Abstract
BACKGROUND
The transcriptional regulation of the human angiotensin receptor subtype 1 (AT1R) gene in pathophysiologies, like the metabolic syndrome, is poorly understood. The human AT1R gene has polymorphisms in its promoter that can be arranged in 2 haplotypes. Variants -810T, -713T, -214A, and -153A always occur together (Hap-I) and variants -810A, -713G, -214C, and -153G form Hap-II. We have hypothesized that high fat diet will alter cellular transcriptional milieu and increase hAT1R gene expression in a haplotype-dependent manner. This will set up an AT1R-mediated feed-forward loop promoting inflammation, oxidative stress, and hypertension in Hap-I mice.
METHOD
Since Hap-I of the human AT1R gene is associated with hypertension in Caucasians, we generated transgenic (TG) mice with Hap-I and Hap-II and studied the physiological significance of high fat diet (HFD) on haplotype specific gene expression. Animals were fed with HFD for 20 weeks followed by blood pressure (BP) analysis and collection of their tissues for molecular and biochemical studies.
RESULTS
After HFD treatment, as compared to Hap-II, TG mice with Hap-I show increased expression of hAT1R gene and higher BP; suppression of antioxidant defenses (HO1, SOD1) and increased expression of IL-6, TNFα, IL-1β, NOX1. In vivo ChIP assay has shown that transcription factors CEBPβ, STAT3, and USF bind more strongly to the chromatin obtained from Hap-I TG mice.
CONCLUSIONS
Taken together, our results suggest, that after HFD treatment, as compared to Hap-II, the TG mice with Hap-I overexpress the AT1R gene due to the stronger transcriptional activity, thus resulting in an increase in their BP.
Keywords: angiotensin receptor type I, blood pressure, high fat diet, hypertension, single nucleotide polymorphism, transgenic
Angiotensin-II (AngII) is central to the pathophysiology of cardiovascular and renal systems. Its excess causes inflammatory/oxidative response, hypertension, endothelial dysfunction, and vascular remodeling; effects mediated by the angiotensin receptor subtype 1 (AT1R). Various studies indicate that increased AT1R gene expression contributes to the onset of hypertension and other associated cardiovascular anomalies. An increase in blood pressure (BP) was observed in female transgenic (TG) mice that overexpress AT1aR.1,2 Whereas, in heterozygous AT1R knockout mice, a 50% reduction in AT1aR expression accompanied by significantly reduced systolic BP was observed.2 Furthermore, enhanced expression of AT1 receptors in cardiac myocytes leads to massive cardiac enlargement, congestive heart failure, and early death in TG mice.3,4
Metabolic syndrome (MetS), characterized by obesity and hypertension, has been shown to be associated with an overactive renin–angiotensin system (RAS) (reviewed by Kalupahana and Moussa).5,6 This association is not surprising as RAS promotes an oxidative/inflammatory milieu that is also the hallmark of MetS.5,7 Reactive oxygen species and inflammatory cytokines including, IL-6 and TNF-α increase insulin resistance (IR) and promote hypertriglyceridemia. These events play key role in bringing about vascular endothelial dysfunction and atherosclerosis, which results in cardio-renal damage in chronic MetS. By promoting pro-oxidative and proinflammatory milieu, AngII is intricately linked to the development and progression of MetS. However, reverse association between MetS and AngII-signaling has not been well studied. Although it has been shown by other investigators that obesity could increase AT1R expression, how MetS could regulate hAT1R expression remains inconclusive. Moreover, transcriptional regulation of the hAT1R gene is also incompletely understood and may be affected by altered transcriptional milieu of the MetS.
In this regard, we have identified two distinct haplotypes of the hAT1R gene, based on linkage disequilibrium of SNPs in its promoter.8 Variants -810T (rs275651), -713T (rs275652), -214A (rs422858), and -153A (rs275653) always occurs together (Hap-I representing SNPs TTAA) and variants -810A, -713G, -214C, and -153G always occur together (haplotype-II or Hap-II representing SNPs AGCG). We have shown that Hap-I of the hAT1R is associated with hypertension in Caucasians. To understand the role of these haplotypes in transcriptional regulation of the hAT1R gene and to examine their role in BP regulation, we have generated TG mice containing either Hap-I or Hap-II of the hAT1R gene. We have found that TG mice containing Hap-I have increased hAT1R expression due to increased binding of transcription factors (TFs) such as, USF and CEBP. This is associated with high BP and elevated inflammatory markers in TG mice containing Hap-I as compared to Hap-II.8
We have hypothesized that high fat diet (HFD) will alter cellular transcriptional milieu and increase hAT1R gene expression in a haplotype-dependent manner. This will set up an AT1R-mediated feed-forward loop promoting inflammation, and oxidative stress, and hypertension in Hap-I mice. Thus, in this communication, we set out to examine the contributions of diet-induced MetS in regulating cellular transcriptional milieu and its effects on the hAT1R expression. We show here that HFD precipitates MetS in TG mice; increases the hAT1R expression via increased binding of USF, CEBP, and STAT3 to the hAT1R promoter; significantly increases BP; and, causes cardio-renal damage. Importantly, all of these effects are significantly more pronounced in Hap-I-TG mice of the hAT1R, as compared to the Hap-II. This is the first report showing MetS-induced regulation of the hAT1R gene, which in turn is haplotype dependent.
METHODS
Association of MetS with the T/G allele in haplotypes of hAT1R gene
We have performed association studies for the hAT1R gene in 183 Caucasian subjects (mean age: 46 ± 8 years). Genomic analysis was performed by polymerase chain reaction (PCR) amplification of 238 bp sequence containing SNP at -713 followed by restriction analysis.8 Forty-seven patients were identified to have MetS by the following clinical criteria: patients with central adiposity and receiving treatment for hypertension and diabetes. Chi-square test was performed to analyze allele frequency and its association with MetS.
TG mice and HFD
All animal experiments were performed according to the National Institute of Health Guide for the care and use of laboratory animals and approved by the institutional IACUC committee at the University of Toledo. The TG mice utilized in this study were generated in our laboratory as described previously.2,8 Genotyping analysis of the tail snips, followed by sequencing, was performed to confirm the genetic lineage of these TG mice. As previously described, the TG mice with Hap-I have variants -810T, -713T, -214A, and -153A, whereas TG mice with Hap-II have variants -810A, -713G, -214C, and -153G. For experimental purpose, these TG mice were housed individually. Twelve-week-old control C57 mice or TG male mice containing Hap-I and Hap-II each were divided into 2 groups (n = 4). For both haplotypes, mice were randomly assigned to either a control diet (TD10819) or a commercially available HFD (contain 41 Kcal% fat with 43 Kcal% carbohydrate, D12079B) for a period of 20 weeks. Diets were purchased from Harlan Lab (WI). Animals were maintained in a 22°C room with a 12-hour light/dark cycles and received water, standard chow, and HFD ad libitum.
Quantitative real-time PCR
Kidneys and heart were harvested at the end of HFD treatment and snap frozen in liquid nitrogen. The extracted tissues were stored at −80°C until utilized for further experiments. RNA was isolated using RNeasy Plus mini kit (Qiagen). One microgram of RNA was reverse-transcribed into cDNA using Revert Aid First Strand cDNA Synthesis Kit (Fermentas), as described in the manufacturer’s protocol. Quantitative real-time PCR was performed using primers for genes of interest including mouse and hAT1R; inflammatory mediators: IL1, IL6, TNFα, CRP; mediators of oxidative stress: NOX1, iNOS; adhesion molecules: iCAM, VCAM; and attenuation of antioxidant pathways: HO1, SOD1, and adiponectin. Primers were obtained from Super Array Bioscience Corporation (SAB, MD) or from Integrated DNA Technologies (IDT, IA). Gene expression was examined using Power SYBR green master mix on ABI 7500 Fast Real-Time PCR system from Life Technologies. Threshold cycles (CT) for 3 replicate reactions were determined and relative enriched DNA abundance calculated following normalization with input DNA.
In vivo ChIP analysis
The chromatin immunoprecipitation (ChIP) assay was performed using the EZ-ChIP assay kit from EMD Millipore, MA. Mice were perfused with normal saline and the kidney and the heart were removed, washed and fixed with 1% formaldehyde for 20 minutes at room temperature. The DNA was fragmented by sonication and 10 µl of the chromatin solution was saved as input. Five microgram of each antibody CEBPβ, STAT3, USF2, Pol2, or rabbit immunoglobulin G were added to the tubes containing 900 µl of the sonicated chromatin solution; the mixture was incubated overnight at 4°C. The antibody complexes were captured with the protein A-agarose beads and subjected to serial washes (as described in manufacturer’s protocol). The chromatin fraction was extracted with SDS buffer and reverse cross-linked at 65°C for 4–6 hours. The DNA was then purified as described in the manufacturer’s protocol. The immunoprecipitated DNA (1 µl) and the input DNA (1 µl) were subjected to quantitative PCR using (i) -213for TCCGCAGGAAATGATACTCC as a forward and -213rev ACGAGGCTCTGTTTTGCATT as a reverse primer when USF2 antibody was used for immunoprecipitation. This amplified 217 bp amplicon spanning the USF binding site located around -213 region of the human AT1R gene promoter (ii) -119for TGATAGTTGACACGGGACGA as a forward primer and -119rev TTTATAGTGAGGGGCGTTGC as a reverse primer when and RNA pol II antibody was used. This spanned 178 bp amplicon containing STAT3, CEBP, and RNA pol II binding region of the human AT1R gene promoter. The fraction enriched by rabbit IgG was used as a negative control for nonspecific binding. Specificity of the PCR was also checked using primers for a nonspecific region in the hAT1R gene that does not have a putative site for the used antibodies. Threshold cycles (CT) for 3 replicate reactions were determined and relative enriched DNA abundance calculated following normalization with input DNA.
Immunoblotting
Protein extracts were prepared from the kidney tissues collected from the control- and HFD-treated TG mice. Protein extracts (25 µg) were separated by SDS-PAGE (12% polyacrylamide) and transferred to 0.45-m PVDF membranes (Millipore, Billerica, MA) for 1 hour. The membranes were incubated with hAT1R antibody (1:2000) (catalog no. sc81671, Santa Cruz Biotechnology); developed using an infrared imaging system (Odyssey, LI-COR Biosciences, and Lincoln, NE); and the results were normalized to mouse β-actin (A2228, Sigma).
BP and heart rate measurement in TG mice
BP and heart rate (HR) was measured in the conscious state by radio telemetry as described previously.8,9 Mean arterial pressure (MAP) was continuously acquired by implantation of telemetric probe PA-C20 into the aorta via the left carotid artery. After 1 week of recovery from the surgical procedure, BP and HR readings were recorded every 10 minutes using Data-Dataquest ART software purchased from Data Science International instrument as described previously.10,11 Baseline BP was collected for 7 days followed by BP measurements in response to 20 weeks of HFD treatment.
Measurement of plasma creatinine and aldosterone by ELISA
Plasma creatinine and aldosterone levels were determined by ELISA assay kits purchased from Cayman Chemical (cat#700460) and Kamiya Biomedical Company (cat#KT-57554), respectively. The creatinine and aldosterone concentrations in the samples were determined directly from the standard curve as described in the manufacturer’s protocol.
IR quantification
To quantify IR in our TG lines, we measured plasma glucose and insulin by ELISA (Glucose colorimetric assay kit, Cayman; Mouse insulin ELISA kit, EMD Millipore) to calculate homeostatic model assessment (HOMA2) score (www.dtu.ox.ac.uk/homacalculator/).
Statistical analyses
Association studies for T/G alleles with MetS were analyzed using the chi-square test. All other experiments were conducted with 4 animals in each group. Data are expressed as the means ± SE. Statistical significance was assessed using 2-way analysis of variance with a Tukey Kramer post hoc analysis. The significance level was set at (P < 0.05).
RESULTS
MetS is associated with the T-allele of Hap-I of hAT1R gene
Chi-square test was performed to analyze allele frequency and its association with MetS. Our results show that the T-allele frequency (Hap-I, 0.69) was almost 2 and half times higher than the frequency of the G-allele (Hap-II, 0.31) (P < 0.05) in patients with MetS (Table 1).
Table 1.
Association of T/G allele of hAT1R haplotypes with metabolic syndrome
| Total subjects | Frequency T allele | Frequency G allele | P | |
|---|---|---|---|---|
| Hypertensive with MetS | 47 | 0.69 | 0.31 | <0.05 |
HFD increases hAT1R expression in kidneys and vasculature of Hap-I TG mice
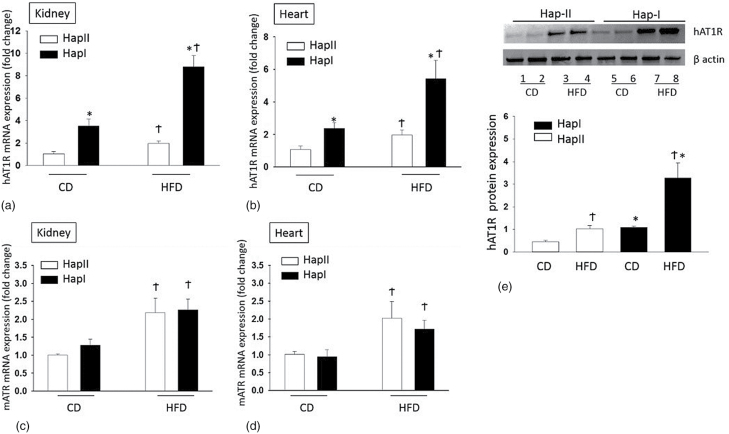
Transcriptional effects of the increased hAT1R were examined by gene expression analysis. Kidney, heart, and the vasculature are the major targets for the hAT1R expression and its physiological actions. Renal and cardiac extracts show increased (P < 0.05) baseline hAT1R mRNA level in the Hap-I TG mice (Figure 1a and b). Importantly, HFD treatment more significantly increased hAT1R transcription in TG mice with Hap-I (4.45 fold in kidney and 2.51-fold in heart after HFD, P < 0.05) (Figure 1a and b). Endogenous mAT1R mRNA is also upregulated in both TG lines by the HFD, as compared to the control diet. Importantly, however, this upregulation is not different in the 2 TG-haplotypes (Figure 1c and d). It is worth noting that the mAT1R gene is exactly the same in the 2 haplotypes as they are both on the same genetic background i.e., C57BL6. Effect of HFD on hAT1R expression was also confirmed by mRNA expression in the aorta (Hap-I shows 3.35-fold increase vs. 1.61-fold in Hap-II) and by immunoblotting in the kidney tissues. Figure 1e show that in comparison to Hap-II the hAT1R is 2.4-fold higher in Hap-I which goes up by 3-folds when treated with HFD.
Figure 1.
Expression of mouse and human AT1R in TG mice by quantitative RT-PCR analysis: (a, b) hAT1R expression and (c, d) mATR expression in the kidney (a, c) and heart (b, d) with CD or HFD. (e) The hAT1R protein expression in the kidney by immunoblotting (top panel) and its densitometry analysis normalized to β actin (n = 4). *P < 0.05 compared with Hap II. †P < 0.05 compared with their respective control diet group. Abbreviations: CD, control diet; HFD, high fat diet; TG, transgenic.
HFD promotes greater binding of TFs to the chromatin of Hap-I mice
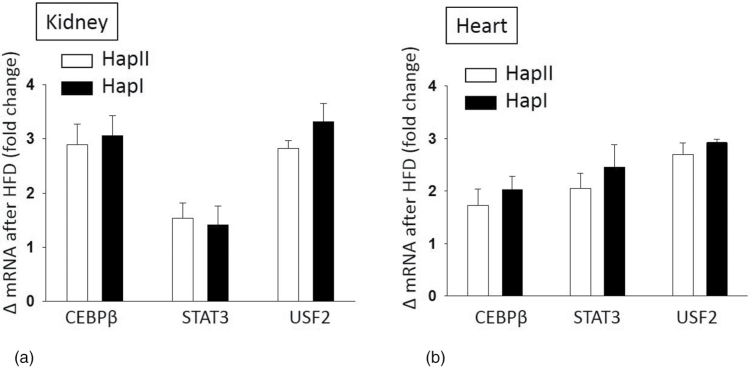
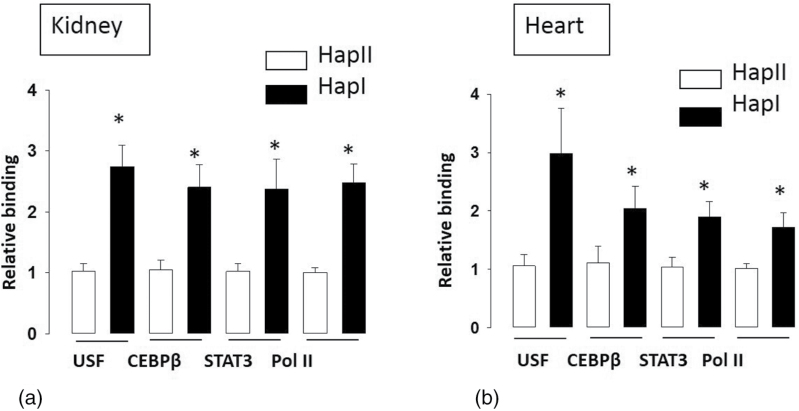
HFD is well documented to promote inflammation and oxidative stress.5,6,12–15 These in turn alter the levels of TFs like STAT3, CEBPβ, and USF. Nucleotide sequence of the hAT1R gene promoter contains binding sites for these TFs. Also, the nucleotide sequence in -214 regions of Hap-I promoter has stronger homology to the USF2 TF.8 This may alter the binding of USF2 and other coactivating TFs to the promoter region and may differentially regulate hAT1R gene expression. Therefore, we examined the chronic effect of HFD (20 weeks) on expression of these TFs and also on their binding to the hAT1R promoter. Our Q-PCR results show that TFs STAT3, CEBPβ, and USF2 were significantly upregulated (P < 0.01) in the kidney and heart tissues from both the TG lines and this effect was independent of their haplotype (Figure 2). To elucidate the role of SNPs in differential binding of TFs, ChIP assays were performed using STAT3, CEBPβ, USF2, and Pol II antibodies on chromatin extracts from the kidney and heart tissues of the TG animals. As shown in Figure 3a, STAT3 (2.37-fold), CEBPβ (2.41-fold), USF2 (2.74-fold), and Pol II (2.48-fold) bind more strongly to the chromatin obtained from renal tissue of TG mice with Hap-I, as compared to Hap-II. Similar results were observed in chromatin isolated from the heart tissue of these TG animals (Figure 3b). These results show increased binding of TFs to the hATR promoter in kidney and heart of TG mice containing Hap-I.
Figure 2.
Effect of HFD on expression of transcription factors in the kidney and heart of transgenic mice: change in mRNA expression for CEBPβ, STAT3, and USF2 in the kidney (a) and heart (b) after HFD treatment. Results are shown as mean ± SE (n = 4). Abbreviations: CD, control diet; HFD, high fat diet.
Figure 3.
ChIP assay using immunoprecipitated DNA from the kidney (a) and the liver (b) of TG mice containing either Hap-I or Hap-II in the presence of HFD. The assay was performed in the presence of antibodies against USF2, CEBPβ, STAT3, and Pol II. Immunoprecipitated DNA was used to amplify the nucleotide sequence encompassing -213 region (USF2 binding region) and -119 region (CEBPβ, STAT3, and PolII binding regions) as described in “Material and Methods”. Quantification of the PCR amplicon of respective antibody-enriched DNA was done by normalizing to their input DNA as performed by Q-PCR. Result shows a significant increase in the high-fat diet-induced binding of respective TFs and Pol II in TG mice with Hap-I. *P < 0.05 compared with Hap II. Results are shown as mean ± SE (n = 4). Abbreviations: AU, arbitrary units; ChIP, chromatin immunoprecipitation; HFD, high fat diet; Q-PCR, quantitative polymerase chain reaction; TFs, transcription factors; TG, transgenic.
Effects of HFD treatment on physiological and MetS-related parameters in the 2 TG lines
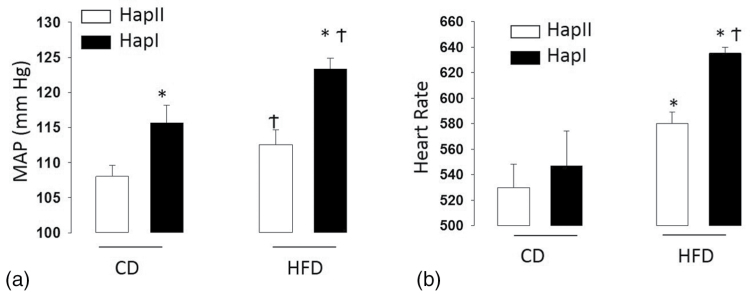
Multiple physiological parameters were assessed to gauge the severity of MetS in our 2 haplotypes. The following were measured after HFD: MAP (Figure 4), oxidative stress (Figure 5), creatinine levels, HOMA II score, and plasma aldosterone levels (Table 2). They are all significantly (P < 0.05) and consistently higher in Hap-I mice as compared to mice with Hap-II.
Figure 4.
Blood pressure (a) and heart rate (b) analysis in TG mice: each bar represents MAP or HR taken from 4 male animals over a period of 24 hours. The hourly mean was taken from the average of 6 MAP or HR data points at an interval of 10 minutes each. *P < 0.05 compared with Hap-II; †P < 0.05 compared with their control diet group. Results are shown as mean ± SE (n = 4). Abbreviations: HFD, high fat diet; HR, heart rate; MAP, mean arterial pressure; TG, transgenic.
Figure 5.
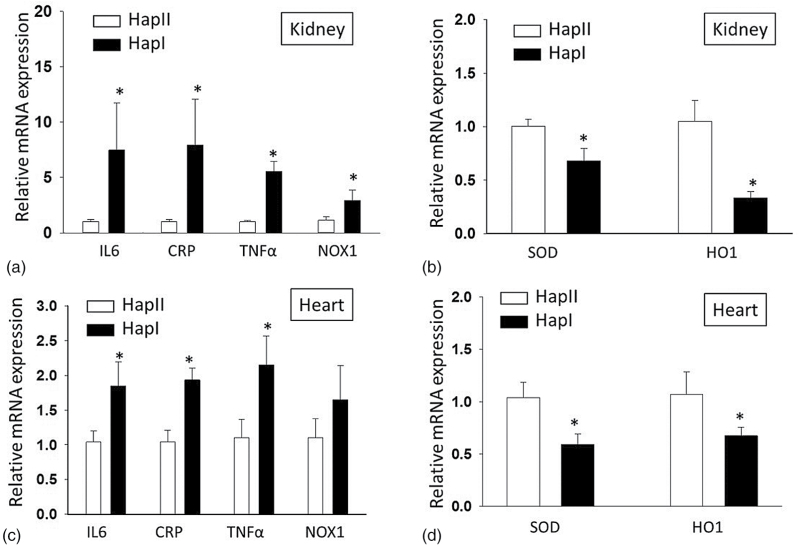
Effect of high fat diet on mRNA expression of proinflammatory markers and antioxidant defense in the kidney and heart of transgenic mice by quantitative RT-PCR analysis: functional relevance of the upregulated hAT1R was assessed by relative mRNA expression analysis of the proinflammatory, pro-oxidative genes including IL6, CRP, TNFα, NOX1 (a, kidney; c, heart), and antioxidant defense markers SOD1 and HO1 (b, kidney; d, heart) after HFD treatment. *P < 0.05 compared with Hap II after HFD. Results are shown as mean ± SE (n = 4). Abbreviations: CRP, C-reactive protein; HFD, high fat diet; RT-PCR, real-time polymerase chain reaction; TNFα, tumor necrosis factor alpha.
Table 2.
Plasma creatinine, aldosterone, and HOMA2 score in transgenic mice
| Parameter | Hap I | Hap II | P |
|---|---|---|---|
| Plasma creatinine (mg/dl) | 1.50 ± 0.06 | 0.76 ± 0.13 | <0.05 |
| Plasma aldosterone (pg/ml) | 634 ± 86 | 341 ± 16 | <0.05 |
| HOMA2 score | 6.06 ± 0.19 | 3.56 ± 0.12 | <0.05 |
Abbreviation: HOMA, homeostatic model assessment.
MAP and HR increased in Hap-I TG mice
We observed that TG mice with Hap-I have significantly increased MAP as compared to Hap-II at resting conditions consistent with our published study.8 We next analyzed the effect of HFD on the MAP and HR of both of our TG mice with Hap-I and Hap-II. Our results show that MAP and HR increases in both the TG lines but more significantly (P < 0.05) in mice containing Hap-I of hAT1R (from 115.7 ± 2.4 to 123.4 ± 1.7 mm Hg; P < 0.05) as compared to Hap-II animals (from 108 ± 1.6 to 112.5 ± 2.1 mm Hg, P < 0.05) (Figure 4). To validate our TG model of HFD-induced hypertension, we also measured BP in C57/BL6 non-TG background mice. We observed that HFD treatment also increased MAP in C57 mice (from 98 ± 1.3 to 105 ± 2.8 mm Hg) which is similar to the AT1R TG animals. This suggests that our ATR-TG model can be used to study the role of HFD in MetS-related comorbidities (especially high BP). Each bar in Figure 5 represents MAP and HR taken from 4 male animals over a period of 24 hours. The hourly mean was taken from the average of 6 MAP and HR data points at an interval of 10 minutes each. It is important to note that there is significant variability in HR tracings, which may have contributed to the elevated BP in either haplotype. Especially, since recent evidence has pointed to the role of AngII and the hAT1R in central sympathetic outflow.16–18 These examinations, however, are beyond the scope of the current study.
TG mice containing Hap-I of hAT1R show an increased expression of proinflammatory and oxidative markers.
Functional relevance of the upregulated hAT1R was assessed by expression analysis of the proinflammatory/oxidative markers including IL6, CRP, TNFα, and NADPH oxidase (NOX1). Complementary experiments were performed to examine the cellular antioxidant defenses including superoxide dismutase (SOD1) and hemoxygenase (HO1). Our results show that HFD treatment results in a significant (P < 0.05) increase in all the indicated inflammatory markers in both the renal (IL6 7.4-fold; CRP 7.9-fold; TNFα 5.5-fold; Nox1 2.94-fold) and cardiac (IL6 1.8-fold; CRP 1.9-fold; TNFα 2.15-fold; Nox1 1.65-fold) tissues from the TG mice containing Hap-I as compared to the Hap-II of the hAT1R gene (Figure 5a and c). Similarly, the expression of genes involved in defense system was significantly downregulated in both cardiac (SOD 0.59-fold; HO1 0.67-fold) and renal (SOD 0.33-fold; HO1 0.68-fold) tissues in Hap-I animals (Figure 5b and d).
Increased plasma creatinine levels in Hap-I TG animals.
Plasma creatinine levels were elevated in both lines after HFD but significantly (P < 0.05) higher in Hap-I TG mice (Hap-II 0.76 ± 0.13 mg/dl vs. Hap-I 1.50 ± 0.06 mg/dl) (Table 2).
Hap-I TG mice show higher HOMA-IR score.
The HOMA2 is significantly (P < 0.05) higher in Hap-I mice after HFD (Hap-II, 3.56 ± 0.12 vs. Hap-I, 6.06 ± 0.19) (Table 2).
TG mice with Hap-I show higher plasma aldosterone.
Plasma aldosterone levels were significantly increased in both TG mice after HFD but more significantly in Hap-I (306 pg in Hap-I vs. 95pg in Hap-II, P < 0.05) (Table 2).
DISCUSSION
RAS is a well-established link between MetS, inflammation, and IR. Most of the physiological actions of the RAS pathway are induced by the AT1R in target cells. Our present study is based on the hypothesis that MetS mediates, allele-specific, transcriptional regulation of the hAT1R gene in cardiac and renal tissues. In our previous studies, we have identified 2 haplotypes (Hap-I and Hap-II) of the hAT1R gene based on SNPs in its promoter. We have shown that (i) Hap-I is associated with hypertension in Caucasians and (ii) TG mice with Hap-I overexpress hAT1R gene in different tissues and have high BP as compared to the TG mice containing Hap-II.8 Our present study highlights the role of MetS in regulating hAT1R expression. This is important since frequency of Hap-I of the hAT1R gene is significantly higher in patients with MetS (Table 1).
The first key finding of our study is that HFD increases hAT1R expression in both TG lines. More importantly, this effect of HFD is haplotype-dependent where, hAT1R expression is significantly more in Hap-I as compared to Hap-II. HFD was purposed to induce a state of MetS in our experimental groups. HFD-induced MetS has been shown previously to cause renal and cardiovascular disorders.6,12,13 Oliveira et al. have shown that systolic BP, hyperglycemia, and cardiac remodeling were induced by a hyper caloric diet for 20 weeks in Wistar–Kyoto rats.19 MetS-induced hAT1R upregulation is not unexpected and is in line with published reports indicating RAS involvement in MetS. Recent studies by Xue et al.15 show that HFD induces central RAS activation, inflammation, and enhanced hypertensive response to AngII which can be abolished using AT1R blocker, irbesartan. However, the novel observation in our study is that the upregulation of hAT1R gene after HFD was significantly higher in TG mice containing Hap-I as compared to TG mice with Hap-II. These results confirm our hypothesis that gene-haplotypes not only govern basal gene regulation but also significantly affect its regulation in pathophysiological settings, like the MetS. This is further confirmed by the observation that the endogenous mAT1R, although upregulated, is not different in the 2 TG-haplotypes. It is well established that HFD induces chronic inflammation,12,13,15 which in turn alters cellular TF expression levels. Our results confirm these observations and show that HFD increases the levels of cardiac and renal TFs including, CEBPβ, STAT3, and USF2 that bind to the promoter of the hAT1R gene. It is noteworthy that increase in the expression of these TFs is haplotype independent.
Although, MetS-induced increase in TFs is not haplotype-dependent, their binding to the hAT1R as determined by ChIP assay depends on the haplotype. Thus, haplotype-dependent, variable TF binding to the hAT1R gene in mice undergoing diet-induced MetS is the second key finding of the study. We have previously shown8 that nucleotide sequence of the Hap-I (containing nucleoside A at -214) has greater homology with E-box (CANNTG) that is recognized by helix-loop-helix (HLH) family of TFs including USF1.20 USF1 binds as a homo-dimer, or as a heterodimer with the related TF USF2 to E boxes and controls the expression of a number of genes.21,22 USF has been shown to increase the expression of genes involved in glucose and lipid metabolism.23 Overexpression of USF2 in TG mice influences metabolic traits such as obesity, lipid profiles, and glucose/insulin ratio.24 Additionally, earlier studies have established SNP-dependent differential gene regulation by USF.25–27 Beside USF, the other TFs which have sequence homology to the proximal promoter region of the hAT1R gene are CEBPβ and STAT3. Their binding affinity to the hAT1R promoter may vary due to their cross-talk with USF1. Role of CEBPβ is well documented in etiology and progression of MetS.28 Previous work has shown that binding of CEBPβ acts like a “pioneer” TF and opens up the chromatin to allow the binding of other TFs (such as glucocorticoid receptor) to the opened chromatin.29 In addition, CEBPβ and –δ are involved in IL6 and TNF induced expression of various genes.30 Also, the genetic variations in STAT3 are correlated with obesity and IR in adult males.31 Thus, this has led us to conclude that the MetS alters cellular transcriptional milieu independent of haplotypes. It is the differential binding of these TFs to the 2 haplotypes that then dictates variable hAT1R expression. This highlights the role of SNPs in the hAT1R promoter in governing its expression during pathophysiological stresses, like the MetS.
Our final key finding of this study is that pathophysiological impact of this differential AT1R regulation is significant. The principal pathophysiological scenario envisioned in this study, diet-induced MetS, is frequently linked with an increased circulating levels of corticoids, proinflammatory milieu and end-organ damage. Although not directly demonstrated in this study, RAS activation is a frequent accompaniment in the setting of MetS. Increased expression of the AT1R has the potential to exacerbate the pathological outcomes of MetS, including increased IR, increased BP, oxidative stress, and cardio-renal remodeling.15,32 Thus, MetS can operate in a vicious cycle with the RAS; where, altered transcriptional milieu up regulates AT1R, in turn, contributing to the pathologies of the MetS. Crucially, however, this MetS-induced hAT1R regulation is haplotype-dependent with clear implications for associated pathophysiology. Higher BP, oxidative stress, exaggerated IR (HOMA2 score), and renal damage (as evidenced by elevated plasma creatinine levels) in Hap-I mice are indicative of the same. These observations are in line with published reports showing a direct correlation between AT1R levels and oxidative stress, primarily via activation of NADPH oxidase, and secretion of proinflammatory/profibrotic cytokines.33 Increased expression of cytokines, including IL-6, TNFα, and IL1β; and increased levels of NADPH oxidase component, NOX1, and suppressed antioxidant defense system confirm the functional role of increased hAT1R activation after HFD treatment in TG mice with Hap-I. Thus, the synergistic interaction between MetS and hAT1R, in Hap-I mice, worsens their cardiac and renal functions with progression to end-organ damage.
In summary, the present study demonstrates that pathophysiological variables like the HFD alters cellular transcriptional environment that in turn variably regulate the expression of hAT1R gene based on its haplotype. In this regard, Hap-I of hAT1R is much more amenable to such modulations due to greater affinity of its promoter to the upregulated TFs. This would potentially make subjects with Hap-I more susceptible to HFD-induced, AT1R-mediated complications such as hypertension and associated cardiovascular-renal damage. Clinically, this is highly significant as it will help identify individuals with risk haplotype undergoing diet-induced MetS. In this setting, identification of “at-risk” haplotypes for the hAT1R will benefit patients by providing timely and targeted therapy so as to prevent long-term complications.
DISCLOSURE
The authors declared no conflict of interest.
ACKNOWLEDGMENT
This work was supported, in whole or in part, by NIH grants HL81752, HL105113, and HL092558 (to A.K.). S.J., N.P., and A.K designed the research. S.J., A.R., M.K., and N.S., B.M. performed the experiments. S.J., N.P., M.K., and A.K. analyzed the data. S.J. and N.P wrote the manuscript.
REFERENCES
- 1. Le TH, Kim HS, Allen AM, Spurney RF, Smithies O, Coffman TM. Physiological impact of increased expression of the AT1 angiotensin receptor. Hypertension 2003; 42:507–514. [DOI] [PubMed] [Google Scholar]
- 2. Ito M, Oliverio MI, Mannon PJ, Best CF, Maeda N, Smithies O, Coffman TM. Regulation of blood pressure by the type 1A angiotensin II receptor gene. Proc Natl Acad Sci USA 1995; 92:3521–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hein L, Stevens ME, Barsh GS, Pratt RE, Kobilka BK, Dzau VJ. Overexpression of angiotensin AT1 receptor transgene in the mouse myocardium produces a lethal phenotype associated with myocyte hyperplasia and heart block. Proc Natl Acad Sci USA 1997; 94:6391–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paradis P, Dali-Youcef N, Paradis FW, Thibault G, Nemer M. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc Natl Acad Sci USA 2000; 97:931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalupahana NS, Moustaid-Moussa N. The renin-angiotensin system: a link between obesity, inflammation and insulin resistance. Obes Rev 2012; 13:136–149. [DOI] [PubMed] [Google Scholar]
- 6. Sweazea KL, Walker BR. High fat feeding impairs endothelin-1 mediated vasoconstriction through increased iNOS-derived nitric oxide. Horm Metab Res 2011; 43:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skov J, Persson F, Frøkiær J, Christiansen JS. Tissue renin-angiotensin systems: a unifying hypothesis of metabolic disease. Front Endocrinol (Lausanne) 2014; 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jain S, Prater A, Pandey V, Rana A, Puri N, Kumar A. A haplotype of angiotensin receptor type 1 associated with human hypertension increases blood pressure in transgenic mice. J Biol Chem 2013; 288:37048–37056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jain S, Tillinger A, Mopidevi B, Pandey VG, Chauhan CK, Fiering SN, Warming S, Kumar A. Transgenic mice with -6A haplotype of the human angiotensinogen gene have increased blood pressure compared with -6G haplotype. J Biol Chem 2010; 285:41172–41186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Butz GM, Davisson RL. Long-term telemetric measurement of cardiovascular parameters in awake mice: a physiological genomics tool. Physiol Genomics 2001; 5:89–97. [DOI] [PubMed] [Google Scholar]
- 11. Jain S, Vinukonda G, Fiering SN, Kumar A. A haplotype of human angiotensinogen gene containing -217A increases blood pressure in transgenic mice compared with -217G. Am J Physiol Regul Integr Comp Physiol 2008; 295:R1849–R1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aoqui C, Chmielewski S, Scherer E, Eissler R, Sollinger D, Heid I, Braren R, Schmaderer C, Megens RT, Weber C, Heemann U, Tschöp M, Baumann M. Microvascular dysfunction in the course of metabolic syndrome induced by high-fat diet. Cardiovasc Diabetol 2014; 13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kobayasi R, Akamine EH, Davel AP, Rodrigues MA, Carvalho CR, Rossoni LV. Oxidative stress and inflammatory mediators contribute to endothelial dysfunction in high-fat diet-induced obesity in mice. J Hypertens 2010; 28:2111–2119. [DOI] [PubMed] [Google Scholar]
- 14. Panchal SK, Poudyal H, Iyer A, Nazer R, Alam MA, Diwan V, Kauter K, Sernia C, Campbell F, Ward L, Gobe G, Fenning A, Brown L. High-carbohydrate, high-fat diet-induced metabolic syndrome and cardiovascular remodeling in rats. J Cardiovasc Pharmacol 2011; 57:611–624. [DOI] [PubMed] [Google Scholar]
- 15. Xue B, Thunhorst RL, Yu Y, Guo F, Beltz TG, Felder RB, Johnson AK. Central renin-angiotensin system activation and inflammation induced by high-fat diet sensitize angiotensin II-elicited hypertension. Hypertension 2016; 67:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zucker IH, Xiao L, Haack KK. The central renin-angiotensin system and sympathetic nerve activity in chronic heart failure. Clin Sci (Lond) 2014; 126:695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsuda K. Renin-angiotensin system and sympathetic neurotransmitter release in the central nervous system of hypertension. Int J Hypertens 2012; 2012:474870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zucker IH, Schultz HD, Patel KP, Wang W, Gao L. Regulation of central angiotensin type 1 receptors and sympathetic outflow in heart failure. Am J Physiol Heart Circ Physiol 2009; 297:H1557–H1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oliveira Junior SA, Padovani CR, Rodrigues SA, Silva NR, Martinez PF, Campos DH, Okoshi MP, Okoshi K, Dal-Pai M, Cicogna AC. Extensive impact of saturated fatty acids on metabolic and cardiovascular profile in rats with diet-induced obesity: a canonical analysis. Cardiovasc Diabetol 2013; 12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 2000; 20:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sawadogo M. Multiple forms of the human gene-specific transcription factor USF. II. DNA binding properties and transcriptional activity of the purified HeLa USF. J Biol Chem 1988; 263:11994–12001. [PubMed] [Google Scholar]
- 22. Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res 1994; 22:427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vallet VS, Casado M, Henrion AA, Bucchini D, Raymondjean M, Kahn A, Vaulont S. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J Biol Chem 1998; 273:20175–20179. [DOI] [PubMed] [Google Scholar]
- 24. Wu S, Mar-Heyming R, Dugum EZ, Kolaitis NA, Qi H, Pajukanta P, Castellani LW, Lusis AJ, Drake TA. Upstream transcription factor 1 influences plasma lipid and metabolic traits in mice. Hum Mol Genet 2010; 19:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dickson ME, Tian X, Liu X, Davis DR, Sigmund CD. Upstream stimulatory factor is required for human angiotensinogen expression and differential regulation by the A-20C polymorphism. Circ Res 2008; 103:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J Mol Med (Berl) 2005; 83:440–447. [DOI] [PubMed] [Google Scholar]
- 27. Zhao YY, Zhou J, Narayanan CS, Cui Y, Kumar A. Role of C/A polymorphism at -20 on the expression of human angiotensinogen gene. Hypertension 1999; 33:108–115. [DOI] [PubMed] [Google Scholar]
- 28. van der Krieken SE, Popeijus HE, Mensink RP, Plat J. CCAAT/enhancer binding protein β in relation to ER stress, inflammation, and metabolic disturbances. Biomed Res Int 2015; 2015:324815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 2013; 32:1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alam T, An MR, Mifflin RC, Hsieh CC, Ge X, Papaconstantinou J. trans-activation of the alpha 1-acid glycoprotein gene acute phase responsive element by multiple isoforms of C/EBP and glucocorticoid receptor. J Biol Chem 1993; 268:15681–15688. [PubMed] [Google Scholar]
- 31. Gianotti TF, Sookoian S, Gemma C, Burgueño AL, González CD, Pirola CJ. Study of genetic variation in the STAT3 on obesity and insulin resistance in male adults. Obesity (Silver Spring) 2008; 16:1702–1707. [DOI] [PubMed] [Google Scholar]
- 32. Boustany CM, Brown DR, Randall DC, Cassis LA. AT1-receptor antagonism reverses the blood pressure elevation associated with diet-induced obesity. Am J Physiol Regul Integr Comp Physiol 2005; 289:R181–R186. [DOI] [PubMed] [Google Scholar]
- 33. Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol 2007; 292:C82–C97. [DOI] [PubMed] [Google Scholar]