Sir,
Dengue, a viral disease transmitted by Aedes mosquitoes, is a serious global public health problem, infecting 50 to 100 million people each year in >100 countries and resulting in at least 20,000 deaths annually1,2. Dengue is caused by one of the four antigenically related dengue virus serotypes (DENV-1 to 4) belonging to the family Flaviviridae3. The viral genome is a single-stranded, positive-sense RNA of about 10.7 kb which encodes three structural and seven non-structural (NS) proteins in the order 5'-C-prM(M)-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3'4. Based on the nucleotide sequence of the envelope (E), E/NS1 or capsid-premembrane (CprM) gene, the four DENV serotypes have been classified phylogenetically into three to five genotypes with certain genotypes being further classified into lineages or clades5.
In India the incidence rate of dengue infection has grown steadily in the past few years6. Though DENV fevers have been recorded in India since the 1940s7, the first reported epidemic of dengue fever was during 1963-1964 from the Eastern Coast of India. Since then many outbreaks have been reported from all over the country8,9. The epidemiology of DENV and its prevalent serotypes has been ever changing in the country. With the increase in DENV cases, the need to understand the genetic nature of circulating DENV has become necessary. There are no reports of in-depth molecular genetic characterization of DENV in the State of Karnataka. CprM gene-based genotyping of DENV has proved to be useful for carrying out molecular epidemiology of these viruses10,11. Phylogenetic trees based on CprM and E gene regions are found to have a similar topology with no discrepancy between genotypes/sublineages. Hence, considering the availability of a larger dataset based on CprM gene sequences of Indian DENV isolates, the present study was undertaken during January 2016-August 2016 to understand the genetic nature of circulating DENV in Karnataka by sequencing the CprM gene junction and comparing 504 bp of this region with other similar Indian DENV sequences reported till date and global reference sequences.
Being the diagnostic laboratory, blood/serum samples from suspected cases of DENV infection were referred to the ICMR-National Institute of Virology, Bengaluru unit, from multiple public health agencies. Samples were transported to the laboratory with the due precautions recommended for handling RNA viruses. Overall, 17 DENV-1 and 58 DENV-2 isolated from different outbreaks in Karnataka during 1993-2005 and three isolates from Tamil Nadu (1997) were used in this study, for sequencing of the CprM gene junction region. In brief, viral RNA was extracted from 140 μl of serum samples, using a QIAamp Viral RNA Mini kit (Qiagen, Germany), according to the manufacturer's instructions using published CprM gene-specific primers12. The PCR products were purified using QIAquick gel extraction Kit (Qiagen) and sequenced using BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA) in an automatic sequencer (ABI PRISM Genetic Analyzer 3100, Applied Biosystems). The CprM sequences of DENV-1 (nucleotide positions 137-640, with respect to DENV-1 sylvatic sequence Malaysia/1972) and DENV-2 (nucleotide positions 137-643, with respect to DENV-2 sylvatic sequence BurkinaFaso/1980) were compared with other CprM sequences of DENV-1 and -2, respectively, from India and other countries retrieved from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/, as on 30 June, 2016). Multiple sequence alignment and phylogenetic trees were constructed using MEGA version 6.013, employing the Neighbour-Joining method14 with the Kimura-2 parameter distance model and 1000 bootstrap replications, to determine the extent of genetic heterogeneity and trace the phylogeny of DENV-1 and DENV-2.
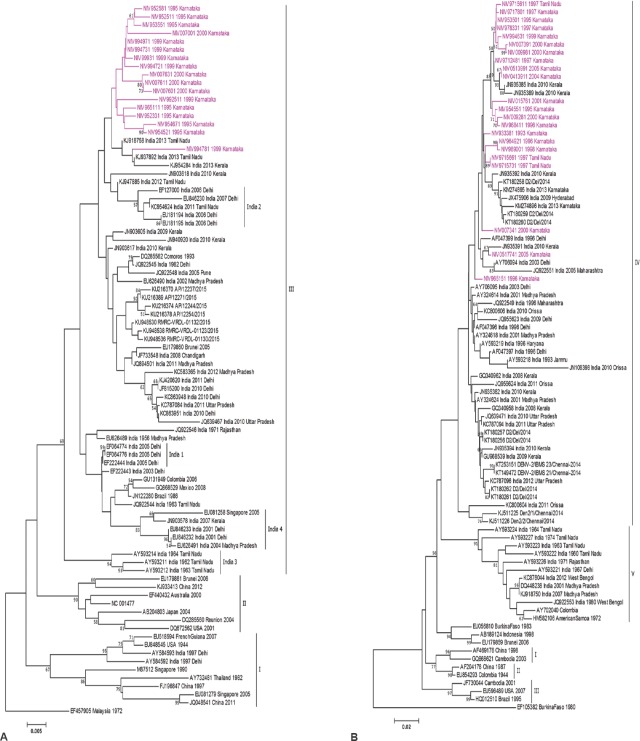
Phylogenetic analyses of the 17 DENV-1 isolates of this study and the other Indian and global sequences revealed that all the Karnataka isolates of the study period clustered in Genotype III (Fig. A). It was noted that these sequences could not be classified into any of the India 1-4 lineages that included the DENV-1 strains of several States in north and south India over the period from 1956 to 201515. The per cent nucleotide divergence (PND) within the sequences from Karnataka was found to be 1.41 per cent. The amino acid alignment showed that there were a few mutations specific to some of the Karnataka isolates, namely, I127V in four isolates of 1995 and R97K in two isolates of 1999 (Data not shown). Analysis of DENV-2 isolates [Figure B] showed that all the isolates of this study (which included three isolates of 1997 from Tamil Nadu) clustered within the Genotype IV (also referred to as the Cosmopolitan genotype) of DENV-2 along with majority of the other Indian isolates. The PND among the sequences of this study was found to be 1.7 per cent. A mutation A102T was found to be unique to many of the Karnataka isolates of the period 1995-2005. A few isolates from Karnataka were found to possess A102V as in some other strains from other parts of India. Co-circulation of both serotypes was noted during 1995-1996 and 1999-2000. At position 104, Karnataka sequences possessed M as in the Tamil Nadu sequences. The sequences of the later period from Kerala, Chennai and north India possessed a substitution M104V.
Figure.
Phylogenetic tree of selected representative sequences of CprM region of (A) DENV-1 and (B) DENV-2 by Neighbour-Joining method.
DENV-1 has been circulating in India since the 1940s7 with earliest outbreaks reported from Vellore, Tamil Nadu, in 1950s-1960s. DENV-1 was also implicated during different outbreaks in Delhi, Rajasthan, Uttar Pradesh and Madhya Pradesh since 1970s8,16,17 and recently (during 2015) in Arunachal Pradesh (GenBank accession numbers: KU216370, KU216374, KU216378 and KU216389). DENV-1 was associated with a dengue haemorrhagic fever (DHF) outbreak in Delhi in 199718. Delineation of this serotype into three major genotypes designated as I-III19 has been noted, with Genotype III (cosmopolitan genotype)20, being the most dominant one. Within this genotype, four distinct Indian sublineages have been identified, designated as India 1-415,20. India-3 is extinct while the other three are the currently evolving lineages21. While the Delhi sequences exist in all the three lineages, during 2007-2008, Kerala also showed the circulation of the India-4 lineage of DENV-122. A study based on the complete E gene has identified some more Indian clusters23. No change in the lineage of the Karnataka DENV-1 isolates has been observed during the period of more than a decade based on this study.
DENV-2 represents six genotypes including the sylvatic genotype5. The Genotype IV24 (Cosmopolitan genotype) having a wide geographical distribution and including majority of Indian DENV-2 strains circulating in northern India such as Delhi, Lucknow and Gwalior8,25; Gujarat and Haryana replaced the Genotype V (American genotype) of DENV-2 that predominantly circulated in India during the pre-1971 period. In Delhi, till 2003, the predominant serotype was DENV-226. DENV-2 has also been reported from Southern India, in Kerala, along with DENV-327 and also in Chennai, Tamil Nadu (GenBank accession numbers: KT149472 and KT253151). The present study showed that the dominant serotype of DENV in Karnataka during the period 1993-2005 was DENV-2 of a single genotype (Genotype-IV). Many of the Karnataka sequences analyzed formed a separate cluster with a high bootstrap support value (82%) that was most likely due to the presence of a conserved amino acid T102. Overall, the present results revealed the involvement of two dengue serotypes, type 1 and 2, in Karnataka. The strains circulating in the State were genetically similar to those circulating in the other States of India during similar time periods. Further studies including continuation of DENV surveillance in the State are warranted to get the complete picture of the circulating serotypes and note any shift in dominance of these serotypes/genotypes.
Acknowledgment
The authors acknowledge financial support provided by the Indian Council of Medical Research (ICMR), Ministry of Health and Family Welfare, Government of India, New Delhi, India.
Footnotes
Conflicts of Interest: None.
References
- 1.World Health Organization. Dengue and severe dengue. [accessed on July 18, 2016]. http://www.who.int/mediacentre/factsheets/fs117/en/
- 2.Centers for Disease Control and Prevention. Dengue: Epidemiology. [accessed on July 18, 2016]. Available from: http://www.cdc.gov/dengue/epidemiology/
- 3.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–42. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 4.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–88. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 5.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–93. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- 6.Shepard DS, Halasa YA, Tyagi BK, Adhish SV, Nandan D, Karthiga KS, et al. Economic and disease burden of dengue illness in India. Am J Trop Med Hyg. 2014;91:1235–42. doi: 10.4269/ajtmh.14-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karamchandani PV. Dengue group of fevers in India. Lancet. 1946;1:92–3. doi: 10.1016/s0140-6736(46)91229-9. [DOI] [PubMed] [Google Scholar]
- 8.Gupta N, Srivastava S, Jain A, Chaturvedi UC. Dengue in India. Indian J Med Res. 2012;136:373–90. [PMC free article] [PubMed] [Google Scholar]
- 9.Cecilia D. Current status of dengue and chikungunya in India. WHO South East Asia J Public Health. 2014;3:22–6. doi: 10.4103/2224-3151.206879. [DOI] [PubMed] [Google Scholar]
- 10.Singh UB, Seth P. Use of nucleotide sequencing of the genomic cDNA fragments of the capsid/premembrane junction region for molecular epidemiology of dengue type 2 viruses. Southeast Asian J Trop Med Public Health. 2001;32:326–35. [PubMed] [Google Scholar]
- 11.Avilés G, Rowe J, Meissner J, Manzur Caffarena JC, Enria D, St. Jeor S, et al. Phylogenetic relationships of dengue-1 viruses from Argentina and Paraguay. Arch Virol. 2002;147:2075–87. doi: 10.1007/s00705-002-0886-3. [DOI] [PubMed] [Google Scholar]
- 12.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Domingo C, Palacios G, Jabado O, Reyes N, Niedrig M, Gascón J, et al. Use of a short fragment of the C-terminal E gene for detection and characterization of two new lineages of dengue virus 1 in India. J Clin Microbiol. 2006;44:1519–29. doi: 10.1128/JCM.44.4.1519-1529.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukreti H, Dash PK, Parida M, Chaudhary A, Saxena P, Rautela RS, et al. Phylogenetic studies reveal existence of multiple lineages of a single genotype of DENV-1 (genotype III) in India during 1956-2007. Virol J. 2009;6:1. doi: 10.1186/1743-422X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra G, Jain A, Prakash O, Prakash S, Kumar R, Garg RK, et al. Molecular characterization of dengue viruses circulating during 2009-2012 in Uttar Pradesh, India. J Med Virol. 2015;87:68–75. doi: 10.1002/jmv.23981. [DOI] [PubMed] [Google Scholar]
- 18.Kurukumbi M, Wali JP, Broor S, Aggarwal P, Seth P, Handa R, et al. Seroepidemiology and active surveillance of dengue fever/dengue haemorrhagic fever in Delhi. Indian J Med Sci. 2001;55:149–56. [PubMed] [Google Scholar]
- 19.Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, et al. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303:110–9. doi: 10.1006/viro.2002.1686. [DOI] [PubMed] [Google Scholar]
- 20.Patil JA, Cherian S, Walimbe AM, Patil BR, Sathe PS, Shah PS, et al. Evolutionary dynamics of the American African genotype of dengue type 1 virus in India (1962-2005) Infect Genet Evol. 2011;11:1443–8. doi: 10.1016/j.meegid.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Walimbe AM, Lotankar M, Cecilia D, Cherian SS. Global phylogeography of dengue type 1 and 2 viruses reveals the role of India. Infect Genet Evol. 2014;22:30–9. doi: 10.1016/j.meegid.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Anoop M, Mathew AJ, Jayakumar B, Issac A, Nair S, Abraham R, et al. Complete genome sequencing and evolutionary analysis of dengue virus serotype 1 isolates from an outbreak in Kerala, South India. Virus Genes. 2012;45:1–13. doi: 10.1007/s11262-012-0756-3. [DOI] [PubMed] [Google Scholar]
- 23.Dash PK, Sharma S, Soni M, Agarwal A, Sahni AK, Parida M, et al. Complete genome sequencing and evolutionary phylogeography analysis of Indian isolates of dengue virus type 1. Virus Res. 2015;195:124–34. doi: 10.1016/j.virusres.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Lewis JA, Chang GJ, Lanciotti RS, Kinney RM, Mayer LW, Trent DW, et al. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197:216–24. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- 25.Dar L, Broor S, Sengupta S, Xess I, Seth P. The first major outbreak of dengue hemorrhagic fever in Delhi, India. Emerg Infect Dis. 1999;5:589–90. doi: 10.3201/eid0504.990427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dar L, Gupta E, Narang P, Broor S. Cocirculation of dengue serotypes, Delhi, India, 2003. Emerg Infect Dis. 2006;12:352–3. doi: 10.3201/eid1202.050767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anoop M, Issac A, Mathew T, Philip S, Kareem NA, Unnikrishnan R, et al. Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South India. Indian J Exp Biol. 2010;48:849–57. [PubMed] [Google Scholar]