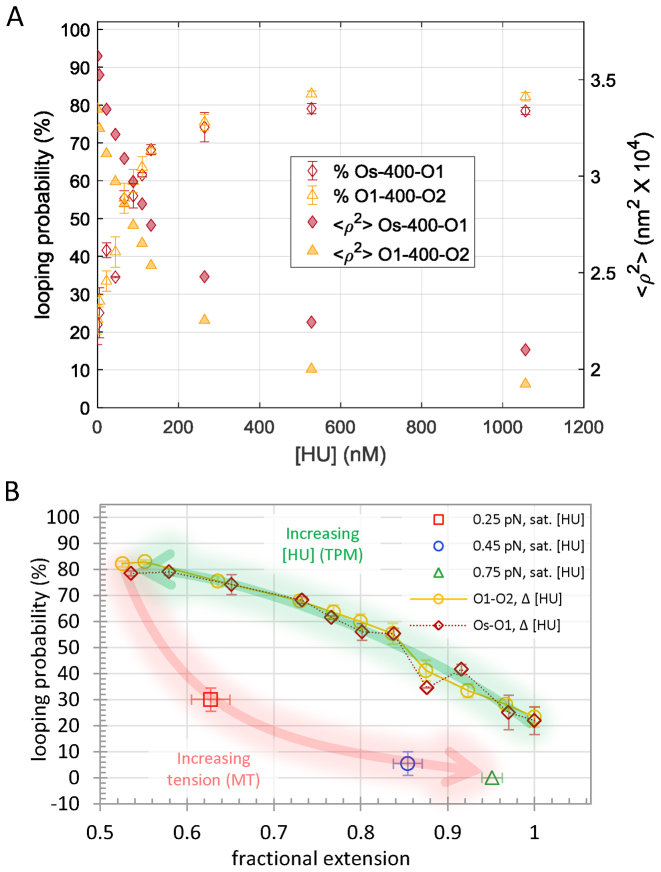
After establishing a concentration of LacI to set a baseline level of looping, increasing amounts of the HU protein were added to determine the effect on looping. In previously published magnetic tweezer experiments, the salt concentration affected the binding of HU to DNA (12). At monovalent salt concentrations in the range between 150 and 200 mM, increasing the HU concentration produced compaction and increased the flexibility of extremely long, 48 kbp, DNA tethers (12). At least 500 nM, HU also contracted 445 bp DNA tethers by 7% and drove LacI-mediated looping of short, roughly 140 bp, DNA segments to maximum levels without altering the binding of LacI (14). To test whether or not HU similarly affects looping of DNA segments longer than a persistence length, the looping probability was measured as HU protein was titrated from 0 to ∼1 μM, a high concentration yet still below maximum estimates for exponentially proliferating E. coli (∼ 50 μM, see materials and methods; (48)). For Os-400-O1 or O1-400-O2 tethers in the presence of LacI, excursions of beads, <ρ2>, tethered by both the looped and unlooped DNA decreased as the HU concentration increased (Figure 2A, Supplementary Figure S4). To establish the associated looping probabilities, combinations of three Gaussian curves were fit to histograms of the excursion values aggregated from recordings in identical conditions (Supplementary Figure S4). As [HU] increased, the DNA tethers contracted to reduce excursions of the attached beads. The fractional extension of the unlooped tether contracted more than the looped tethers (Supplementary Figure S5). Since HU should bind randomly along the tether, this is most likely due to the greater contraction of centrally located, HU-induced bends compared to those in the flanking ends. This effect has been used previously in circular permutation assays to discover the location and extent of protein-induced bends in DNA segments (52). The looping probability was calculated as the area of the Gaussians for the looped states divided by the area of the combination of three Gaussians representing all the states. As HU gradually contracted the DNA tether and decreased excursions, the looping probability increased from 25 to a plateau near 80% (Figure 2A). Apparent contour lengths of the DNA tethers were determined from measurements of the mean square excursions of beads (Supplementary Table S3), which in λ buffer are related to the contour length of the DNA tether by: <ρ2> = 100.89 × L + 3445 (42). Looping probabilities were plotted as a function of the fractional extension at a given HU concentration to that without HU, 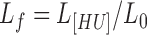 . Figure 2B shows that HU shifted the looped/unlooped equilibrium to 80% looped when the fractional extension of the DNA dropped by 50%.
. Figure 2B shows that HU shifted the looped/unlooped equilibrium to 80% looped when the fractional extension of the DNA dropped by 50%.
Figure 2.
HU promotes looping. (A) Increasing HU concentration progressively decreased the mean squared extension values of Os-400-O1 or O1-400-O2 tethers from 100 to 50% (right hand Y-axis) simultaneously driving the looping probability from 25 to 80% (left hand Y-axis). (B) Plotting the looping probability as a function of the HU-modified extension of the DNA reveals an inverse relationship for DNA without tension. Application of slight tension reversed the HU-induced contraction and lowered the looping probability. The lower looping probability of tethers under tension in a magnetic tweezer may reflect a lower probability of the tether to adopt conformations with writhe when the bead was rotationally constrained.
Applying tension opposed the HU-driven contraction restoring the extended state and reducing the looping probability. At 1 μM HU concentration, which produced 80% looping in DNA without tension, 0.25, 0.45 or 0.75 pN of tension reduced the probability of looping to 30, 5 or 0% (Figure 2B). Although the contraction that was induced by HU (Figure 2B, green arrow) was essentially reversed by tension (Figure 2B, red arrow), for a given extension, the percentage of looping depended on the concentration of HU and the tension. Thus, the induction of looping by HU can be reversed with tension and appears to be related to the contraction of DNA. A reduction of the overall extension of the DNA does not indicate that the DNA is shorter, because the contour length is unchanged. It indicates random coiling. We have compared the extension of rotationally unconstrained DNA to the looping probability, because coiling creates juxtaposed binding sites that can be linked by the lac repressor to form loops. The HU protein stabilizes large bends in DNA and may induce coiling in rotationally free DNA tethers like those in the TPM experiments described in the manuscript. However, even slight tension opposes coiling and reverses the enhancement of looping by HU. The looping probability drops sharply with slight tension, so that a DNA tether that is partially saturated with HU exhibits a much higher probability of looping without tension than a similarly extended, HU-saturated DNA tether under 0.25 pN of tension. The difference is that HU binding induces kinks in a DNA tether under no tension that reduce the overall extension by 45% and promote random coiling. Slight tension extends the kinked tether which reduces the frequency of juxtaposition of binding sites required for lac repressor to secure a loop.
Supercoiling decreased tether lengths and induced LacI-mediated looping
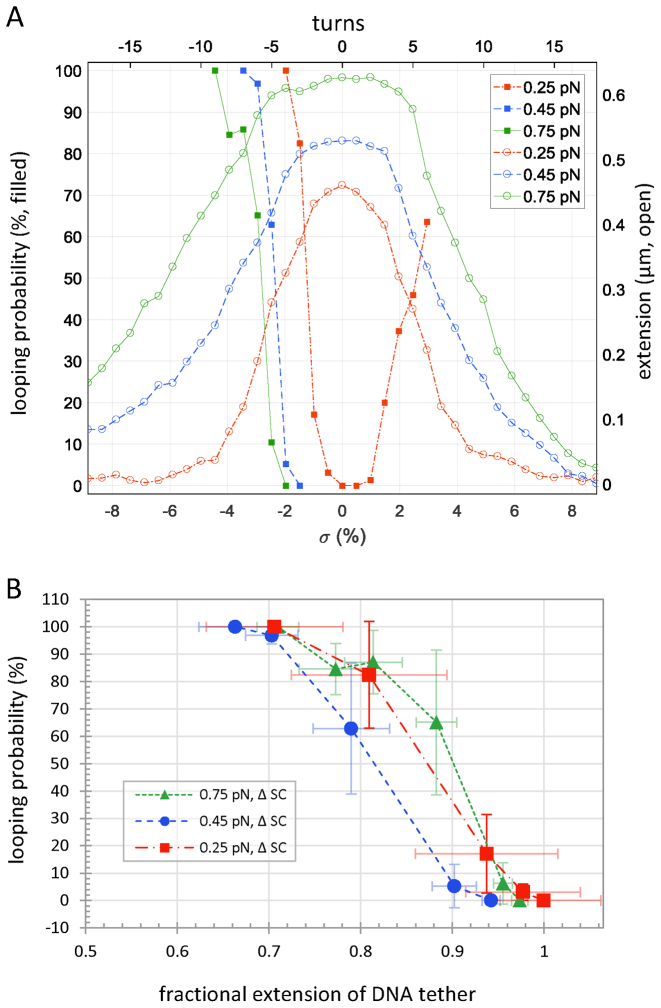
Supercoiling can also contract DNA when the torsional strain exceeds the buckling threshold and the molecule forms plectonemes. Supercoiling was recently shown to enhance λ repressor (CI)-mediated looping in both linear and circular DNA (24,25). It influences gene expression (53,54) and is a ubiquitous feature of genomes. To quantitatively characterize the impact of supercoiling on LacI-mediated looping, the probability of the looped state was characterized while gently stretching and twisting 2115 bp DNA tethers using a magnetic tweezer (see schematic in Figure 1C). Experiments on tethers with a centrally located O1-400-O2 loop sequence (Supplementary Figure S1) were carried out at 0.25, 0.45 and 0.75 pN, in the presence of 1 nM LacI. No looping occurred without LacI (Figure 1D, blue recordings) or with LacI acting on torsionally relaxed tethers under tension (Figure 3A), because such tension precludes juxtaposition of operators separated by 400 bp, ∼130 nm, along the tether (55,56). However, superhelical densities between –1.9 and –3.4% induced switching between looped and unlooped states (Figure 1D, black recordings). Under low tension, DNA tethers buckle randomly to form plectonemes that rapidly slither or hop to different locations along the length (57). LacI has the opportunity to secure a loop when the binding sites slither into juxtaposition. In this conformation plectonemic supercoils are constrained inside the loop. Dwell times in the looped and unlooped states were calculated as described above to determine the looping probabilities shown in Figure 3A. Loops persisted several tens of seconds as reported previously (58,59) and significantly longer than those of the extended and plectonemic states observed as a twisted DNA tether begins to buckle (60).
Figure 3.
Supercoiling promotes looping. (A) No looping was observed in tethers under slight tension in a magnetic tweezer. However, over- or underwinding O1-400-O2 DNA tethers under 0.25 pN of tension produced plectonemes that reduced extension (open red circles) and increased the looping probability (filled red squares). Higher tensions (green and blue data) extended the tether further and required more negative supercoiling to produce comparable looping. For each tension, supercoiling decreased the extension and drove the looping probability from 0 to 100%! (B) Although achieving equivalent looping probabilities required more negative supercoiling in tethers under higher tension, the looping probabilities as a function of fractional extension were similar for the three different levels of tension tested. When the supercoil-induced plectoneme contracted the tether to 90–95% of the extended length, a loop intermittently formed and became the dominant conformation as further unwinding reduced the extension to 70%.
Supercoil-driven, LacI-mediated looping exhibited several noteworthy features. First, at each tension, looping probability increased from 0 to 100% as superhelical density became more negative (Figure 3A). The superhelical density of the tethers is σ = (Lk – Lk0)/Lk0 = n/Lk0,where Lk and Lk0 are the linking numbers of twisted and relaxed DNA respectively, and n is the number of mechanically introduced twists. Lk0 is given by the total length in base pairs divided by 10.4 bp per turn. Additional superhelicity lowered the free energy, ΔG, for looping at each tension (Supplementary Figure S6) and negative superhelicity was more effective than positive. Second, the free energy change for LacI-induced looping as a function of supercoiling was compared with data for the formation of a 393 bp DNA loop by the λ repressor protein (CI) (25), which secures looping via cooperative interactions amongst multiple dimers. Negative supercoiling decreased the free energy for LacI- more than for CI-mediated looping (Supplementary Figure S6). These free energy measurements support theoretical investigations indicating that the free energy change for 400 bp LacI-mediated looping in vivo is –10 kcal/mol (-4.4 kT) or less (61). Third, overlay of the looping probability versus twist measurements with the DNA extension versus twist curves (Figure 3A), revealed that the onset of loop formation induced by negative supercoiling coincided with the onset of plectoneme formation. Indeed, in DNA tethers under tension, plectoneme formation was essential for looping, and more extreme (negative) supercoiling densities were required at higher tensions to achieve similar looping probabilities.
The coincidence of the initial increase in looping probability and plectoneme formation suggested that contraction juxtaposes binding sites for LacI looping. Graphically comparing the contraction of a DNA tether and the associated LacI-mediated looping probability shows that the percentage of looping resembles the percentage of contractions below a threshold that is one loop length shorter than the full-length, unlooped tether, such as that produced when LacI secures a loop. After introducing -2% negative supercoiling during an extension-vs.-twist experiment at 0.45 pN, momentary tether lengths (Supplementary Figure S7 grey points) occasionally dip below a level that is one loop segment shorter (Supplementary Figure S7 red dashed line) and the looping probability begins to rise. With additional negative supercoiling, more measurements fall below that level and the looping probability increases, finally reaching 100%. This is also reflected in extension versus time records which display increases in the frequency and duration of intermittent looping as a DNA tether is negatively supercoiled (Figure 1D), until the looped state becomes perpetual.
If contraction of the DNA were indeed the mechanism by which supercoiling induced looping, then it would be expected to dictate the looping probability independently of the tension. To quantitatively examine this relationship, the fractional extension was defined as the tether extension at a given supercoiling density divided by that of torsionally relaxed DNA, Lf= Lσ/L0. In Figure 3B, plots of the looping probability associated with different levels of negative supercoiling at three levels of tensions are shown. The looping probability began to rise after supercoiling induced contraction to 0.9–0.95 of the torsionally relaxed extension. Thereafter, the looping probability rapidly increased, and tethers contracted to 0.7 of the torsionally relaxed extension were 100% looped for all three levels of tension tested.
DISCUSSION AND CONCLUSION
Contraction, especially with writhe, enhances loop formation
The conformation of DNA stretched and twisted to the buckling point under different tensions rapidly fluctuates between plectonemic and extended conformations (60). Comparison of the onset of loop formation in Os-400-O1 DNA constructs under different tensions (Figure 3B), shows that LacI-mediated loops begin to form just beyond the buckling threshold at which plectonemes form. The contraction of DNA caused by the initial plectonemic gyre has been studied as a function of tension using an optical tweezer with high temporal resolution. This gyre of DNA is about 130 nm-long under 0.45 pN of tension (60). The LacI-induced loop is similarly sized, and this coincidence indicates that lac repressor captures and stabilizes looped configurations produced by writhe. Indeed, Supplementary Figure S7 shows that the looping probability correlates with the probability that a tether exhibits an instantaneous extension with at least one loop length of slack. This is the only way in which LacI, which is not a DNA translocase, can establish DNA loops.
HU also seems to promote looping by reducing the fractional extension. It is noteworthy that saturating HU promoted looping more effectively in DNA tethers without tension. Although equivalent levels of contraction were produced in tethers exposed to various concentrations of HU without tension and other tethers exposed to saturating levels of HU and varying levels of tension, the looping probabilities observed for tethers under tension were much lower (Figure 2B). This is likely due to the fact that tension opposes juxtaposition of the protein binding sites and looping even in a DNA tether with a saturating level of HU-induced kinks.
Tension is likely to be physiologically relevant for at least three reasons: First, models for the hierarchical packaging of DNA into chromatin include loops ranging from 15 kb segments in Caulobacter crescentus (62) to hundreds of kbps in mammals (63), and supercoiling appears to be critical for these models. The Meiners laboratory has shown that sub-piconewton tensions can prevent the formation of 305 bp loops (34), and even femtonewton-scale tensions would significantly interfere with loop formation in much larger segments and interfere with hierarchical packaging. Indeed, novobiocin treatment of Caulobacter crescentus cells, which inhibits DNA gyrase and lessens negative supercoiling, altered interactions between segments separated by 20–800 kbp (62). Our finding that supercoiling rescues loop formation in DNA under tension, suggests that DNA in vivo is under tension and that supercoiling is required to condense chromatin through plectoneme formation and looping. Second, in vitro experiments have shown that the translocation velocity of RNA polymerase is unaffected by up to 20 pN of opposing tension on the DNA (35), and that the nucleic acid translocation enzyme that packages DNA into the p29 bacteriophage exerts as much as 50 pN of tension on the DNA (64). It is unlikely that these enzymes would have evolved such capabilities unless they operate against mechanical tension. Third, the Belmont and Wang labs have recently demonstrated that the tension exerted by forces applied to microscopic beads attached to the surface of a cell can be transmitted to the nucleus where it draws segments of chromatin apart and simultaneously enhances transcription from genes in the intervening DNA (65). This demonstration of enhanced transcription by stretching chromatin in a live cell suggests that cells in mechanically distorted tissues enduring may respond transcriptively to the stress.
Supercoiling lowers the free energy of DNA looping in a protein-dependent manner
Once the loop closes the DNA is partitioned into different topological domains. Previous work from our lab demonstrates that the lambda CI loop serves as a barrier for supercoiling diffusion (25), and work in this paper shows that LacI behaves similarly. Key features of this interplay are that supercoiling can become sequestered within a loop, but also that supercoiling modulates the looping probability (26). Supplementary Figure S6 is a plot of the energy change between looped and unlooped states under different supercoiling densities. Both positive and negative supercoiling lowered the free energy of LacI-mediated loop formation, but negative supercoiling was more effective. In addition, the response of different loop-mediating proteins, LacI and λ CI negative was different. The free energy dropped more rapidly for LacI-mediated looping as negative supercoiling increased. This may reflect different binding interfaces with DNA, or their structures which may have different tolerances for changes in binding site phasing.
Comparing the effects of HU and supercoiling on DNA looping via the J factor
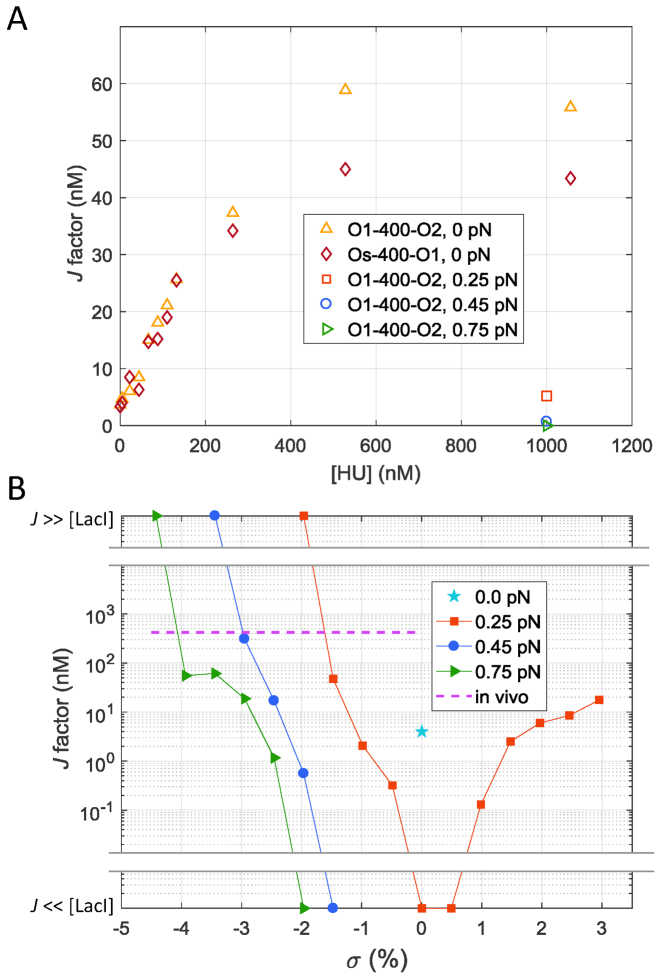
The J factor is a succinct parameter for comparing the effects of HU and supercoiling on DNA looping. It represents the effective concentration driving the binding of a protein at one site along a DNA molecule while simultaneously bound to another site along the same DNA tether. This factor accounts for the energy changes involved in forming a DNA loop independently of the affinity of protein linkages. The J factor can be calculated (10) from the ratio of the unlooped and looped state probabilities, 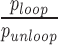 , the dissociation constant from the secondary operator, Kd, and the concentration of lac repressor, [LacI], using the equation
, the dissociation constant from the secondary operator, Kd, and the concentration of lac repressor, [LacI], using the equation 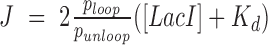 (10). Increasing the HU concentration surrounding torsionally relaxed DNA tethers drove J factors to plateau at 11–15 times the initial values, and slight tension reversed the gains (Figure 4A). In contrast, supercoiling, especially negatively, increased J factors by several orders of magnitude from far below the free LacI concentration (1 nM), for which there was no looping under tension. Negative supercoiling drove the concentration of lac repressor binding sites to extremely high levels corresponding to J factors far greater (effectively infinite) than those reported for a 400 bp loop in vivo (Figure 4B; purple dashed line). This produced perpetual loops which were not observed with HU protein alone even in tethers under no tension.
(10). Increasing the HU concentration surrounding torsionally relaxed DNA tethers drove J factors to plateau at 11–15 times the initial values, and slight tension reversed the gains (Figure 4A). In contrast, supercoiling, especially negatively, increased J factors by several orders of magnitude from far below the free LacI concentration (1 nM), for which there was no looping under tension. Negative supercoiling drove the concentration of lac repressor binding sites to extremely high levels corresponding to J factors far greater (effectively infinite) than those reported for a 400 bp loop in vivo (Figure 4B; purple dashed line). This produced perpetual loops which were not observed with HU protein alone even in tethers under no tension.
Figure 4.
J factors decrease as DNA tethers shorten especially when the DNA becomes negative supercoiled. (A) Increasing the HU concentration surrounding Os-400-O1 (diamonds) or O1-400-O2 (upright triangles) tethers drives the J factors to saturation at multiples of 11–15. Slight tension lowered the HU-improved J factors back toward their original values (left and right-pointing triangles). (B) The J factor measured for O1-400-O2 in TPM experiments (with negligible tension) was 4 nM (cyan star). Slight tension precluded looping (J << [LacI] = 1 nM) until the DNA became sufficiently supercoiled (Figure 1B). Indeed, supercoiling, especially negative, increased J factors by several orders of magnitude to levels (J>> [LacI] = 1 nM) beyond that reported for a 400 bp loop in vivo (purple dashed line).
CONCLUSION
Although HU binding promoted the formation of 400 bp, LacI-mediated loops in DNA molecules without tension, supercoiling was essential to drive significant looping of molecules under slight tension. While direct measurements of tension in genomic DNA have not been reported, the mechanical fact that DNA translocases can operate against as much as tens of pN of tension and the tension sensitivity of looping described above suggest that supercoiling is critical for looping in vivo. Simultaneously, a cellular perspective of the prominent role that protein-mediated loops play in chromatin condensation and transcriptional regulation, and the ubiquitous genetic requirement for topoisomerases that regulate supercoiling, reveal the importance of supercoiling in DNA biochemistry. Indeed, supercoiling directly reflects the current state of cellular metabolism and is a fundamental genomic parameter. Future studies are likely to show that, similarly to proteins that act at very specific sites, supercoiling can be constrained within loops and plectonemes to control dispersion and locally regulate DNA transactions.
Supplementary Material
ACKNOWLEDGEMENTS
Kathleen Matthews generously provided the LacI protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health [GM084070, 1R15GM109254]. Funding for open access charge: National Institutes of Health [GM084070].
Conflict of interest statement. None declared.
REFERENCES
- 1. Brennan L.D., Forties R.A., Patel S.S., Wang M.D.. DNA looping mediates nucleosome transfer. Nat. Commun. 2016; 7:13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hnisz D., Day D.S., Young R.A.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duderstadt K.E., Geertsema H.J., Stratmann S.A., Punter C.M., Kulczyk A.W., Richardson C.C., van Oijen A.M.. Simultaneous real-time imaging of leading and lagging strand synthesis reveals the coordination dynamics of single replisomes. Mol. Cell. 2016; 64:1035–1047. [DOI] [PubMed] [Google Scholar]
- 4. Qian Z., Trostel A., Lewis D.E.A., Lee S.J., He X., Stringer A.M., Wade J.T., Schneider T.D., Durfee T., Adhya S.. Genome-wide transcriptional regulation and chromosome structural arrangement by GalR in E. coli. Front. Mol. Biosci. 2016; 3:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker N.A., Peters J.P., Lionberger T.A., Maher L.J.. Mechanism of promoter repression by Lac repressor–DNA loops. Nucleic Acids Res. 2013; 41:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hao N., Sneppen K., Shearwin K.E., Dodd I.B.. Efficient chromosomal-scale DNA looping in Escherichia coli using multiple DNA-looping elements. Nucleic Acids Res. 2017; 45:5074–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Priest D.G., Cui L., Kumar S., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of the DNA tethering effect in long-range DNA looping in vivo and in vitro using the Lac and λ repressors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mulligan Peter J., Chen Y.-J., Phillips R., Spakowitz Andrew J.. Interplay of protein binding interactions, DNA mechanics, and entropy in DNA looping kinetics. Biophys. J. 2015; 109:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 2001; 26:733–740. [DOI] [PubMed] [Google Scholar]
- 10. Han L., Garcia H.G., Blumberg S., Towles K.B., Beausang J.F., Nelson P.C., Phillips R.. Concentration and length dependence of DNA looping in transcriptional regulation. PLoS One. 2009; 4:e5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kar S., Edgar R., Adhya S.. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16397–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao B., Johnson R.C., Marko J.F.. Modulation of HU–DNA interactions by salt concentration and applied force. Nucleic Acids Res. 2010; 38:6176–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lal A., Dhar A., Trostel A., Kouzine F., Seshasayee A.S., Adhya S.. Genome scale patterns of supercoiling in a bacterial chromosome. Nat. Commun. 2016; 7:11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boedicker J.Q., Garcia H.G., Johnson S., Phillips R.. DNA sequence-dependent mechanics and protein-assisted bending in repressor-mediated loop formation. Phys. Biol. 2013; 10:066005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dorman C.J. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 2013; 41:542–547. [DOI] [PubMed] [Google Scholar]
- 16. Ferrándiz M.-J., Martín-Galiano A.J., Arnanz C., Camacho-Soguero I., Tirado-Vélez J.-M., de la Campa A.G.. An increase in negative supercoiling in bacteria reveals topology-reacting gene clusters and a homeostatic response mediated by the DNA topoisomerase I gene. Nucleic Acids Res. 2016; 44:7292–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D.. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013; 20:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N.. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013; 20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh L.S., Rouviere-Yaniv J., Drlica K.. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J. Bacteriol. 1991; 173:3914–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Workum M., van Dooren S.J., Oldenburg N., Molenaar D., Jensen P.R., Snoep J.L., Westerhoff H.V.. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol. Microbiol. 1996; 20:351–360. [DOI] [PubMed] [Google Scholar]
- 21. Broyles S.S., Pettijohn D.E.. Interaction of the Escherichia coli HU protein with DNA: Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986; 187:47–60. [DOI] [PubMed] [Google Scholar]
- 22. Rouvière-Yaniv J., Yaniv M., Germond J.-E.. E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell. 1979; 17:265–274. [DOI] [PubMed] [Google Scholar]
- 23. Rovinskiy N., Agbleke A.A., Chesnokova O., Pang Z., Higgins N.P.. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012; 8:e1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Norregaard K., Andersson M., Sneppen K., Nielsen P.E., Brown S., Oddershede L.B.. DNA supercoiling enhances cooperativity and efficiency of an epigenetic switch. Proc. Natl. Acad. Sci. U.S.A.. 2013; 110:17386–17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding Y., Manzo C., Fulcrand G., Leng F., Dunlap D., Finzi L.. DNA supercoiling: a regulatory signal for the λ repressor. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fulcrand G., Dages S., Zhi X., Chapagain P., Gerstman B.S., Dunlap D., Leng F.. DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli. Sci. Rep. 2016; 6:19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lia G., Bensimon D., Croquette V., Allemand J.F., Dunlap D., Lewis D.E.A., Adhya S.C., Finzi L.. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11373–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leng F., Chen B., Dunlap D.D.. Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:19973–19978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krämer H., Amouyal M., Nordheim A., Müller-Hill B.. DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988; 7:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du Q., Smith C., Shiffeldrim N., Vologodskaia M., Vologodskii A.. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5397–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker N.A., Kahn J.D., Maher L.J.. Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007; 35:3988–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson S., Lindén M., Phillips R.. Sequence dependence of transcription factor-mediated DNA looping. Nucleic Acids Res. 2012; 40:7728–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mogil L.S., Becker N.A., Maher L.J. III. Supercoiling effects on short-range DNA looping in E. coli. PLoS One. 2016; 11:e0165306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y.F., Milstein J.N., Meiners J.C.. Femtonewton entropic forces can control the formation of protein-mediated DNA loops. Phys. Rev. Lett. 2010; 104:048301. [DOI] [PubMed] [Google Scholar]
- 35. Forde N.R., Izhaky D., Woodcock G.R., Wuite G.J., Bustamante C.. Using mechanical force to probe the mechanism of pausing and arrest during continuous elongation by Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11682–11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whitson P.A., Hsieh W.T., Wells R.D., Matthews K.S.. Influence of supercoiling and sequence context on operator DNA-binding with Lac repressor. J. Biol. Chem. 1987; 262:14592–14599. [PubMed] [Google Scholar]
- 37. Voros Z., Yan Y., Kovari D.T., Finzi L., Dunlap D.. Proteins mediating DNA loops effectively block transcription. Protein Sci. 2017; 26:1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fernández-Sierra M., Shao Q., Fountain C., Finzi L., Dunlap D.. E. coli gyrase fails to negatively supercoil diaminopurine-substituted DNA. J. Mol. Biol. 2015; 427:2305–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ucuncuoglu S., Schneider D.A., Weeks E.R., Dunlap D., Finzi L.. Spies M, Chemla RY. Methods in Enzymology. 2017; 582:Academic Press; 415–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kovari D.T., Yan Y., Finzi L., Dunlap D.. Tethered particle motion: an easy technique for probing DNA topology and interactions with transcription factors. Methods Mol. Biol. 2018; 1665:317–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han L., Lui B.H., Blumberg S., Beausang J.F., Nelson P.C., Phillips R.. Benham CJ, Harvey S, Olson WK, Sumners DWL, Swigon D. Mathematics of DNA Structure, Function and Interactions. 2009; 150:NY: Springer; 123–138. [Google Scholar]
- 42. Kumar S., Manzo C., Zurla C., Ucuncuoglu S., Finzi L., Dunlap D.. Enhanced tethered-particle motion analysis reveals viscous effects. Biophys. J. 2014; 106:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Priest D.G., Kumar S., Yan Y., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of interactions between two DNA loops demonstrates loop domain insulation in E. coli cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E4449–E4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V.. The elasticity of a single supercoiled DNA molecule. Science. 1996; 271:1835–1837. [DOI] [PubMed] [Google Scholar]
- 45. Kriegel F., Ermann N., Lipfert J.. Probing the mechanical properties, conformational changes, and interactions of nucleic acids with magnetic tweezers. J. Struct. Biol. 2017; 197:26–36. [DOI] [PubMed] [Google Scholar]
- 46. Manzo C., Finzi L.. Nils GW. Methods In Enzymology. 2010; 475:Academic Press; 199–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watkins L.P., Yang H.. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B. 2005; 109:617–628. [DOI] [PubMed] [Google Scholar]
- 48. Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A.. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999; 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Azam T.A., Ishihama A.. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999; 274:33105–33113. [DOI] [PubMed] [Google Scholar]
- 50. Johnson S., van de Meent J.W., Phillips R., Wiggins C.H., Linden M.. Multiple LacI-mediated loops revealed by Bayesian statistics and tethered particle motion. Nucleic Acids Res. 2014; 42:10265–10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wong O.K., Guthold M., Erie D.A., Gelles J.. Interconvertible lac repressor-DNA loops revealed by single-molecule experiments. PLoS Biol. 2008; 6:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu H.M., Crothers D.M.. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984; 308:509–513. [DOI] [PubMed] [Google Scholar]
- 53. Matsumoto K., Hirose S.. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J. Cell Sci. 2004; 117:3797–3805. [DOI] [PubMed] [Google Scholar]
- 54. Travers A., Muskhelishvili G.. DNA supercoiling - a global transcriptional regulator for enterobacterial growth?. Nat. Rev. Microbiol. 2005; 3:157–169. [DOI] [PubMed] [Google Scholar]
- 55. Blumberg S., Pennington M.W., Meiners J.C.. Do femtonewton forces affect genetic function? A review. J. Biol. Phys. 2006; 32:73–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen Y.-F., Milstein J.N., Meiners J.-C.. Protein-mediated DNA loop formation and breakdown in a fluctuating environment. Phys. Rev. Lett. 2010; 104:258103. [DOI] [PubMed] [Google Scholar]
- 57. van Loenhout M.T.J., de Grunt M.V., Dekker C.. Dynamics of DNA supercoils. Science. 2012; 338:94–97. [DOI] [PubMed] [Google Scholar]
- 58. Finzi L., Gelles J.. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995; 267:378–380. [DOI] [PubMed] [Google Scholar]
- 59. Vanzi F., Broggio C., Sacconi L., Pavone F.S.. Lac repressor hinge flexibility and DNA looping: single molecule kinetics by tethered particle motion. Nucleic Acids Res. 2006; 34:3409–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Forth S., Deufel C., Sheinin M.Y., Daniels B., Sethna J.P., Wang M.D.. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008; 100:148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saiz L., Vilar J.M.G.. DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 2006; 16:344–350. [DOI] [PubMed] [Google Scholar]
- 62. Le T.B.K., Imakaev M.V., Mirny L.A., Laub M.T.. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013; 342:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dixon Jesse R., Gorkin David U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith D.E., Tans S.J., Smith S.B., Grimes S., Anderson D.L., Bustamante C.. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001; 413:748–752. [DOI] [PubMed] [Google Scholar]
- 65. Tajik A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A.S., Wang N.. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016; 15:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.