Abstract
The position 34 of a tRNA is always modified for efficient recognition of codons and accurate integration of amino acids by the translation machinery. Here, we report genomics features of a deep-sea gut symbiotic Spiroplasma, which suggests that the organism does not require tRNA(34) anticodon modifications. In the genome, there is a novel set of tRNA genes composed of 32 species for recognition of the 20 amino acids. Among the anticodons of the tRNAs, we witnessed the presence of both U34- and C34-containing tRNAs required to decode NNR (A/G) 2:2 codons as countermeasure of probable loss of anticodon modification genes. In the tRNA fragments detected in the gut transcriptome, mismatches expected to be caused by some tRNA modifications were not shown in their alignments with the corresponding genes. However, the probable paucity of modified anticodons did not fundamentally change the codon usage pattern of the Spiroplasma. The tRNA gene profile that probably resulted from the paucity of tRNA(34) modifications was not observed in other symbionts and deep-sea bacteria, indicating that this phenomenon was an evolutionary dead-end. This study provides insights on co-evolution of translation machine and tRNA genes and steric constraints of codon-anticodon interactions in deep-sea extreme environment.
INTRODUCTION
Translation emerged during the early steps of the origin of the first cells, with some RNAs (the future genes/mRNAs) used as templates to direct synthesis of peptides from amino acids loaded on the ancestors of tRNAs. This was still a quite inaccurate operation. The process was carried over on a large RNA-based catalytic structure (the ancestor of the ribosome) (1). Accuracy progressed, with a need to associate a specific amino acid to a specific anticodon. As supportive handles of metabolic pathways (2), ancestral RNAs were not only likely to be loaded with amino acids but also to be the scaffold for a variety of metabolic processes. This resulted in the presence of modified nucleotides at the most exposed regions of the tRNA molecules. In extant living organisms, more than 100 nucleotide modifications tag tRNA molecules (3). Some have evolved to stabilize its tridimensional structure. Indeed tRNA loops are metal and nuclease sensitive, but the most important modifications are involved in maintaining accuracy of codon-anticodon interactions. Among the 64 codons, 61 specify an amino acid in general. Yet, the number of decoding tRNAs is smaller (ranging from 22–46 (4)) because some wobble is allowed between the first base of the anticodon (N34 in standard nomenclature) and the third base of a codon (N3) (5). Decoding accuracy is context-dependent and several amino acids can be decoded by a set of four codons with the third base being any (N) nucleotide. In other cases the 4-codon box is split into several boxes. NAN codons, for example, split into NAR and NAY (R for purine; Y for pyrimidine) doublets. Accuracy of decoding therefore requires that purines and pyrimidines are stringently decoded by the first base (N34) of a specific tRNA anticodon. Surprisingly, recent structural work has shown that there is a very specific obstacle posed by formation of G•U wobble pairs: within the ribosome decoding center they are not symmetrical. The pairing between the first anticodon and the third codon positions G3•U34 is stronger than U34•G3 on the base of isostericity and the need of a Watson-Crick-like geometries within the A-site of the decoding center of the ribosome (6).
Despite this structural constraint, a single tRNA species is used to decode both NAR codons (CAR = Gln; AAR = Lys; UAR = stop; GAR = Glu) in all extant organisms of all three domains of life, but the U34 anticodon position is heavily modified to allow for this situation (7,8). To a lesser ubiquitous extent, the same is true for NAY codons. How is this achieved? Specific modification of base N34 appears to be omnipresent. To recognize NAY codons, G34 is modified by an exchange with a complex 7-deazapurine derivative, queuine (9). However, because the G34•U3 wobble pair in the ribosomal decoding center is still fairly stable, this modification, while extremely widespread, is not ubiquitous (it is generally absent, for example, from Hemiascomycetes and from Mollicutes (4), as well as from a variety of plants such as Arabidopsis). By contrast the U34 modification enables U34-bearing tRNAs to serve as the only acceptor of synonymous codons and therefore has been found to be ubiquitous. Specifically, a complex modification pattern of U34, that may vary between different organisms, allows the modified anticodon to pair with codons in 4-codon boxes containing synonymous codons with any base at the third position (7,8). The uracil base is modified both at its carbon C5 and further modified at C2 with sulfur containing groups into thiouridine derivatives, sometimes very complex ones (10). These modifications are essential for decoding of A- and G-ending NAR codons but also NNR in 2-codon boxes (or 2:2 codons for which NNY and NNR are assigned for different amino acids). By contrast, G34 may be used to decode C- and U-ending codons (NNY) easily (6). This accounts for the observation that U34 modifications at the first anticodon position of a tRNA appeared as an essential feature of the translation machinery (11,12).
It is often believed that codons were originally G+C-rich, making modifications on the N34 base of tRNA not yet mandatory (7). As more A/U-containing codons were used to specify additional amino acids, genes progressively evolved to allow modification of U34 for recognition of NNR in 2:2 codons with a single tRNA species (7). Previous studies have described a variety of posttranscriptional modifications at the N34 position of the tRNAs, usually strengthened by modifications at the N37 position, next to context-sensitive anticodons. These modifications required a variety of genes, sometimes reflecting convergent evolution (7,8,13). Despite the burden of keeping functional genes across many generations, there was apparently a considerable negative selection pressure against loss of U34 modifications. As a case in point, a recent study has demonstrated the maintenance of U34 modification genes in the highly reduced genome of symbiotic Buchnera aphidicola (14). Furthermore, this happened despite the fact that, in line with genomic reduction pressure, the symbiosis process selectively removed isoacceptor tRNA genes (8), again emphasizing the importance of U34 modifications.
Several further anticodon nucleotides, in particular C34 and A34 of prokaryotic tRNAs, are also modified. In higher eukaryotes, the C32 and G34 nucleotides are further modified by 2′-O-ribose methylase (15–17). tRNA positions 35 and 36 are so far known to be modified mostly in eukaryotes (7). However, existence of pseudouridine modification at position 35 of tRNAs for tyrosine has been recently reported in Escherichia coli (18). Overall, the modifications at position U34 are much more extensive than others on tRNA anticodons (7). As stressed previously the requirement for translation accuracy imposes a considerable genetic burden to keep the U34 modification genes. This prompted us to explore the situation in organisms with genomic decay. The bacteria in Tenericutes are of particular interest, because they lack the sulfur relay system essential for some U34 modifications (7,19,20). s2U34 is almost ubiquitous in U34-bearing tRNAs coding for Gln/Lys/Glu (NAR codon) of majority of bacteria due to its vital role in efficient and accurate codon reading (7). What would be the situation in the absence of U34 modifications? In their absence, the only obvious solution is that C34-containing tRNAs would be present in parallel with the ones with U34, a somewhat costly countermeasure as it requires the presence of more tRNA genes. Whether this alternative solution exists in extant organisms has not yet been evaluated. In fact, the complete loss of U34 modifications was seldom reported and the simultaneous presence of C34 and U34 for NAR codons has not yet been observed in genomes. In the present study, the highly reduced genome of a deep-sea endosymbiotic Spiroplasma was analyzed for its tRNA genes. In this genome, U34 modification genes were not detected. Remarkably this resulted in a completely novel tRNA gene profile for codon recognition.
MATERIALS AND METHODS
Analysis of tRNA genes
The genome of ‘Candidatus Spiroplasma holothuricola’ was obtained from the hindgut of a sea cucumber recently (21). Coding sequences (CDSs) were predicted with Prodigal (v2.6) (22) and manually refined in some cases. The deduced proteins of all the CDSs were probed against the NCBI-nr database with BLASTp program with e-value cutoff of 1e–4 and seed size of 3. tRNA genes in the genome were searched using tRNAscan (23). The genes for potential tRNA posttranscriptional modifications and their targets were predicted with tRNAmodpred (24). Phylogeny-based filtration and NCBI SmartBLAST were performed to examine the output of tRNAmodpred.
Measurement of codon usage
The codon usage bias of the CDSs was evaluated by measurement of Relative Synonymous Codon Usage (RSCU) and Codon Adaptation Index (CAI) (25,26). RSCU is a relative frequency that each synonymous codon is used to encode a particular amino acid. The CAI of each species was calculated as a geometric mean of the CAI weights associated with all codons of the species (26). The CAImax for the most favorable codon usage of a species was also obtained for each species by using 18 housekeeping genes that encode reference ribosomal proteins, translation factors and transcription antitermination factor NusA (Supplementary Table S1). The first codons and stop codons were trimmed from the CDSs before the calculation. Genomes of the closest relatives in Spiroplasma, Mycoplasma and Ureaplasma clades were downloaded from the NCBI INSDC entry point, along with three highly reduced genomes (Supplementary Table S2). The codon usage biases and tRNA genes were investigated with the same method.
Detection of tRNAs in transcriptome
Small RNA molecules (<200 bp) were extracted from the hindgut of the ethanol-preserved sea cucumber using mirVana miRNA Isolation Kit (Life Technologies, USA), and were subsequently treated with DNase (Life Technologies, USA). The quality of the RNA sample was measured with a Qubit 2.0 Fluorometer (Life Technologies, USA).
Approximately 25 ng of small RNA sample was used for library preparation. Firstly, cDNA synthesis was directly performed with NEBNext RNA Library Prep Kit for Illumina (NEB, UK). Then, the double-stranded cDNA was purified using MinElute Reaction Cleanup Kit (QIAGEN, Germany). Finally, a DNA sequencing library was prepared by Ovation Ultralow System V2 Kit (NuGEN, USA), and immediately sequenced on an Illumina Miseq platform.
Overall quality of 20 Gbp raw reads was visualized using FastQC v0.10.1 (http://www.bioinformatics.babraham.ac.uk). Low-quality bases (quality threshold was set to 5) at both ends were trimmed using Trimmomatic-0.36 (27). The reads with an average quality lower than 15 were also removed by Trimmomatic-0.36. All the trimmed reads were retained for subsequent analysis without a further length filtration. Paired-end reads were merged using PEAR v0.9.5 (28) with a minimum assembly length of 10 bp. The resulted sequences along with unpaired reads were searched against tRNA sequences of ‘Ca. Spiroplasma holothuricola’ using BLASTn alignment (Blast+ 2.2.29) with megablast task for sequences longer than 50 bp or with blastn-short task for those shorter than 50 bp, and with an e-value threshold of 1e-5. The fragments with a similarity of >95% to the tRNAs were collected and re-examined by searching against the NCBI nt database.
RESULTS AND DISCUSSION
‘Ca. Spiroplasma holothuricola’ is a gut endosymbiont of Zygothuria oxysclera captured in the Mariana trench at a depth of 6140m and at a temperature of 1.6°C. The complete ‘Ca. Spiroplasma holothuricola’ genome is composed of two chromosomes 143947 bp and 280592 bp, respectively in size (21). Given its genomic reduction, it is remarkable that the genome still harbors a huge set of clustered regularly interspaced short palindromic repeats (CRISPRs, making a total of 5053 bp for 76 spacers), presumably involved in immunity against viruses. In the genome, we identified three tRNA modification systems, coded by genes tsaBDE and trmD that are responsible for A37-N6-threonylcarbamoyl modification and G37-N1 methylation, respectively. However, all the essential tRNA (34) modification genes such as those encoding GidA (uridine 5-carboxymethylaminomethyl modification or cmnm5U34) were absent from the ‘Ca. Spiroplasma holothuricola’ genome. A possible trmK gene responsible for A22 methylation was also identified, but likely was a false positive according to the phylogeny-based examination by the tRNAmodpred (24). However, we cannot exclude possible transport of tRNA modifying enzymes encoded by the sea cucumber host to the endoensymbiont. Also, while unlikely because of the small size of genes of unknown function in the genome there still could be unknown genes coding for tRNA modifications developed through convergent evolution in the isolated niche.
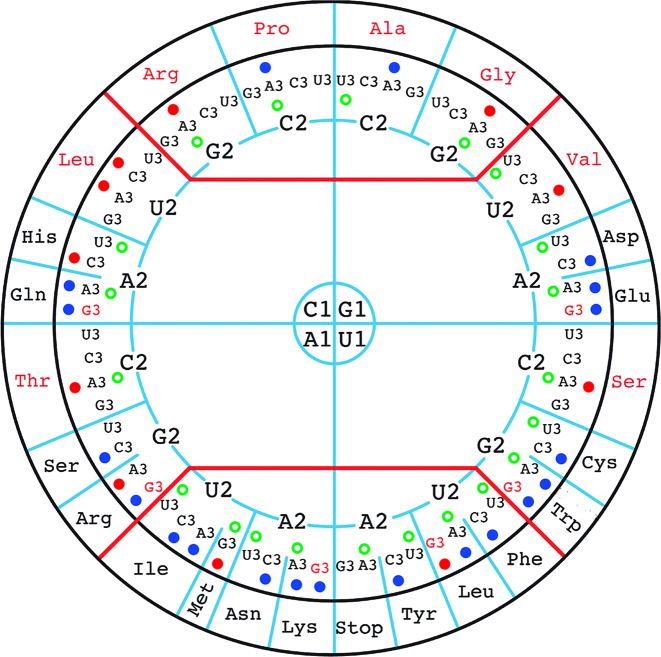
In agreement with the possible loss of anticodon modification genes in the ‘Ca. Spiroplasma holothuricola’ genome, we noticed the remarkable presence of a unique tRNA gene profile in the genome (Supplementary Table S3). In contrast to the organisms that have a fully modified U34 base in their tRNAs and can accommodate a very small set of tRNAs in streamlined genomes, extra tRNA genes are present in the ‘Ca. Spiroplasma holothuricola’ genome. In particular there are genes allowing the organism to use two tRNA species for each of the glutamate, glutamine and lysine amino acids, depending whether their codons end with a U or a C (Supplementary Table S3). Remarkably, this apparent countermeasure extends to all amino acids decoded by NNR 2:2 codons (Figure 1). In contrast, the NNY 2:2 codons seem to be deciphered by only one tRNA with a G34 at the first anticodon position, as is the case in Saccharomyces cerevisiae, for instance (Figure 1). This observation provides strong phylogenetic support to the previously reported asymmetrical pairing between G34•U3 and U34•G3 (7). All things considered, three tRNA species are required for accurate recognition of 2:2 codons in ‘Ca. Spiroplasma holothuricola’. While this is also the fact for most bacteria, this is generally not happening in G/C-poor bacteria with a reduced genome. Devoted to cope with G3-ending codons in G/C-poor bacteria, chemical modifications at U34 allow the use of U34-containing tRNAs for decoding both A3- and G3-ending codons (7). As a consequence, this leads to sparing of C34-bearing tRNA genes in these bacterial genomes. Our observation in the ‘Ca. Spiroplasma holothuricola’ genome, maintaining six C34-bearing tRNA genes, goes against the rule. Therefore lack of recognizable genes for U34 modifications is a potential explanation to the observation of redundant C34-bearing tRNA genes in such a tiny G/C-poor (G+C content 29.6%) genome.
Figure 1.
Codons that are recognized by tRNA genes in ‘Ca. Spiroplasma holothuricola’ genome. The bases with 1, 2 and 3 correspond to the first, second and third positions of codons, respectively. In the outermost circle, there are amino acids that are encoded by N1N2N3 (N: A/G/C/U) codons. A solid dot (deep blue or red) adjacent to a N3 indicates that there is a tRNA gene with an anticodon perfectly matching to the codon. The tRNA genes for the codons with a deep blue dot were identified in the ‘Ca. Spiroplasma holothuricola’ genome; those for the codons with a red dot were further detected in the transcriptome. The green blank circle refers to the most frequently used codon for an amino acid. The original figure was adapted from Grosjean and Westhof (7) with permission of the authors.
Furthermore, we were also unsuccessful in picking out genes that code for modification of the tRNA’s position 35, suggesting that ‘Ca. Spiroplasma holothuricola’ has consistently abolished anticodon modifications. In contrast, we detected a gene responsible for modification at tRNA position 37 in the organism. It must be noted that this tRNA (37) modification is essential for maintaining accurately the translational reading frames, a translation-related feature that is likely to be key for survival (29). This allows us to argue that modification at this position is perhaps more critical for living organisms than are anticodon modifications.
The 4-codon boxes of ‘Ca. Spiroplasma holothuricola’ were read by a single tRNA species with U34, except for that of leucine associated with the two tRNAs bearing GAG and UAG anticodons (Figure 1). Note that the GAG-bearing tRNALeu is not used by the representative Tenericutes while it is preserved in two symbiotic genomes (Figure 2). This suggests that unmodified U34 can meet some structural constrain to stably fit with C3 when U2 is present in a codon. However, this was not predicted in a recent calculation of the energy involved in codon-anticodon interactions (7).
Figure 2.
Distribution of anticodons and CAI-weight of codons. A total of 14 species were selected for a survey of anticodons in tRNAs and codon usage bias. The anticodons of the tRNAs in their genomes were displayed with a red block. CAI-weight of codons was calculated using all the CDSs of the individual species and was converted to different green depths. Codon usage bias was assessed with CAI. Mycoplasma sp. Bg1 and Bg2 (PRJNA309720), B. aphidicola (NC_002528), ‘Ca. Moranella endobia’ (NC_015735), and ‘Ca. Nasuia deltocephalinicola’ (CP013211 and CP013212) are symbiotic bacteria. The stop codon ‘TGA’ in universal codon table was decoded into amino acid ‘W’ and was thus shaded.
Finally, the ‘Ca. Spiroplasma holothuricola’ genome lacks a key gene, tilS, that is involved in modification of CAU anticodon-bearing tRNAIle by forming a lysidine at position 34 (30). In this way, the codon specificity of tRNAIle with anticodon CAU is shifted from AUG to AUA for integration of an isoleucine rather than a methionine (31). However, mutants without tilS gene are not completely lethal and some microbes in nature do not own a TilS modification system (31,32). How can ‘Ca. Spiroplasma holothuricola’ recognize AUA codon without TilS? In Mycoplasma mobile, this hurdle was solved by introduction of a tRNA gene bearing a UAU anticodon that recognizes AUA codon. This tRNA feature also required the involvement of an IleRS enzyme of a novel type (32). Likewise, we identified a tRNAIleUAU gene in the ‘Ca. Spiroplasma holothuricola’ genome, indicating that the symbiont adopted a similar strategy to cope with the loss of tilS gene. Counting the two context-dependent ATG codons for methionine (initiation and elongation), the ‘Ca. Spiroplasma holothuricola’ genome harbors a set of 32 tRNA genes for 20 amino acids. Such a phenomenon has not been observed in any of the Spiroplasma, Mycoplasma and other bacterial genomes to date (Figure 2) (7).
In summary, it appears that ‘Ca. Spiroplasma holothuricola’ has to manipulate two different tRNAs to decode two NNR 2:2 codons rather than one single tRNA for the synonymous codons. This is likely to affect translation speed by slackening its pace. Interestingly, the lack of EF-P gene in the ‘Ca. Spiroplasma holothuricola’ genome may further slow down the translation process (21), because EF-P as a key factor may help resolve stalling at polyproline stretches (33). The simultaneous occurrence of EF-P depletion and special tRNA gene profile indicates a slow metabolism for the spiroplasmas dwelling in the gut cell of the hadal sea cucumber. At present, it is very difficult to determine which of the above events came first.
We examined the anticodon list of tRNA genes for consistency among the species that maintain anticodon modifications (Figure 2). The Spiroplasma and Ureaplasma tRNAs analyzed in the present study have more or less the same anticodons. However, there is a difference in employment of tRNA species to read arginine 4-codon box. As observed in a previous study, all the Tenericutes species lack anticodon CCG-harboring tRNA gene specialized for arginine codon CGG in the other bacteria. This results from the loss of the tadA gene (34). The consequence is that some Tenericutes depend on tRNA that has ACG anticodon to decipher the remaining three arginine codons in the 4-codon box before acquisition of tRNA with UCG anticodon for full recognition of CGN codons (34). Our work shows that ‘Ca. Spiroplasma holothuricola’, along with the two Ureaplasma species and Mycoplasma suis, have gained the anticodon UCG-bearing tRNA, while the anticodon ACG-bearing tRNA is still crucial in some spiroplasmas (Figure 2). Six species in Figure 2 are symbiotic bacteria. As a rule, a number of rare anticodons are present in the symbiotic genomes. Anticodon CUC was only found in the ‘Ca. Spiroplasma holothuricola’ genome; anticodons CUG and CCU were present only in ‘Ca. Spiroplasma holothuricola’ and ‘Candidatus Moranella endobia’. These symbiotic genomes probably experienced deviation of their tRNA contents in parallel with protein-coding ‘gene sparing’ (35), leading to invention of new tRNA species in a context where the nutrient supply appears to be rich in nucleotides (21). Redundant tRNA species emerged even in the species of ‘Ca. Moranella endobia’ and ‘Ca. Nasuia deltocephalinicola’ that are equipped the gidA and the associated mnmE genes involved in U34 modification in their genomes. Perhaps it has not been noticed that the expansion and diversification of tRNA species provide the basis for fitness of the individuals that evolved toward reducing their tRNA (34) modification capacity. We propose that the new tRNA species were obtained prior to the possible malfunction of the modification genes as the sudden loss of these genes will lead to disastrous translational error rates. Removal of tRNA (34) modifications would probably have associated with positive selection of tRNA species that may compensate the declined codon recognition efficiency. This may be the way that the novel tRNA gene profile was achieved in the ‘Ca. Spiroplasma holothuricola’. The highly simplified life mode of the endosymbiotic spiroplasmas probably meets some constrains in synthesis of secondary metabolites necessary for tRNA modifications. This is likely the driving force for the spiroplasmas to select against the tRNA modifications as a strategy for deciphering of synonymous codons. We must also emphasize that ‘Ca. Spiroplasma holothuricola’ is confronted both by low temperature and extreme high pressure that has been shown to influence ribosomal RNA structure (36). However, we do not know whether the Spiroplasma had obtained novel genes that enable modifications of tRNA (34) with unknown chemical groups synthesized using molecules imported from the host. A similar stepwise evolutionary process has been proposed to explain the evolvement of tRNA with UCG anticodon in Tenericutes that lost the tadA gene responsible for decoding CGG codon for arginine (34).
Since ‘Ca. Spiroplasma holothuricola’ was a hadal symbiont, we examined whether this remarkable finding could be the result of endosymbiosis in the deep-sea environment. We searched for U34 modification genes in five other symbionts (see Figure 2), and identified the presence of at least one modification gene, gidA(mnmA), in their genomes. The GidA enzyme catalyzes cmnm5U modification on tRNA(U34), which is required for reliable recognition of NNR codons (37). MnmE that forms a heterotetramer with GidA could also be encoded by the genomes (38). Using single anticodons of their tRNA genes for decoding NNR 2:2 codons is direct evidence for the presence of the gidA and mnmE genes, even in the smallest genomes of ‘Ca. Moranella endobia’ and ‘Ca. Nasuia deltocephalinicola’ (Figure 2). The Mycoplasma suis genome has gidA but not mnmEG (20). This may be the reason to its abnormal tRNA gene profile when compared with other species (Figure 2). Perhaps also due to the malfunction of cmnm5U34 modification system, M. suis and ‘Ca. Spiroplasma holothuricola’ both have a rare tRNA species: anticodon AAG-harboring tRNA for Lys. The genomes of a Ureaplasma from a hadal snailfish (7100m depth) (unpublished) and two Mycoplasma species (39) from a deep-sea isopod (900m depth) were also searched for U34 modification genes. Again, we detected the presence of gidA and associated mnmE genes in these deep-sea genomes (40). Therefore, the elimination of tRNA (34) modification genes is not widespread in deep-sea organisms or symbionts.
Interestingly, the anticodons of the ‘Ca. Spiroplasma holothuricola’ tRNAs do not always correspond to the most frequently used codons. For example, the GCA and GUA codons are associated the highest RSCU value among codons for Val and Ala but they are not decoded by a tRNA with perfectly matching anticodon (Figure 1). However, all the selected AT-rich genomes prefer A/U-ending codons, suggesting less importance of the third codon position in correct positioning of tRNAs in the ribosome (Figure 2). We calculated the CAI weight of the codons to reflect the dominant codon usage bias in this organism. The result was that the CAI weights of the codons did not much differ from the values observed for most of the other species (Figure 2). In contrast, the codon usage bias of ‘Ca. Moranella endobia’ is inconsistent with that of the other species as indicated by the relatively homogeneous codon weights of synonymous codons. CAI of most of the selected species was also rather consistent, ranging between 0.69 and 0.73. Exceptions are the relatively high CAI values between 0.76 and 0.78 for ‘Ca. Moranella endobia’ and ‘Ca. Nasuia deltocephalinicola’, which probably resulted from relaxation of selection for pathogenic endosymbionts (41). To some extent, it is surprising that the special tRNA gene profile did not affect the codon usage bias of the ‘Ca. Spiroplasma holothuricola’ that is also supposed to be an endosymbiont (21).
To validate the dearth of anticodon modifications, we examined the transcriptome of the sea cucumber hindgut where the ‘Ca. Spiroplasma holothuricola’ was found. As expected, most of the small RNA transcripts were derived from the host, which resulted in a low output of tRNA transcripts from the spiroplasmas. Within a total of 12.5 Gbp clean transcriptomic data, we could identify 20 tRNA fragments for the symbiotic spiroplasmas (Supplementary Table S4). After removal of duplicates, 11 were retained, as displayed in Figure 1. One of them deciphers the UUG codon for Leu, a rare codon among the Tenericutes (Figure 2). Although not complete, all the sequenced tRNA transcripts were identical to the aligned part of the corresponding tRNA genes. If there were inosine(A) and cmnm5s2(U) tRNA (34) modifications, added by TadA and GidA/IscS/MnmEG, respectively, we would have expected mismatches in the alignment between the sequenced cDNAs and the genes (14,42). However, we did not observe any mismatch. Other modifications such as cmnm5Um and k2C would not lead to mismatches (14), and would have thus remained undetectable in our transcriptomic data. Transfer RNA molecules are easily degraded at codon-anticodon pairing positions (43). This made it particularly difficult to reversely transcribe the tRNAs into cDNA fragments comprising anticodons. In addition, because of the way the samples were collected (a single specimen, domination of the host RNAs and difficulty in preserving RNA under deep sea sampling conditions) the evidence collected from the transcriptome analysis is obviously limited. Despite considerable efforts we could not obtain more complete sequences for the tRNA transcripts of the ‘Ca. Spiroplasma holothuricola’ symbiont. Our biochemical observations therefore only weakly substantiate that tRNA is seldom modified in this organism as suggested by our in silico study of its genome. Nevertheless, the ‘Ca. Spiroplasma holothuricola’ tRNA set is unusually large for such a small genome. It was likely a result of the selection pressure allowing the organism to drop most tRNA modifications genes. Perhaps tRNA anticodon modifications are not critical for the spiroplasmas surviving at 1.6°C and high pressure. Such a low temperature coupled with very high pressure (61MPa) will certainly modify the codon–anticodon interactions as it seems that steric constraints, rather than hydrogen bonds, are dominating the interactions (44).
Phylogenetic studies indicate that ‘Ca. Spiroplasma holothuricola’ was derived from an ancient spiroplasma lineage neighboring Spiroplasma ixodetis (21). In this study, the bacterium stands out as a probably unique case of paucity of nucleotide modifications in anticodons of the whole tRNA population. This was possibly a result of long-term niche-specific endosymbiosis, which might have occurred and retained in the isolated hadal trenches probably as an evolutionary dead-end. In the hadal trenches, there are far more unknowns that were not uncovered due to our exploration limitations.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Science Foundation of China [41476104, 31460001]; The Strategic Priority Research Program B of Chinese Academy of Sciences [XDB06010201, XDB06010103]; National Key Research and Development Program of China [2016YFC0302500]. Funding for open access charge: NSFC of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Noller H.F. The parable of the caveman and the Ferrari: protein synthesis and the RNA world. Philos. Trans. R Soc. Lond. B Biol. Sci. 2017; 372:20160187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong J.T. A co-evolution theory of the genetic code. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:1909–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grosjean H., Marck C., de Crécy-Lagard V.. The various strategies of codon decoding in organisms of the three domains of Life: evolutionary implications. Nucleic Acids Symp. Ser. 2007; 51:15–16. [DOI] [PubMed] [Google Scholar]
- 5. Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. [DOI] [PubMed] [Google Scholar]
- 6. Westhof E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett. 2014; 588:2464–2469. [DOI] [PubMed] [Google Scholar]
- 7. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gustilo E.M., Vendeix F.A., Agris P.F.. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008; 11:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harada F., Nishimura S.. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry. 1972; 11:301–308. [DOI] [PubMed] [Google Scholar]
- 10. Haruehanroengra P., Vangaveti S., Ranganathan S.V., Wang R., Chen A., Sheng J.. Nature's selection of geranyl group as a tRNA modification: the effects of chain length on base-pairing specificity. ACS Chem. Biol. 2017; 12:1504–1513. [DOI] [PubMed] [Google Scholar]
- 11. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. New structural insights into translational miscoding. Trends Biochem. Sci. 2016; 41:798–814. [DOI] [PubMed] [Google Scholar]
- 12. Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G.. A new understanding of the decoding principle on the ribosome. Nature. 2012; 484:256–259. [DOI] [PubMed] [Google Scholar]
- 13. Grosjean H., de Crécy-Lagard V., Marck C.. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010; 584:252–264. [DOI] [PubMed] [Google Scholar]
- 14. Hansen A.K., Moran N.A.. Altered tRNA characteristics and 3′ maturation in bacterial symbionts with reduced genomes. Nucleic Acids Res. 2012; 40:7870–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Motorin Y., Grosjean H.. Transfer RNA modification. eLS. 2001; 2001:doi:10.1038/npg.els.0000528. [Google Scholar]
- 16. Torres A.G., Pineyro D., Filonava L., Stracker T.H., Batlle E., Ribas de Pouplana L.. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 2014; 588:4279–4286. [DOI] [PubMed] [Google Scholar]
- 17. Pintard L., Lecointe F., Bujnicki J.M., Bonnerot C., Grosjean H., Lapeyre B.. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002; 21:1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Addepalli B., Limbach P.A.. Pseudouridine in the anticodon of Escherichia coli tRNA(Tyr(Q Psi A)) is catalyzed by the dual specificity enzyme RluF. J. Biol. Chem. 2016; 291:22327–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Danchin A., Fang G.. Unknown unknowns: essential genes in quest for function. Microb. Biotech. 2016; 9:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barre A., Yoshizawa S., Fourmy D., de Crecy-Lagard V. et al. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 2014; 10:e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He L.-S., Zhang P.-W., Huang J.-M., Zhu F.-C., Danchin A., Wang Y.. The enigmatic genome of an obligate ancient Spiroplasma symbiont in a hadal holothurian. Appl. Environ. Microbiol. 2018; 84:e01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lowe T.M., Eddy S.R.. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Machnicka M.A., Dunin-Horkawicz S., de Crecy-Lagard V., Bujnicki J.M.. tRNAmodpred: A computational method for predicting posttranscriptional modifications in tRNAs. Methods. 2016; 107:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharp P.M., Cowe E.. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991; 7:657–678. [DOI] [PubMed] [Google Scholar]
- 26. Sharp P.M., Li W.-H.. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987; 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J., Kobert K., Flouri T., Stamatakis A.. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014; 30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenner L.B., Demeshkina N., Yusupova G., Yusupov M.. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010; 17:555–560. [DOI] [PubMed] [Google Scholar]
- 30. Soma A., Ikeuchi Y., Kanemasa S., Kobayashi K., Ogasawara N., Ote T., Kato J.-i., Watanabe K., Sekine Y., Suzuki T.. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003; 12:689–698. [DOI] [PubMed] [Google Scholar]
- 31. Kohrer C., Mandal D., Gaston K.W., Grosjean H., Limbach P.A., Rajbhandary U.L.. Life without tRNAIle-lysidine synthetase: translation of the isoleucine codon AUA in Bacillus subtilis lacking the canonical tRNA2Ile. Nucleic Acids Res. 2014; 42:1904–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taniguchi T., Miyauchi K., Nakane D., Miyata M., Muto A., Nishimura S., Suzuki T.. Decoding system for the AUA codon by tRNAIle with the UAU anticodon in Mycoplasma mobile. Nucleic Acids Res. 2013; 41:2621–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ude S., Lassak J., Starosta A.L., Kraxenberger T., Wilson D.N., Jung K.. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013; 339:82–85. [DOI] [PubMed] [Google Scholar]
- 34. Yokobori S.-i., Kitamura A., Grosjean H., Bessho Y.. Life without tRNAArg–adenosine deaminase TadA: evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes. Nucleic Acids Res. 2013; 41:6531–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stapleton M., Carlson J.W., Celniker S.E.. RNA editing in Drosophila melanogaster: New targets and functional consequences. RNA. 2006; 12:1922–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lauro F.M., Chastain R.A., Blankenship L.E., Yayanos A.A., Bartlett D.H.. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 2007; 73:838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi R., Villarroya M., Ruiz-Partida R., Li Y., Proteau A., Prado S., Moukadiri I., Benítez-Páez A., Lomas R., Wagner J. et al. Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme. J. Bacteriol. 2009; 191:7614–7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yim L., Moukadiri I., Björk G.R., Armengod M.E.. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 2006; 34:5892–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Huang J.M., Wang S.L., Gao Z.M., Zhang A.Q., Danchin A., He L.S.. Genomic characterization of symbiotic mycoplasmas from the stomach of deep-sea isopod bathynomus sp. Environ. Microbiol. 2016; 18:2646–2659. [DOI] [PubMed] [Google Scholar]
- 40. Lo W.S., Huang Y.Y., Kuo C.H.. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol. Rev. 2016; 40:855–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Botzman M., Margalit H.. Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 2011; 12:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iida K., Jin H., Zhu J.K.. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009; 10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shigematsu M., Ogawa T., Kitamoto H.K., Hidaka M., Masaki H.. Specific phase arrest of cell cycle restores cell viability against tRNA cleavage by killer toxin. Biochem. Biophys. Res. Commun. 2012; 420:750–754. [DOI] [PubMed] [Google Scholar]
- 44. Khade P.K., Shi X.Y., Joseph S.. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. J. Mol. Biol. 2013; 425:3778–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.