Abstract
Noncoding RNAs (ncRNA) include a diverse range of functional RNA species—microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) being most studied in pathophysiology. Cardiovascular morbidity is associated with differential expression of myriad miRNAs; miR-21, miR-155, miR-126, miR-146a/b, miR-143/145, miR-223, and miR-221 are the top 9 most reported miRNAs in hypertension and atherosclerotic disease. A single miRNA may have hundreds of messenger RNA targets, which makes a full appreciation of the physiologic ramifications of such broad-ranging effects a challenge. miR-21 is the most prominent ncRNA associated with hypertension and atherosclerotic disease due to its role as a “mechano-miR”, responding to arterial shear stresses. “Immuno-miRs”, such as miR-155 and miR-223, affect cardiovascular disease (CVD) via regulation of hematopoietic cell differentiation, chemotaxis, and activation in response to many pro-atherogenic stimuli. “Myo-miRs”, such as miR-1 and miR-133, affect cardiac muscle plasticity and remodeling in response to mechanical overload. This in-depth review analyzes observational and experimental reports of ncRNAs in CVD, including future applications of ncRNA-based strategies in diagnosis, prediction (e.g., survival and response to small molecule therapy), and biologic therapy.
Keywords: atherosclerosis, biomarker, blood pressure, hypertension, microRNA, miRNA, myocardial infarction, ncRNA, noncoding RNA
Hypertension and cardiovascular disease (CVD) have well-documented genetic, epigenetic, and environmental effectors. While approximately 25 mutations and 53 single nucleotide polymorphisms have been associated with hypertension,1,2 environmental factors such as diet and the increased prevalence of obesity are predominant drivers of disease.3 Obesity is most prevalent in countries where food-related expenditures represent a low percentage of per capita gross domestic product, such as the United States. Of the 85 countries analyzed, the United States has the lowest percentage (6.4%) of total consumer spending on food and 23.5% of deaths from CVD (CDC data), compared to Nigeria at 56.4%,4 with 7% of mortality attributed to CVD (WHO data). Fittingly, bariatric surgery (e.g., Roux-en-Y gastric bypass) showed durable remittance of hypertension and dyslipidemia extending 12 years post intervention.5 In attempts to better understand CVD etiology, noncoding RNAs (ncRNAs) have been widely assayed and shown to have differential expression in cardiovascular and metabolic disease, when examined in a variety of samples. In vivo interventions blocking microRNA (miRNA) activity have been shown to remediate CVD disease in animal models. This review will focus on the main ncRNAs involved in CVD, based on current literature.
NONCODING RNAS: BIOGENESIS AND FUNCTION
The human genome is 98.5% non–protein-coding DNA. Some of this non–protein-coding DNA is transcribed to a heterogeneous group of functional RNA species broadly called ncRNA.6,7 ncRNAs are categorized into 3 classes based on size: small (~19–25 nt.), intermediate-sized (~20–200 nt.), and long (>200 nt.); Please see Table 1. miRNAs are small (21–25 nucleotides) single-stranded RNAs, which can be highly conserved across species8 and are the mostly widely studied functional ncRNA. miRNAs may occur in the genome in a polycistronic organization [1 primary transcriptional event yields multiple mature miRNAs from a single primary miRNA strand, this primary strand can also function as an long ncRNAs (lncRNAs) with repressor functions, and may be further processed to miRNAs. These polycistronic miRNA genes are often referred to as a “cluster” of microRNAs, however, clusters may also—simply—refer to miRNA genes with <10 kb between them. The primary miRNA (product of RNA polymerase II) may then form multiple stem loop structures that are spliced by Drosha, DGCR8 RNase II complex into multiple pre-miRNAs within the nucleus before being exported (exportin 5) to the cytoplasm where they are cleaved (Dicer-TRBP complex) and associate with argonaute 2 (Ago2) protein to become part of the microRNA-induced silencing complex (miRISC), referred to from now as a “mature” microRNA. mRNA knockdown with siRNAs essentially utilize this endogenous miRISC pathway to knockdown targets (Figure 1). It is important to note, however, that a family of miRNAs may be coded by different chromosomes but have the same or very similar seed sequences and similar targets and physiologic roles. The miRNA expression atlas by Landgraf et al. is a seminal contribution to our understanding of the variable abundance of miRNA species across tissue compartments in vivo.9
Table 1.
Noncoding RNA classifications
| Class | Size in bases | Location | Functional role | Example |
|---|---|---|---|---|
| Small106–108 | miRNAs (~19–24) | Intracellular, extracellular | Post-transcriptional regulation of gene expression by degradation or translation repression of target mRNAs | miRNA-21 |
| siRNA (~21–23) | Exogenous | Silencing gene expression by degradation of target mRNAs | TTR-siRNA109 | |
| piRNAs (~26–31) | Repression of reproductive cell-specific gene expression | piRNAs targeting LINE1 elements | ||
| Intermediate107,110,111 | snoRNAs (~20–200) | Intra-nucleolus | Modification and processing of rRNA precursors | SNORD |
| snRNAs | Intra-nuclear | Nuclear maturation of primary transcripts of mRNAs, RNA splicing | U snRNA | |
| Long106,112 | >200 | Nucleus, cytoplasm, mitochondria | Maintaining inactive chromatin as scaffold by repressing genes including HOXD genes | HOTAIR (>200) |
| X-chromosome inactivation | XIST (~17 kb) | |||
| Repression of XIST (antisense transcript of XIST) | TSIX (>200) |
Abbreviations: HOTAIR, (HOX) transcript antisense RNA; HOX, a subset of homeotic genes; miRNA, microRNA; mRNA, messenger RNA; piRNA, piwi-interacting RNA; rRNA, ribosomal RNA; siRNA, small interfering RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; TSIX, XIST antisense RNA; XIST, X-inactive specific transcript.
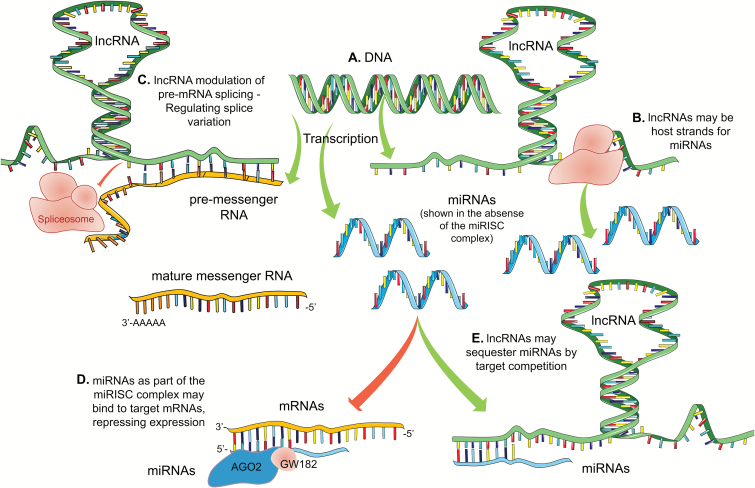
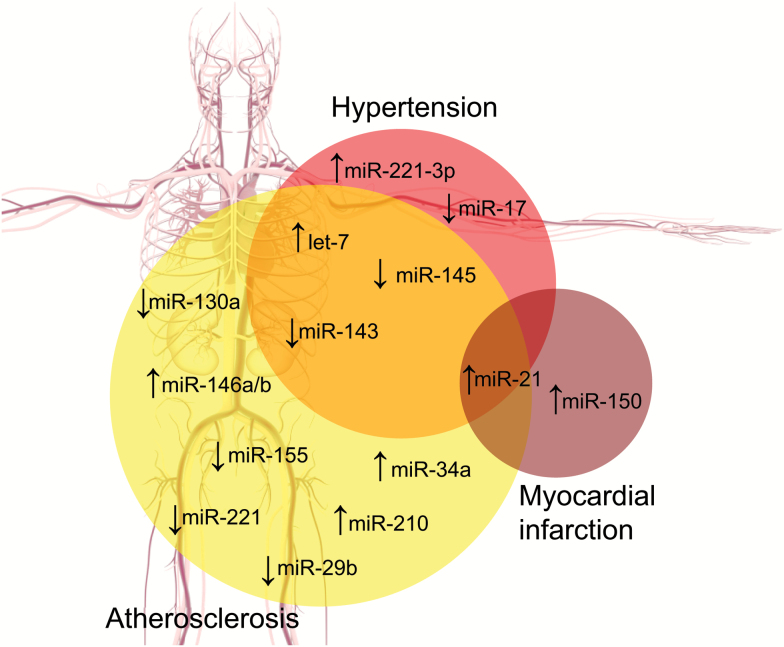
Figure 1.
Dysregulation of miRNA levels in human CVD. Venn diagram depicts how miRNAs change in 3 main categories, hypertension, atherosclerosis, and MI. The arrow that precedes the miRNA represents the authors summary of the literature and is not conclusive; there may be conflicts in reports but the authors have attempted to generalize the findings to the best of their abilities. miR-21 is one of the only miRNAs that has a reasonable history of being reported in human hypertension, atherosclerosis, and MI. miR-150 is the species most associated with acute MI in humans. Many miRNAs are associated with hypertension and atherosclerosis and these will be reviewed in the text. Abbreviations: CVD, cardiovascular disease; MI, myocardial infraction; miRNA, microRNA.
miRNA genes may also be present in the introns between exons of a coding gene and are often referred to as intronic miRNAs. Intronic primary miRNAs are transcribed at approximately a 1:1 ratio with pre-mRNAs, with 1 transactivation of the “gene” potentially resulting in a mature miRNA and a mature mRNA; a context-appropriate example of this is miR-21 (MIRN21 gene) which is intronic within the mRNA coding TMEM49/VMP1 (vacuole membrane protein-1) gene.10 Intronic ncRNAs (lnc or mi-RNA) may have a negative feedback loop with the host mRNA directly, or the pathway in which the host gene (“host RNA”) plays a role. Exemplary intronic miRNA-mRNA feedback relationships include miR-33a/b (intronic) with SREBF2/SREBF1 pathways. miR-33b represses ABCA1 expression, the main cholesterol efflux protein within cells, while the host RNA SREBP2 functions to increase transcription of cholesterol synthetic and uptake genes HMGCR, LDLR, and HMGCS. miR-33a represses fatty acid degradation genes CPT1A, CROT, and HADHB, while SREBP1 increases fatty acid synthetic genes FASN, SCD, and ACC.11,12 Other reports of intronic miRNA-host mRNA pathway feedback loop includes but are not limited to miR-579-ZFR, miR-26b-CTDSP2, miR-301-SKA2, and miR-338-AATK pathway,13–18 making first-line investigation of the immediate pathways around the host RNA reasonable for an intronic ncRNA in question. While the TMEM49 protein product is putatively involved with autophagy mechanisms, no clear link has been verified between miR-21 and TMEM49. Data support a transcriptional schema in which miR-21 is semi-independent of TMEM49-transcription through an extra-ordinary mechanism.19,20 An example of an intergenic miRNA gene (between protein coding genes) pertinent to CVD is miR-223 (MIRN223). A pri-miR-223 strand produces no coding mRNA and MIRN223 is present once in the human (and mouse) genome. Also, the pri-miR-223 transcript may function as a lncRNA—linc-223—with effects on interferon regulatory factor 4 (IRF4) mRNA, as studied in the context of leukemogenesis by Mangiavacchi et al.21 The role of miR-223 in CVD is discussed further in the text.
Alternative host mRNA adenylation is a mechanism by which a ncRNA-host mRNA feedback loop can be differentially regulated.13,22 More specifically, the length of a mRNA 3′UTR may be changed due to alternative polyadenylation (APA; shortened vs. the maximal length variant) removing miRNA binding sites effectively producing mature mRNA transcript variants that may not be targeted by an miRNA or ncRNA. This is obviously case specific, though >50% of human genes yield APA-derived transcript variants,23 and may account for some of the inaccuracy in miRNA-mRNA target prediction. Currently, links between APA, ncRNAs, and CVD are not numerious,24–26 though reports will likely increase as RNA-Seq-based analysis required for such studies becomes more commonplace.
NONCODING RNAS: LOCALIZATION AND DETECTION STRATEGIES
miRNA species may be present in bodily fluids at exceedingly low levels, that require quantitative polymerase chain reaction (qPCR)-based methods for their detection. Those found in serum or plasma require qPCR detection and are commonly called “circulating” miRNAs and can be associated: with albumin, as a part of lipoprotein particles (high-density lipoprotein cholesterol, low-density lipoprotein cholesterol), or within submicroscopic vesicles [microvesicles (100–1000 nm), exosome (30–100 nm)] of varying sizes.27–29 Due to the complex and not fully understood compartmental enrichment of miRNAs, it is important to reference the primary work to fully understand the methodology used to isolate miRNA, as findings will vary due to sample processing and the association of the mature miRNA species to other complexes. These caveats are especially relevant to the development of an extracellular miRNA or signature of miRNAs as a clinical biomarker.30 Processing can have dramatic effects on miRNA levels, including contamination with red blood cell–derived ncRNAs due to hemolysis. Additionally, platelet contamination can affect plasma-derived miRNA levels. Anticoagulated blood should be spun at sufficient speed to remove platelets, and care should be taken when separating the plasma following centrifugation. It should be noted that heparin is not compatible with qPCR-based methods used for biofluid (e.g., plasma, urine, tears, etc.) miRNA detection, and whole blood should be collected in EDTA containing vacutainers. Plasma miRNAs are quite stable, and RNA preservation compounds added to the vacutainer are not needed at the time of blood collection, making retrospective or freezer-studies acceptable upon peer scrutiny. Small RNA-Seq and array-based detection methods can be used when miRNAs and lncRNAs are isolated from sources of abundant levels, such as cells or tissue.
A default PubMed search at the time of writing for “((microRNA) AND hypertension)” resulted in 528 publications—7 clinical trials including work by our group on renin-sensitive microRNAs in humans.31 All publications are for primary literature not indexed within PubMed as a review article. A “((microRNA) AND atherosclerosis)” search returned 617 publications with 4 indexed as clinical trials. “((lncRNA) AND atherosclerosis)” yielded only 49 publications (2 clinical trials) and “((lncRNA) AND hypertension)” only 44 (0 clinical trials). Thus, this review will focus primarily on microRNA (miRNA) species. Table 2 contains a miRNA species-specific search, which dictated which miRNAs were focused on in the text. Tables 3 and 4 are not limited by the number of reports on a miRNA and are intended to be broad and inclusive.
Table 2.
PUBMED database search returns by miRNA species
| RNA name | AND (hypertension) | AND (atherosclerosis) | Combined | Family/cluster |
|---|---|---|---|---|
| miR-21 | 44 | 49 | 93 | |
| miR-155 | 17 | 48 | 65 | |
| miR-126 | 17 | 46 | 63 | |
| miR-146a/b | 19 | 37 | 50 | |
| miR-145 | 16 | 23 | 39 | A |
| miR-27a/b | 19 | 15 | 34 | |
| miR-143 | 12 | 19 | 31 | A |
| miR-223 | 12 | 19 | 31 | |
| miR-221 | 8 | 22 | 30 | |
| miR-27a/b | 18a | 12a | 30 | |
| miR-17 | 18 | 11 | 29 | B |
| miR-210 | 21 | 8 | 29 | |
| miR-125 | 6 | 15 | 21 | |
| miR-29 | 14 | 4 | 18 | |
| let-7 | 9 | 8 | 17 | |
| miR-222 | 6 | 11 | 17 | |
| miR-130a | 10 | 4 | 14 | |
| miR-122 | 5 | 8 | 13 | |
| miR-1 | 9 | 3 | 12 | C |
| miR-34a/b | 4 | 7 | 11 | |
| miR-106a/b | 3 | 7 | 10 | B |
| miR-16 | 8 | 2 | 10 | |
| miR-499 | 8 | 2 | 10 | |
| miR-133 | 5 | 2 | 7 | C |
| miR-200a/b | 3 | 3 | 6 | |
| miR-206 | 4 | 2 | 6 | C |
| miR-10a/b | 1 | 4 | 5 | |
| miR-217 | 2 | 2 | 4 | |
| miR-361 | 2 | 2 | 4 | |
| miR-365 | 0 | 4 | 4 | |
| miR-101 | 1 | 3 | 4 | |
| miR-362 | 2 | 1 | 3 | |
| miR-1185 | 0 | 2 | 2 | |
| miR-15 | 2 | 0 | 2 | |
| miR-483 | 1 | 1 | 2 | |
| miR-188 | 0b | 0b | 0 |
miRNAs with a/b variants were searched in the following manner: (miR-XXa AND hypertension) OR (miR-XXb AND hypertension); (miR-XXa AND atherosclerosis) OR (miR-XXb AND atherosclerosis).
Mishra113– does not return with search parameters in pubmed.org.
Table 3.
Differential microRNA expression in human hypertension and atherosclerosis
| miRNA | Patient population | Tissue source | Findings | Author |
|---|---|---|---|---|
| miR-17miR-15 | Hypertensives with CKD vs. hypertensive controls (w/o CKD); mean age 77 years, African Americans | Whole blood (PAXgene Blood RNA tubes, BD, USA) | ↓ miR-17↓ miR-15in CKD | Nandakumar (2017)114 |
| let-7 | Hypertensives, carotid intima-thickness (CIT) | Plasma RNA | let-7 positively correlated with CIT | Huang (2017)115 |
| miR-361-5pmiR-362-5p | Salt-sensitive hypertensives vs. salt-resistant normotensive, N = 91 | Plasma RNA | ↓ in both miRNAs | Qi (2017)116 |
| miR-221-3p | PAH patients (n = 10) vs. control (n = 10) | Human lung tissuePulmonary artery smooth muscle cells | ↑ miR-221-3p in lung tissues | Nie (2017)117 |
| miR-1185 | Northern Chinese population, N = 406 | Plasma RNA | miR-1185 positively correlated with arterial stiffening, VCAM1, and E-Selectin | Deng (2017)118 |
| miR-143miR-145 | Hyperhomocysteinaemia (Hhcy), Hhcy + atherosclerosis, atherosclerosis alone vs. healthy controls, N = 100 | Plasma RNA (total) | ↑ miR-143↑ miR-145in disease cohort vs. control | Liu (2017)73 |
| miR-221, miR-130a, and miR-155 | Coronary atherosclerosis patients of different ethnicities, (N = 932, 681 M, 251 F), 587 Han, 146 Uygur, 91 Kazakh, 67 Hui, and 41 “other” | Plasma RNA | ↓ Plasma miR-221, miR-130a, and miR-155 | Jia (2017)63 |
| These miRNAs are potential risk factors for CHD | ||||
| miR-155 | Eplerenone (MR antagonism) in older adults (N = 16) | Serum RNA | ↑ miR-155 positively correlated with decrease in SBP | DuPont (2016)42 |
| miR-365 | Atherosclerosis (n = 21 M, 17 F) patients vs. healthy (n = 18 M, 8 F) control | Arterial plaque tissue and adjacent endometrial tissue; peripheral blood monocytes, serum | ↓ miR-365 | Lin (2016)119 |
| ↑ IL-6 in coronary plaque and blood monocytes | ||||
| miR-143let7fmiR-1miR-29b | Atherosclerosis, N = 39 (n = 14 asymptomatic, n = 10 symptomatic carotid) vs. control (n = 15 nonatherosclerotic mammary artery) | Carotid endarterectomy | ↑ miR-143↓ let7f↓ miR-1↓ miR-29b | Markus (2016)74 |
| miR-143-3pmiR-222-3p | Primary human coronary artery smooth muscle cells (HCASMC, normal donors) exposed to plasma from familial hypercholesterolemic vs. normolipidemic donors (n = 24) | HCASMC-derived microparticle- associated RNA | ↓ miR-143-3p↓ miR-222-3pin microparticles, no change in HCASMC cells | de Gonzalo-Calvo (2016)120 |
| miR-21 | Hypertensives vs. normotensives (N = 56), with measures for carotid intima media thickness (CIMT) | Plasma RNA | ↑miR-21, miR-21 positively correlated with hypertension and negatively correlated with eNOS and NOx | Cengiz (2015)121 |
| miR-143, miR-145, miR-133miR-21, miR-1 | Hypertension (n = 60) vs. healthy (n = 29) | PBMC RNA | ↓ miR-143, miR-145, and miR-133↑ miR-21, miR-1 | Kontaraki (2014)75 |
| miR-223 | Acute ischemic stroke (n = 79) vs. nonstroke age matched controls (n = 75) | PBMC RNA | ↑ miR-223 post stroke | Wang (2014)87 |
| ↑miR-16, miR-27a; ↓miR-101, miR-150 | Post acute MI (N = 150) | Plasma total RNA | Changes in these miRNAs helped predict impaired LV contractility | Devaux (2013)32 |
| miR-146a, miR-499 | Hypertension (n = 340) vs. healthy (n = 515) | Unclear | miR-146a C>G polymorphism and allelic combinations, affect susceptibility to hypertension | Jeon (2013)69 |
| miR-122-3pmiR-519e-5pmiR-200b-5p | HF-REF vs. healthy, ~70% hypertension in both groups | Whole blood (PAXgene Blood RNA tubes, BD) | miR-519e-5p negatively correlated with survival, 8 miRNA signature biomarker for SHF | Vogel (2013)122 |
| miR-145 | Hypertension (n = 15) vs. control (n = 7) | Atherosclerotic plaques from carotid artery | ↑ miR-145 | Santovito (2013)76 |
| miR-21miR-210miR-34amiR-146a/b | Atheroma samples from 3 locations vs. left internal thoracic nonatherosclerotic artery (N = 50) | Carotid plaqueAortic plaqueFemoral plaqueControl arteries | Ten miRNAs differentially expressed: ↑ miR-21, -34a, -146a, -146b-5p, and -210 | Raitoharju (2011)56 |
Abbreviations: CHD, coronary heart disease; CKD, chronic kidney disease; eNOS, endothelial nitric oxide synthase; HF-REF, non-ischemic heart failure with reduced ejection fraction; PAH, pulmonary arterial hypertension; F, female; M, male; MR, mineralocorticoid receptor; N, total study sample size; n, group-specific sample size; NOx, nitric oxide; PBMCs, peripheral blood mononuclear cells; SHF, systolic heart failure.
Table 4.
microRNAs in animal and cell culture models
| miRNA | Model | Intervention | Finding | Target/pathway | Author |
|---|---|---|---|---|---|
| Animal Models | |||||
| Dicer KO (↓ all mature miRNA— AGO2 complexes) | Mouse, Dicer KO | Genetic ablation of Dicer (needed for maturation of pre-miRNAs / active mature miRNAs) | ↓ Systemic blood pressure in Dicer-KO | 5-HT2A | Dahan (2017)81 |
| miR-30c targets 5-HT2A in primary mVSMCs | |||||
| miR-34a | Rat, Myocardial ischemia- reperfusion (IR) injury model, Proximal LAD Coronary ligation | miR-34a mimic | ↑ Apoptosis | SIRT1 | Fu (2017)123 |
| miR-31 | Rat, MI model LAD (left anterior descending) coronary ligation | AntagomiR-31 (LNA-based) | ↓ Infarct size | NR3C2 TIMP4TNNT2E2F6 | Martinez (2017)38 |
| 10% improvement in LEF (left ejection fraction) | |||||
| miR-34b | Rat, spontaneously hypertensive (SHR) vs. Wister Kyoto | Mimeticsi-CDK6 | ↑ miR-34b | CDK6 | Yang (2017)124 |
| miR-34a | Rat, chronic hypoxia model | Hypoxia, miR-34 mimic and inhibitor | ↓ miR-34a in hypoxic lung tissue and pulmonary artery | PDGFá | Wang (2016)125 |
| miR-155 | Mouse, WT vs. smooth muscle cell— mineralocorticoid receptor (MR) KO mice | ↑ Age-related blood pressure | ↓ miR-155 with aging, MR-KO rescued miR-155 levels in mesenteric resistance vessels | Cav1.2AgtR1 | DuPont (2016)42 |
| miR-222 | Mouse (C57BL/6 and Tg-miR-222) | Coronary ligation and reperfusionExercise models | miR-222 is protective↓ Cardiac remodeling↓ Ischemic injury↑ Cardiomyocyte growth | HMBOX1 p27HIPK1/2 Vinculin | Liu (2015)126 |
| miR-21 | C57BL6/J mice | 4% NaCl-induced hypertension, antagomiR-21 (LNA-based) vs. control oligo | ↓ Blood pressure with antagomiR-21 | JAG1 | Kriegel (2015)127 |
| miR-143miR-145 | miR-143, miR-145, LDLR triple knockout (KO) vs. LDLR KO | Western diet, 16 weeks | ↓ Plaque volume in triple KO mice↓Total plasma cholesterol | ↑ABCA1 | Sala (2014)78 |
| miR-126-5p | Mouse, miR-126−/− Apoe−/− | Wire injury or partial carotid ligation and high-cholesterol diet | ↓ Atherosclerotic lesion formation | DLK1 (Notch1 inhibitor) | Schober (2014)128 |
| miR-21 | Rat, acute MI model | Left coronary artery ligation, adenoviral miR-21 overexpression | Differential expression of miR-21: ↓ Infarcted areas, ↑ Border areas; ↓ Ischemia-induced cell apoptosis | PDCD4 | Dong (2009)129 |
| miR-21 | Rat, carotid artery balloon injury model | AntagomiR-21 | ↓ Cell proliferation;↑ apoptosis | PTEN and Bcl-2 | Ji (2007)130 |
| In-vitro Models | |||||
| miR-222 | Primary, rat pulmonary arterial smooth muscle cells | miR-222 mimic | ↑ Proliferation of pulmonary arterial smooth muscle cells | TIMP3 and P27 | Xu (2017)131 |
| miR-34a | Chronic hypoxic rat model, human pulmonary artery smooth muscle cells (HPASMC), human embryonic kidney (HEK-293) cell line | Hypoxia, miR-34 mimic and inhibitor | Promotes proliferation of human pulmonary artery smooth muscle cells | PDGFα | Wang (2016)125 |
| miR-483-3p | Human embryonic kidney (HEK-293) clonal cell lines, rat HA-AT1R, rat HA-AT2R cell lines; primary human aortic smooth muscle cells | AT1R-specific blockers [Sar1] AngII or losartan/candesartan 24 hours | VSMC-specific 22 miRNAs modulated by AngII, and AT1R regulate 17 of these miRNA; miR-483-3p specifically modulate AT1R-regulated expression of ACE-1 in VSMCs | Renin–angiotensin system (RAS) | Kemp (2014)39 |
| Rat aortic smooth muscle cell line | |||||
| miR-155 | Primary human endothelial cells (26 individuals) | miR-155 mimic and inhibitor | eNOS is a direct target of miR-155TNFα-mediated ↓ eNOS via miR-155 | eNOS | Sun (2012)65 |
| TNFα-eNOS axis | |||||
| miR-21 | HUVECs, HeLa, and THP-1 (human monocyte) cell lines | miR-21 mimic, oscillatory shear stress | ↑ AP-1 activation | ↓ PPARα | Zhou (2011)58 |
| ↑ expression of adhesion molecules, ↑ VCAM-1, and MCP1 | |||||
| miR-365 | HUVEC cell line | oxLDL-induced endothelial cell apoptosis | miR-365 promotes endothelial cells apoptosis; ↑ Atherosclerosis | Bcl-2 | Qin (2011)132 |
| miR-663 | HUVEC cell line | Oscillatory shear stress | ↑ Atherosclerosis↑ Inflammation | IL8, ATF3, and KLF4 | Ni (2011)133 |
| miR-21 | HUVEC cell line | miR-21 mimic | ↓ Apoptosis↑ eNOS phosphorylation↑ nitric oxide production | PTEN | Weber (2010)57 |
| Uni-directional shear stress | |||||
| miR-125a- 5p/125b-5p | H5V-murine cardiac microvascular endothelial cell line | anti-miR-125a and anti-miR-125b | Anti-atherosclerotic | ET-1 | Li (2010)134 |
| Inhibit endothelial dysfunction | |||||
| miR-10a | Primary endothelial cells harvested from swine arterial tissues; swine aortas and aorto-renal bifurcations from, the inner curvature of the aortic arch and nearby descending thoracic aorta, and the cranial wall and caudal wall of the aorto-renal branches and the distal renal artery | Differential endothelial miRNA expression in athero-susceptible and athero-protected regions of aorta and renal arteries | ↓ miR-10a athero-susceptible regions↓ Atherosclerosis↓ Inflammation | MAP3K7 TRC | Fang (2010)135 |
| miR-188 | HL-1 (cardiac muscle) cell line | Homocysteine-induced cardiac remodeling | ↓ miR-188 | Dicer and miR-188 are involved in Hcy-induced cardiac remodeling | Mishra (2009)113 |
| miR-217 | HUVEC, HAEC, and HCAEC cell lines | Cell senescence | Pro-atheroscleroticAnti-angiogenic | SIRT1 | Menghini(2009)136 |
| miR-221 miR-222 | HUVEC cell line, HEK 293T | Wound healing assay, transfection of miR-221 and miR-222 | Inhibit angiogenesisAnti-atherosclerotic | c-Kit STAT5A | Poliseno (2006)137 |
Abbreviations: ATF3, cyclic AMP-dependent transcription factor ATF-3; Bcl-2, B-cell CLL/lymphoma2; DLK1, delta-like 1 homolog; ET-1, endothelin 1; HAEC, human aortic endothelial cells; HCAEC, human coronary artery endothelial cells; Hcy, homocysteine; HIPK1/2, homeodomain interacting protein kinase 1/2; HMBOX1, homeobox containing 1; HUVEC, Human umbilical vein endothelial cells; JAG1, jagged 1; LAD, left anterior descending; MAP3K7, mitogen-activated protein kinase kinase 7; MCP1, monocyte chemotactic protein-1; oxLDL, oxidized low-density lipoprotein; PDGFα, platelet-derived growth factor receptor alpha; PTEN, phosphatase and tensin homolog; KLF4, Kruppel-like factor 4; PDCD4, programmed cell death 4; PPARα, peroxisome proliferator-activated receptor alpha; SIRT1, silent information regulator 1; STAT5A, signal transducer and activator of transcription 5A; TIMP, tissue inhibitor of metalloproteinase; VSMCs, vascular smooth muscle cells.
NCRNAS ASSOCIATED WITH MYOCARDIAL INFARCTION, FOCUS ON MIR-150
ncRNAs reported in relation to myocardial infraction (MI) will be addressed briefly before the roles of individual miRNAs are reviewed. A 4-plasma-miRNA signature [miR-16, miR-27a, miR-101, and miR-150 (odds ratio = 0.08)] was associated with left ventricular (LV) dysfunction from a robust cohort of AMI patients (N = 150).32 Decreased miR-150 in heart tissue itself had been previously reported in MI vs. normal heart tissue.33 Patients (n = 90) with the lowest levels of plasma miR-150 had increased rates of cardiac remodeling at follow-up compared to patients with higher miR-150 levels,34 and miR-150 outperformed Nt-proBNP for predicting LV remodeling in the patient cohort. The authors go on to hypothesize that this is due to miR-150-mediated repression of adrenoceptor beta 1 (ADRB1), C-reactive protein, and tumor necrosis factor (TNF) receptor-associated factor 2,34 genes associated with heart remodeling.
Qin et al. found that the expression of miR-150 was significantly upregulated in human umbilical cord vein endothelial cells (HUVECs) during oxidized low-density lipoprotein (ox-LDL)-induced apoptosis, and inhibition of miR-150 partially reduced cell death, suggesting potential therapeutic function of antagomiR-150 in hyperlipidemia-induced endothelial dysfunction.35
Knockdown of a cardio-related lncRNAs, ZFAs1 reduced infarct size in a rat model of acute MI.36 miR-150 and ZFAs1 bind each other, with ZFAs1 acting as a miRNA-sponge reducing the net efficacy of the miR-150 pool within a cell to repress mRNA targets. Overexpression of miR-150 had a similar infarct-reducing effects as knockdown of ZFAs1.36
MIRNA REGULATION OF THE RENIN–ANGIOTENSIN–ALDOSTERONE SYSTEM
Angiotensin II (AngII) signaling through AGTR1 contributes etiologically to the development and progression of hypertension, vascular hypertrophy, and atherosclerosis via increased mineralocorticoid secretion by the adrenal cortex and via direct, deleterious effects on vascular smooth muscle cells (VSMCs).37 miR-31was upregulated in rats post-MI and antagomiR-31 treatment reduced infarct size and improved post-MI left ejection fraction, while rescuing miR-31-mediated suppression of NR3C2 (mineralocorticoid receptor gene).38 miRNA-488-3p directly targets multiple components of the renin–angiotensin–aldosterone system when examined in human and rat cells, including smooth muscle and kidney.39 Also, AngII treatment of rat aortic SMCs increased miR-21 expression.39 Angiotensin II receptor blocker (ARB) and angiotensin converting enzyme (ACE) inhibition in humans has been reported to alter circulating miRNA levels; however, the physiological significance of this is uncertain.40 miR-146a/b was significantly higher in CAD patients before randomization to ARB/ACEI (plus statin therapy) for 12 months, after which ARB + statin decreased miR-146a/b levels more than ACEI + statin, though miR-146a/b was decreased from baseline in both groups. Furthermore, miR-146a/b levels at baseline predicted cardiac events in the top tertile (third) over the 12-month study.41 We remind the reader that miR-155 upregulation correlated with the response (decrease in blood pressure) after eplerenone.42 Overexpression of miR-155 or miR-221/222 abrogated AngII-stimulated adhesion of T cells to HUVECs.43 Clearly, miRNA regulation of renin–angiotensin–aldosterone system is complex and understanding of the intricacies may lead to novel therapeutics for hypertension.
LONG NCRNAS AND CVD
LncRNAs are longer than 200 nucleotides and lack functional open reading frames. They are involved in cellular differentiation, proliferation, DNA damage response, and chromosomal imprinting.44 There are an estimated 30,000 lncRNAs, with the large majority lacking functional delineation. LncRNAs function post-transcriptionally to regulate RNA splicing, mRNA degradation, and protein translation (Figure 1).45 The first publication reporting a role for lncRNAs in CVD appeared in 1996, with the identification of H19 expression in human atherosclerosis.46 H19 is a developmental gene that is most highly expressed during muscle during development, with undetectable levels in healthy, adult vascular smooth muscle tissue. In the study, H19 became “re-expressed” in the thickened intima of human atheromas.
Vascular remodeling is an important pathological feature of hypertension.47 The Natarajan group was the first to identify 24 novel, AngII-modulated lncRNAs in rat VSMCs, including lnc-Ang32 which promotes VSMC proliferation.48 Lnc-Ang362-mediates VSMC proliferation in part by encoding pre-miR-221/222.48 LincRNA-p21, a p53-induced lncRNA, represses smooth muscle cell proliferation and is increased in human CVD.49 Inhibition of lincRNA-p21 resulted in intimal hyperplasia following carotid wire injury in mice. LncRNA-GAS5 (growth arrest–specific 5) is another novel regulator of hypertension-induced vascular remodeling.50 LncRNA-GAS5 knockdown (KD) in SHR rats increased both systolic and diastolic pressures. KD rats had increased intimal thickness measures with evident worsening of vascular remodeling. The findings were attributed to GAS5 regulation of the β-catenin pathway.50 Gopalakrishnan et al. reported 273 differentially expressed lncRNAs in the kidney of Dahl salt-sensitive vs. salt-resistant rats.51 The group has not yet published mechanistic pathways by which these lncRNAs alter blood pressure. Myosin heavy chain-associated RNA transcript (Mhrt) is a cardiac-specific lncRNA that is intergenic between the genes Myh6 and Myh7 (myosin heavy chain).52 LncRNA-Mhrt is cardioprotective, preventing activity of the chromatin-binding protein, Brg1. Brg1 is part of a chromatin suppressor complex (Brg1-Hdac-Parp) that mediates cardiac remodeling and hypertrophy. The endothelium-expressed and hypoxia-induced lncRNAs-MALAT1 promoted endothelial proliferation. Furthermore silencing of MALAT1 promoted endothelial cell migration.53
Interestingly, miR-150 (its protective role in post-MI LV remodeling discussed previously) is known to target lncRNA-MIAT. MIAT which is positively associated with diabetes-related microvascular dysfunction and endothelial dysfunction.54 MIAT competes with other mRNA targets for miR-150 binding, effectively reducing funtional miR-150 levels. Thus, increased MIAT may mediate its deleterious effects via a sponge-effect on miR-150.
MIR-21 AND CVD
miR-21 is one of the first mammalian microRNAs identified and is one of the most studied miRNAs in CVD (Table 1, position 1, Figure 2). miR-21 is considered by some to be a “mechano-miR”—an miRNA that is abundant in the vessel wall and responds with differential expression upon shear or mechanical stress to the vessel.10 In humans, miR-21 is expressed in most cell types and is highly expressed in podocytes, dendritic cells, and CD14+ monocytes but is nearly undetectable in kidney cells (Supplementary Table S5 from Landgraf et al.).9 miRNA-21 is widely expressed in multiple types of cancers and in CVD. It is one of the most dynamically regulated miRNA in various pathophysiological process (cellular survival, apoptosis, and cell invasiveness).55 It was found to be upregulated in human atherosclerotic plaque.56 Weber et al. reported that miRNA-21 expression on endothelial cells is significantly upregulated by shear stress treatment and that it also causes an anti-apoptotic effect by directly targeting PTEN tumor suppressor gene.57 Another study using similar methods in endothelial cells showed that miRNA-21 overexpression inhibits the expression of peroxisome proliferator-activated receptor-alpha (PPARα), resulting in upregulation in expressions of VCAM-1 and MCP-1.58 Recently, it was reported that hydrogen peroxide and lipopolysaccharide (LPS) differentially affect the expression of several microRNAs, including miR-21, in endothelial cells before and after co-culture with monocytes.59 These studies demonstrate an important role of miR-21 in the development of atherosclerosis (athero); however, the effect of increased miR-21 expression in endothelial cells in the context of atherosclerosisneeds further examination.
Figure 2.
Summary of ncRNA biogenesis and functions within the gene expression machinery. This figure focuses on how miRNAs and lncRNAs affect each other and messenger RNA expression, but it is not exhaustive. mRNAs, lncRNAs, and miRNAs arise from the transcription of DNA by RNA polymerase II (a). (b) LncRNAs may be the host strand of miRNAs—lncRNAs may be processed into active miRNAs. Lnc and miRNAs may both be transcribed as an intronic segment of a premessenger RNA or as a product of their own transcription events, independent of mRNA synthesis. (c) LncRNAs may regulated the splicing of pre-mRNAs, effecting abundance of mature mRNAs as wells as the relative levels of mRNA splice variants within the cell. (d) The “seed sequence” at the 5′ end of the miRNA binds to complementary sequences of a target mRNA, often (but not exclusively) in the 3′-UTR. (e) LncRNAs may complementarily bind with miRNAs resulting in miRNA sequestration. This competitive inhibition of miRNA function reduces the number of mature miRNAs that are free to bind target mRNAs, potentially resulting in a net increase in gene expression. ncRNAs are also capable of reducing mRNA translation rates via interactions with ribosomes (not depicted). Abbreviations: lncRNA, long noncoding RNA; mRNA, messenger RNA; miRNA, microRNA; ncRNA, noncoding RNA.
MIR-155 AND CVD
miR-155 is the prototypical “immuno-miR,” with a primary role in almost all immune cells (innate and adaptive), and is the second most-studied miRNA in respect to atherosclerosis and hypertension. miR-155 promotes B cell-related immunoglobulin production, T cell proliferation in response to antigen, cytokine production, and is significantly expressed in CD34+ cells (hematopoietic stem cells).60 Substantial evidence supports a role for mineralocorticoid stimulation in hypertension; Dupont et al. found that those patients who upregulated serum miR-155 in response to MR blockage (eplerenone) had a significantly greater reduction in systolic blood pressure vs. those who did not upregulate miR-155.42 Further, miR-155 levels were found to be dramatically downregulated in the aortas of aged WT mice, whereas SMC-MR knockout mice had a modest increase in miR-155 with age. SMC-MR-KO mice are resistant to age- and aldosterone-induced hypertension via dilatory effects in resistance vessels (renal and mesenteric etc).61 Plasma miR-155 was negatively correlated with severity of coronary atherosclerosis(Gensini score62) in a large, multiethnicity study of 932 patients in China.63 In mice, lack of miR-155 in bone marrow-derived cells increased atherosclerosisin LDLR-KO mice.64 In regards to mRNA targets, miR-155 partially mediated the effect of inflammatory stimuli (TNFα)-induced endothelial nitric oxide synthase downregulation in HUVEC cells.65 Simvastatin pretreatment ameliorated TNFα (20 ng/ml)-induced miR-155 expression in HUVECs, further clarifying how statin modulation of the mevalonate-geranylgeranyl-pyrophosphate-RhoA signaling pathway has anti-atherosclerotic effects.65 Abundant in vitro studies clearly demonstrate that miR-155 is proinflammatory; however, observational human studies measuring plasma miR-155 show that low circulating levels may be predictive of adverse outcomes. Furthermore, miR-155-KO mice have increased atherosclerosis. The precise reason for this disparity, in the context of the inflammatory theory of CVD, is unclear.
MIR-146 AND CVD
Another “immuno-miR,” miR-146 functions primarily in innate immune cells to negatively regulate the production of proinflammatory cytokines.66 miR-146a is on chr5 in humans, and miR-146b is on chr 10, and the 5p sequence of mature hsa-miR-146a/b is the most abundant.67,68 miR-146a is also one of the few instances of allelic variance in humans, with a C>G polymorphism called miR-146aG is positively correlated with incidence of ischemic stroke.69 Severe preeclampsia, requiring termination of pregnancy, was associated with a decrease in maternal whole blood miR-146a.70 Raitoharju et al. (n = 50) examined miRNA expression in the arterial wall of atheroma-containing and normal vessels and found that miR-146a, miR-146b, and miR-21 levels were elevated in plaque-burdened arteries.56 miR-146a has been shown to be the mediator by which ApoE suppresses myeloid cell inflammation (NF- κB activation).71 ApoE functions to enhance normophysiologic removal of circulating triglyceride-rich particles (e.g., VLDL); allelic variance (e4) or mutations that impair apoE function result in a pro-atherogenic environment with enhanced myeloid cell and vessel wall lipid accumulation. Reports favor a strong anti-inflammatory and atheroprotective role for the miR-146 family.
MIR-143/145 AND CVD
miR-143 and miR-145 are found in close proximity on human chr5, forming a classical “cluster” (149428918-149429023 [+] and 149430646-149430733 [+], respectively on GRCh38). Though part of the same cluster, miR-143 levels tend to be higher than miR-145 due to an unknown mechanism(s).72 Human clinical data suggesting differential expression of miR-143/145 in CVD is substantial (see Table 2–3).73–77 A large study (N = 100) by Liu et al. examining plasma miRNA levels found that miR-143/145 were both significantly higher in subjects with hyperhomocysteinemia (Hhcy) vs. healthy.73 Cohorts examined included Hhcy without carotid atherosclerosis, Hhcy with carotid atherosclerosis, and carotid atherosclerosis without Hhcy. Plasma miR-143/145 negatively correlated most to total plasma cholesterol and LDL levels, but did not correlate well with measures of plaque volume.73 Similarly, peripheral blood mononuclear cell miR-143/145 was significantly lower in hypertensives vs. healthy subjects, with miR-143/145 negatively correlating with blood pressure. miRNA levels were measured in endarterectomy samples from 2 cohorts of subjects: carotid stenosis with a history of ischemia or stroke, or asymptomatic stenosis. Data from both of these endarterectomy samples were compared to each other and to data from nonatherosclerotic control (mammary) arteries from the same endarterectomy patients.74 The authors found that miR-143 was elevated in the asymptomatic cohort vs. mammary artery miR-143 levels. Surprisingly, this study did not assay miR-145 levels in samples. In contrast, miR-145 was found to be elevated in carotid atheromas (endarterectomy) from hypertensive patients vs. carotid atherosclerosiswithout hypertension.76Ex vivo studies of human coronary smooth muscle cells found that treatment with plasma from subjects with familial hypercholesterolemia decreased miR-143 levels vs. plasma from normolipidemic individuals.77 miR-143/145/LDLR triple KO mice were protected from plaque vs. single (LDLR) KO mice.78
Endothelial cells increased miR-143/145 in response to increasing laminar flow, and it is believed that endothelial-derived vesicles enriched in miR-143/145 may be exported to and taken up by smooth muscle cells potentiating vasorelaxation.79 Vesicles collected in vitro were shown to decrease atherosclerosisprogression in an ApoE-KO model, which conflicts with the miR-143/145/LDLR-KO mouse findings. miR-143/145 are highly expressed in smooth muscle cells and SMC lineage cells.80,81 Bottega et al. clearly demonstrated that miR-143/145-KO mice had deregulated blood pressure, due to a shift in smooth muscle cells from a contractile phenotype to a synthetic phenotype (61% decrease in potassium-induced contraction vs. WT). VSMC of KO mice have increased membrane-bound ACE-1, resulting in angiotensin resistance (miR-145 represses ACE-1 via 3′-UTR binding) and lower blood pressure when challenged with AngII-infusion. Neointimal lesions were found in the femoral arteries of 18-month-old miR-143/145 KO mice, while WT controls lacked lesions. These findings support a role for the miR-143/145 cluster in the phenotypic change of VSMC toward a synthetic, pro-atherosclerotic phenotype.
MIR-223 AND CVD
Similar to miR-155, miR-223 is a prototypical immuno-miR, with highest expression in myeloid cells and downregulation required from monocyte-to-macrophage differentiation.82,83 Thus, a role in inflammation-related diseases is not unexpected; our group found that miR-223 was elevated in the visceral adipose of obese humans in the absence of hyperlipidemia and hypertension.84 Increased miR-223 is positively correlated with incidence of acute ischemic stroke.85 Upregulation of miR-223 maybe be due, in part, to hypomethylation of the miR-223 promoter. miR-223 promoter hypomethylation was found to be elevated in atherosclerotic cerebral infarction patients vs. healthy controls (N = 55) and peripheral blood mononuclear cell miR-223 levels were found to be higher in the atherosclerotic cerebral infarction cohort, as expected.86 In a Han-Chinese population of stroke patients (n = 79), miR-223 was elevated in total PMBC RNA from acute ischemic stroke victims (<72 hours after stroke) compared to blood leukocytes from older age- and sex-matched nonstroke controls (n = 75).87 Interestingly, an increase in miR-223 has been shown to have an anti-inflammatory effect in vitro,88 and MIR223-KO predisposed mice to a proinflammatory state with high-fat diet feeding.89 It should be noted, however, that there are a number of human studies showing a decrease in tissue and plasma miR-223 levels in diabetes and CVD.90 The authors are of the opinion that an increase in miR-223 in inflammatory disease is a protective, compensatory response, and that miR-223 mimetics would have a beneficial effect on CVD and inflammation in vivo, though this has not been reported.
MIR-1, MIR-133, AND CVD
In humans, there are 2 distinct microRNA genes, miR-1-1 (chr20) and miR-1–2 (chr18), which produce an identical mature sequence collectively referred to as miR-1. This genomic repetitiveness is not unique and there are instances of 3 genomic locations for a single miRNA (i.e., miR-133 on chr 18, 20, and 6 in humans). Both miR-1 and miR-133a/b, often called “myo-miRs”, are transcribed in a bicistronic fashion and have pivotal roles in heart development; whole genome miR-133 deletion results in murine embryonic lethality.91,92 Similarly, whole body Dicer deletion is embryonically lethal in mice and conditional dicer deletion in the myocardium resulted in massive cardiac remodeling.93 miR-1 is an important regulator of heart adaptation after ischemia or ischemic stress, and it is upregulated in the myocardium of patients after MI, along with miR-150 and miR-133a/b.94 miR-1, like all miRNAs, has a -3p and a -5p strand. The miR-1-3p strand is more abundant based on RNA-Seq data (reads per million) than the -5p strand in human samples.94 Most studies do not distinguish the chromosomal origin of mature miR-1. Elevated peripheral blood mononuclear cell -derived miR-1, along with miR-21 and miR-133, were found in hypertensives and correlated with 24-hour ambulatory blood pressure.75 A follow-up study by the authors again reported a negative correlation between miR-1 and miR-133 levels and LV mass in hypertensives.95 A combination of miRNA-mimics (miR-1, miR-133, miR-208, and miR-499) changed the epigenetic status in cultured fibroblasts, reduced promoter methylation, and upregulated the expression of cardiogenic genes. This has implications for cardiac muscle regeneration after ischemia-reperfusion injury.96 Finally, plasma miRNAs from 444 acute MI patients were examined and analyzed against outcome and plasma high sensitivity troponin T (hsTnt) levels.97 The authors found that miR-133a levels were able to discriminate survivors from nonsurvivors but did not enhance the discriminatory ability of hsTnt, at 6 months post event. Thus, hsTnt remains the definitive myonecrosis marker, superior to plasma miR-133 or miR-1 in this study. Plasma miR-1 was not associated with 6-month post-event mortality, but was significantly elevated vs. an angina control cohort.97
NCRNA-BASED THERAPEUTICS
It has been estimated that the elimination of hypertension would reduce CVD mortality by 30.4% to 38.0%.98 Accomplishing this will likely require next generation biologics including oligonucleotide-based approaches targeting pathologic ncRNAs, mRNAs, and proteins. Antagonizing pathologic ncRNAs is an approach that has been used with success in animal models, including nonhuman primates (as outlined in Table 4).99,100 However, effective in vivo mimetics are not available due in large part to immunologic-reactivity. Targeting proteins using aptamer technology (RNA or DNA-aptamers that bind proteins) is a promising approach which has to-date focused on eye-related disease such as macular degeneration (NCT02686658—complement (C5) binding aptamer, Zimura; aptamer to vascular endothelial growth factor (VEGF)—Macugen/Pegaptanib, FDA approval 2004), though potential uses are vast. The authors believe that aptameric approaches have great potential for the future of biologics as these complex conjugates allow for a large array of modifications and are better adapted to high throughput screening than minimally modified oligonucleotides. Aptamers perform a similar function to monoclonal antibody-based therapies, without requiring animal products/production of immunoglobulins resulting in a reduced risk of toxin contamination and a lower potential for immunogenicity. Aptamers can be synthesized entirely using chemistry-based modifications, increasing purity and stability beyond that of mAbs, at a lower production cost. Aptamer-based screening of human plasma offers a high throughput and highly precise means of proteomic analysis of clinical samples, with great relevance to CVD.101
Stability was the primary barrier to the use of nucleotide-based drugs, as naked RNA and DNAs with a serum half-life of minutes for RNAs102; however, great strides have been made to improve stability, making them suitable therapeutics. Modifications can be made in order to improve or modify binding properties, many of which were developed during the production of aptamers over the last 3 decades. Aptamer technology is maturing and is based on a DNA or RNA backbone, with the aptamer targeting a protein (e.g., IL-6, VEGF, Factor IXa). Development of an aptamer involves screening a target (protein) with an aptamer library; similar to small molecule drug screening. New methods (e.g., SELEX) have identified many useful aptamer-protein target interactions.103 To date, aptamers have had limited success many due to immune-reactivity, as was found with the aptamer to factox IXa. A significate benefit of an aptamer-based approach is that aptamers can have antidotes allowing for near-immediate cessation of aptameric effects, something that is not available for mAb-based therapies. Aptamer pursuits helped lead to the development of modifiers (sugar backbone and phosphate backbone modifications) that stabilize RNA- and DNA-based injectable. Newly developed mutant polymerases (Y639L, H784a, Y639F, etc.) allow for the addition of “pre-modified” (e.g., 2’OMe bases) reactants to the polymer (as opposed to post-synthesis, polymer modification) allowing for more efficient production of stable oligos. Also, high throughput of 50 base long oligos using proprietary solid phase phosphoramidite chemistry allows for synthesis of gram quantities at GMP+ purity needed for human therapeutics.104
siRNAs/shRNA to mRNAs are particularly effective at targeting liver gene expression. Localization of oligonucleotide-based therapeutics to the liver is similar to other drugs and is a limitation that needs improvement for potency in peripheral tissues. One clinical use that takes advantages of hepatic accumulation is for the treatment of familial transthyretin amyloidosis leading to cardiomyopathy or neuropathy. siRNAs, as previously mentioned, utilize the RISC assembly (native to miRNAs) to prevent the translation and production of mutated TTR, reducing accumulation of the mutant protein. RNAi-based approaches are currently being tested for use in various conditions including cancer and for further information, we suggest the review article by Sullenger et al.103
The least investigated but most exciting potential use is in RNA-guided therapies using endoncucleases (e.g., CRISPR-Cas9) for in vivo gene editing, essentially ncRNA-mediated genetic engineering.103,105 Clinical applications are still far off but are in development.
ncRNAS AS BIOMARKERS IN CLINICAL MEDICINE
Blood tests for circulating ncRNA levels measured or other bodily fluids will become part of standard of care in the clinic for diagnosis as well as to measure response to therapy. The main hurdle to this is standardization of sample processing, identification of the most relevant component of the blood or biofluid to assay (Blood-lipoprotein-associated ncRNAs, albumin, peripheral blood mononuclear cells, platelets, etc), streamlining of ncRNA assays, and sufficient prospective clinical testing. Plasma miR-133a/b was examined as a putative marker of MI and was found to be sensitive, though inferior to hsTnT measures.97 Furthermore, combining miR-133 and hsTnt data did not increase the ability to discriminate between MI survivors and nonsurvivors vs. hsTnT alone.
The future of ncRNA-based biomarkers and therapeutics is bright and has been expanding as opposed to contracting. Thus, the authors believe that we will see more such approaches to human disease reach the clinic.
DISCLOSURE
The authors declared no conflict of interest.
Acknowledgements
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases by a K01 award (DK099475) to J.D. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res 2015; 116:937–959. [DOI] [PubMed] [Google Scholar]
- 2. Wain LV., Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O’Reilly PF, Cabrera CP, Warren HR, Rose LM, Verwoert GC, Hottenga J-J, Strawbridge RJ, Esko T, Arking DE, Hwang S-J, Guo X, Kutalik Z, Trompet S, Shrine N, Teumer A, Ried JS, Bis JC, Smith AV, Amin N, Nolte IM, Lyytikäinen L-P, Mahajan A, Wareham NJ, Hofer E, Joshi PK, Kristiansson K, Traglia M, Havulinna AS, Goel A, Nalls MA, Sõber S, Vuckovic D, Luan J, Del Greco MF, Ayers KL, Marrugat J, Ruggiero D, Lopez LM, Niiranen T, Enroth S, Jackson AU, Nelson CP, Huffman JE, Zhang W, Marten J, Gandin I, Harris SE, Zemunik T, Lu Y, Evangelou E, Shah N, de Borst MH, Mangino M, Prins BP, Campbell A, Li-Gao R, Chauhan G, Oldmeadow C, Abecasis G, Abedi M, Barbieri CM, Barnes MR, Batini C, Beilby J, Blake T, Boehnke M, Bottinger EP, Braund PS, Brown M, Brumat M, Campbell H, Chambers JC, Cocca M, Collins F, Connell J, Cordell HJ, Damman JJ, Davies G, de Geus EJ, de Mutsert R, Deelen J, Demirkale Y, Doney ASF, Dörr M, Farrall M, Ferreira T, Frånberg M, Gao H, Giedraitis V, Gieger C, Giulianini F, Gow AJ, Hamsten A, Harris TB, Hofman A, Holliday EG, Hui J, Jarvelin M-R, Johansson Å, Johnson AD, Jousilahti P, Jula A, Kähönen M, Kathiresan S, Khaw K-T, Kolcic I, Koskinen S, Langenberg C, Larson M, Launer LJ, Lehne B, Liewald DCM, Lin L, Lind L, Mach F, Mamasoula C, Menni C, Mifsud B, Milaneschi Y, Morgan A, Morris AD, Morrison AC, Munson PJ, Nandakumar P, Nguyen QT, Nutile T, Oldehinkel AJ, Oostra BA, Org E, Padmanabhan S, Palotie A, Paré G, Pattie A, Penninx BWJH, Poulter N, Pramstaller PP, Raitakari OT, Ren M, Rice K, Ridker PM, Riese H, Ripatti S, Robino A, Rotter JI, Rudan I, Saba Y, Saint Pierre A, Sala CF, Sarin A-P, Schmidt R, Scott R, Seelen MA, Shields DC, Siscovick D, Sorice R, Stanton A, Stott DJ, Sundström J, Swertz M, Taylor KD, Thom S, Tzoulaki I, Tzourio C, Uitterlinden AG, Völker U, Vollenweider P, Wild S, Willemsen G, Wright AF, Yao J, Thériault S, Conen D, Attia J, Sever P, Debette S, Mook-Kanamori DO, Zeggini E, Spector TD, van der Harst P, Palmer CNA, Vergnaud A-C, Loos RJF, Polasek O, Starr JM, Girotto G, Hayward C, Kooner JS, Lindgren CM, Vitart V, Samani NJ, Tuomilehto J, Gyllensten U, Knekt P, Deary IJ, Ciullo M, Elosua R, Keavney BD, Hicks AA, Scott RA, Gasparini P, Laan M, Liu Y, Watkins H, Hartman CA, Salomaa V, Toniolo D, Perola M, Wilson JF, Schmidt H, Zhao JH, Lehtimäki T, van Duijn CM, Gudnason V, Psaty BM, Peters A, Rettig R, James A, Jukema JW, Strachan DP, Palmas W, Metspalu A, Ingelsson E, Boomsma DI, Franco OH, Bochud M, Newton-Cheh C, Munroe PB, Elliott P, Chasman DI, Chakravarti A, Knight J, Morris AP, Levy D, Tobin MD, Snieder H, Caulfield MJ, Ehret GB. Novel blood pressure locus and gene discovery using Genome-Wide Association Study and expression data sets from blood and the KidneyNovelty and significance. Hypertension 2017; 70:e4–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biino G, Parati G, Concas MP, Adamo M, Angius A, Vaccargiu S, Pirastu M. Environmental and genetic contribution to hypertension prevalence: data from an epidemiological survey on Sardinian Genetic Isolates. Clarke R, ed. PLoS One. 2013; 8:e59612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. United States Department of Agriculture Economic Research Service. <https://www.ers.usda.gov/data-products/food-expenditures.aspx>. Accessed 1 October 2017.
- 5. Adams TD, Davidson LE, Litwin SE, Kim J, Kolotkin RL, Nanjee MN, Gutierrez JM, Frogley SJ, Ibele AR, Brinton EA, Hopkins PN, McKinlay R, Simper SC, Hunt SC. Weight and metabolic outcomes 12 years after gastric bypass. N Engl J Med 2017; 377:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev 2007; 17:139–144. [DOI] [PubMed] [Google Scholar]
- 7. Claverie JM. Fewer genes, more noncoding RNA. Science 2005; 309:1529–1530. [DOI] [PubMed] [Google Scholar]
- 8. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408:86–89. [DOI] [PubMed] [Google Scholar]
- 9. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. miRBase accession for hsa-miR-21. <http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000077>. Accessed 25 September 2017.
- 11. Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010; 328:1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010; 328:1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hinske LC, Galante PAF, Limbeck E, Möhnle P, Parmigiani RB, Ohno-Machado L, Camargo AA, Kreth S. Alternative polyadenylation allows differential negative feedback of human miRNA miR-579 on its host gene ZFR. Jin D-Y, ed. PLoS One 2015; 10:e0121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev 2012; 26:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics 2010; 11:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA 2010; 16:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun 2010; 396:978–982. [DOI] [PubMed] [Google Scholar]
- 18. Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res 2008; 36:5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ribas J, Ni X, Castanares M, Liu MM, Esopi D, Yegnasubramanian S, Rodriguez R, Mendell JT, Lupold SE. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res 2012; 40:6821–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 2008; 378:492–504. [DOI] [PubMed] [Google Scholar]
- 21. Mangiavacchi A, Sorci M, Masciarelli S, Larivera S, Legnini I, Iosue I, Bozzoni I, Fazi F, Fatica A. The miR-223 host non-coding transcript linc-223 induces IRF4 expression in acute myeloid leukemia by acting as a competing endogenous RNA. Oncotarget 2016; 7:60155–60168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell 2011; 43:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 2005; 33:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kraynik SM, Gabanic A, Anthony SR, Kelley M, Paulding WR, Roessler A, McGuinness M, Tranter M. The stress-induced heat shock protein 70.3 expression is regulated by a dual-component mechanism involving alternative polyadenylation and HuR. Biochim Biophys Acta 2015; 1849:688–696. [DOI] [PubMed] [Google Scholar]
- 25. Yang Z, Kaye DM. Mechanistic insights into the link between a polymorphism of the 3’UTR of the SLC7A1 gene and hypertension. Hum Mutat 2009; 30:328–333. [DOI] [PubMed] [Google Scholar]
- 26. Creemers EE, Bawazeer A, Ugalde AP, van Deutekom HW, van der Made I, de Groot NE, Adriaens ME, Cook SA, Bezzina CR, Hubner N, van der Velden J, Elkon R, Agami R, Pinto YM. Genome-wide polyadenylation maps reveal dynamic mRNA 3’-end formation in the failing human HeartNovelty and significance. Circ Res 2016; 118:433–438. [DOI] [PubMed] [Google Scholar]
- 27. Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem 2014; 15:923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noferesti SS, Sohel MM, Hoelker M, Salilew-Wondim D, Tholen E, Looft C, Rings F, Neuhoff C, Schellander K, Tesfaye D. Controlled ovarian hyperstimulation induced changes in the expression of circulatory miRNA in bovine follicular fluid and blood plasma. J Ovarian Res 2015; 8:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, Xing Q, Jin L, He L, Wu L, Wang L. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab 2013; 98:3068–3079. [DOI] [PubMed] [Google Scholar]
- 30. Ray P, Le Manach Y, Riou B, Houle TT. Statistical evaluation of a biomarker. Anesthesiology 2010; 112:1023–1040. [DOI] [PubMed] [Google Scholar]
- 31. Deiuliis J, Mihai G, Zhang J, Taslim C, Varghese JJ, Maiseyeu A, Huang K, Rajagopalan S. Renin-sensitive microRNAs correlate with atherosclerosis plaque progression. J Hum Hypertens 2014; 28:251–258. [DOI] [PubMed] [Google Scholar]
- 32. Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, Wagner DR, Squire IB. A Panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. Rouet P, ed. PLoS One. 2013; 8:e70644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zidar N, Boštjančič E, Glavač D, Stajer D. MicroRNAs, innate immunity and ventricular rupture in human myocardial infarction. Dis Markers 2011; 31:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Devaux Y, Vausort M, McCann GP, Zangrando J, Kelly D, Razvi N, Zhang L, Ng LL, Wagner DR, Squire IB. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet 2013; 6:290–298. [DOI] [PubMed] [Google Scholar]
- 35. Qin B, Shu Y, Xiao L, Lu T, Lin Y, Yang H, Lu Z. MicroRNA-150 targets ELK1 and modulates the apoptosis induced by ox-LDL in endothelial cells. Mol Cell Biochem. 2017; 0:–0. [DOI] [PubMed] [Google Scholar]
- 36. Wu T, Wu D, Wu Q, Zou B, Huang X, Cheng X, Wu Y, Hong K, Li P, Yang R, Li Y, Cheng Y. Knockdown of long non-coding RNA-ZFAS1 protects cardiomyocytes against acute myocardial infarction via anti-apoptosis by regulating miR-150/CRP. J Cell Biochem 2017; 118:3281–3289. [DOI] [PubMed] [Google Scholar]
- 37. Manrique C, Lastra G, Gardner M, Sowers JR. The renin angiotensin aldosterone system in hypertension: roles of insulin resistance and oxidative stress. Med Clin North Am 2009; 93:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez EC, Lilyanna S, Wang P, Vardy LA, Jiang X, Armugam A, Jeyaseelan K, Richards AM. MicroRNA-31 promotes adverse cardiac remodeling and dysfunction in ischemic heart disease. J Mol Cell Cardiol 2017; 112:27–39. [DOI] [PubMed] [Google Scholar]
- 39. Kemp JR, Unal H, Desnoyer R, Yue H, Bhatnagar A, Karnik SS. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J Mol Cell Cardiol 2014; 75:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Satoh M, Takahashi Y, Tabuchi T, Tamada M, Takahashi K, Itoh T, Morino Y, Nakamura M. Circulating Toll-like receptor 4-responsive microRNA panel in patients with coronary artery disease: results from prospective and randomized study of treatment with renin-angiotensin system blockade. Clin Sci (Lond) 2015; 128:483–491. [DOI] [PubMed] [Google Scholar]
- 41. Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M, Takeda K, Kaisho T, Akira S, Satoh M, Shimoda Y, Maesawa C, Akatsu T, Ishikawa Y, Minami Y, Itoh T, Nakamura M, Satoh M, Shimoda Y, Akatsu T, Ishikawa Y, Minami Y, Nakamura M, Bartel DP, Kim VN, Taganov KD, Boldin MP, Chang KJ, Baltimore D, Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H, Sonkoly E, Ståhle M, Pivarcsi A, Libby P, Hornig B, Landmesser U, Kohler C, Ahlersmann D, Spiekermann S, Christoph A, Tatge H, Drexler H, O’Driscoll G, Green D, Rankin J, Stanton K, Taylor R, Dasu MR, Riosvelasco AC, Jialal I, Strandberg TE, Vanhanen H, Tikkanen MJ, Grothusen C, Bley S, Selle T, Luchtefeld M, Grote K, Tietge UJ, Drexler H, Schiffer B, Pawlowski N, Kaplan G, Hamill AL, Cohn ZA, Scott WA, Livak KJ, Schmittgen TD, Satoh M, Ishikawa Y, Minami Y, Takahashi Y, Nakamura M, Methe H, Kim JO, Kofler S, Weis M, Nabauer M, Koglin J, Ishikawa Y, Satoh M, Itoh T, Minami Y, Takahashi Y, Akamura M, Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, Akira S, Rajavashisth TB, Arditi M, Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR, Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Liu G, Zhao Y, Pedersen I, David M, Huang Y, Li T, Sane DC, Li L, Engel D, Seijkens T, Poggi M, Sanati M, Thevissen L, Beckers L, Wijnands E, Lievens D, Lutgens E, Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y, Zhou W, Xiong B, Zeng Q, Schmieder RE, Hilgers KF, Schlaich MP, Schmidt BM, Ji Y, Liu J, Wang Z, Liu N, Dandona P, Kumar V, Aljada A, Ghanim H, Syed T, Hofmayer D, Mohanty P, Tripathy D, Garg R, Catar RA, Müller G, Heidler J, Schmitz G, Bornstein SR, Morawietz H, Methe H, Kim JO, Kofler S, Nabauer M, Weis M, Edfeldt K, Swedenborg J, Hansson GK, Yan ZQ, Xu XH, Shah PK, Faure E, Equils O, Thomas L, Fishbein MC, Luthringer D, Xu XP, Rajavashisth TB, Yano J, Kaul S, Arditi M, Kobayashi A, Hayashi H, Kisamori K, Ishizaka K, Yamazaki N, Smith SC, Allen J, Blair SN, Bonow RO, Brass LM, Fonarow GC, Grundy SM, Hiratzka L, Jones D, Krumholz HM. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 2010; 119:395–405. [DOI] [PubMed] [Google Scholar]
- 42. DuPont JJ, McCurley A, Davel AP, McCarthy J, Bender SB, Hong K, Yang Y, Yoo JK, Aronovitz M, Baur WE, Christou DD, Hill MA, Jaffe IZ. Vascular mineralocorticoid receptor regulates microRNA-155 to promote vasoconstriction and rising blood pressure with aging. JCI Insight 2016; 1:e88942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu N, Zhang D, Chen S, Liu X, Lin L, Huang X, Guo Z, Liu J, Wang Y, Yuan W, Qin Y. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 2011; 215:286–293. [DOI] [PubMed] [Google Scholar]
- 44. Zhou T, Ding JW, Wang XA, Zheng XX. Long noncoding RNAs and atherosclerosis. Atherosclerosis 2016; 248:51–61. [DOI] [PubMed] [Google Scholar]
- 45. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han DK, Khaing ZZ, Pollock RA, Haudenschild CC, Liau G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J Clin Invest 1996; 97:1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leung A, Stapleton K, Natarajan R. Functional Long Non-coding RNAs in Vascular Smooth Muscle Cells. In: Current Topics in Microbiology and Immunology. Vol 394. Springer International Publishing: Switzerland, 2015, pp 127–141. [DOI] [PubMed] [Google Scholar]
- 48. Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 2013; 113:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014; 130:1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang YN, Shan K, Yao MD, Yao J, Wang JJ, Li X, Liu B, Zhang YY, Ji Y, Jiang Q, Yan B. Long noncoding RNA-GAS5: a novel regulator of hypertension-induced vascular remodeling. Hypertension 2016; 68:736–748. [DOI] [PubMed] [Google Scholar]
- 51. Gopalakrishnan K, Kumarasamy S, Mell B, Joe B. Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension 2015; 65:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014; 514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014; 114:1389–1397. [DOI] [PubMed] [Google Scholar]
- 54. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015; 116:1143–1156. [DOI] [PubMed] [Google Scholar]
- 55. Jazbutyte V, Thum T. MicroRNA-21: from cancer to cardiovascular disease. Curr Drug Targets 2010; 11:926–935. [DOI] [PubMed] [Google Scholar]
- 56. Raitoharju E, Lyytikäinen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kähönen M, Karhunen PJ, Laaksonen R, Lehtimäki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011; 219:211–217. [DOI] [PubMed] [Google Scholar]
- 57. Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun 2010; 393:643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, Li JY, Chien S. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA 2011; 108:10355–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Talepoor AG, Kalani M, Dahaghani AS, Doroudchi M. Hydrogen peroxide and lipopolysaccharide differentially affect the expression of microRNAs 10a, 33a, 21, 221 in endothelial cells before and after coculture with monocytes. Int J Toxicol 2017; 36:133–141. [DOI] [PubMed] [Google Scholar]
- 60. Seddiki N, Brezar V, Ruffin N, Lévy Y, Swaminathan S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 2014; 142:32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McCurley A, Pires PW, Bender SB, Aronovitz M, Zhao MJ, Metzger D, Chambon P, Hill MA, Dorrance AM, Mendelsohn ME, Jaffe IZ. Direct regulation of blood pressure by smooth muscle cell mineralocorticoid receptors. Nat Med 2012; 18:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol 1983; 51:606. [DOI] [PubMed] [Google Scholar]
- 63. Jia QW, Chen ZH, Ding XQ, Liu JY, Ge PC, An FH, Li LH, Wang LS, Ma WZ, Yang ZJ, Jia EZ. Predictive effects of circulating miR-221, miR-130a and miR-155 for coronary heart disease: a multi-ethnic study in China. Cell Physiol Biochem 2017; 42:808–823. [DOI] [PubMed] [Google Scholar]
- 64. Donners MM, Wolfs IM, Stöger LJ, van der Vorst EP, Pöttgens CC, Heymans S, Schroen B, Gijbels MJ, de Winther MP. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS One 2012; 7:e35877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun HX, Zeng DY, Li RT, Pang RP, Yang H, Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, Yan GJ, Zhou JG. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012; 60:1407–1414. [DOI] [PubMed] [Google Scholar]
- 66. Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 2008; 180:5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. miRBase accession for hsa-miR-146b. <http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0003129>. Accessed 29 September 2017.
- 68. miRBase accession for hsa-miR-146a. <http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000477>. Accessed 29 September 2017.
- 69. Jeon YJ, Kim OJ, Kim SY, Oh SH, Oh D, Kim OJ, Shin BS, Kim NK. Association of the miR-146a, miR-149, miR-196a2, and miR-499 polymorphisms with ischemic stroke and silent brain infarction risk. Arterioscler Thromb Vasc Biol 2013; 33:420–430. [DOI] [PubMed] [Google Scholar]
- 70. Hromadnikova I, Kotlabova K, Hympanova L, Krofta L. Gestational hypertension, preeclampsia and intrauterine growth restriction induce dysregulation of cardiovascular and cerebrovascular disease associated microRNAs in maternal whole peripheral blood. Thromb Res 2016; 137:126–140. [DOI] [PubMed] [Google Scholar]
- 71. Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ Res 2015; 117:e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. miRBase accession for hsa-miR-145. <http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000461>. Accessed 25 September 2017.
- 73. Liu K, Xuekelati S, Zhang Y, Yin Y, Li Y, Chai R, Li X, Peng Y, Wu J, Guo X. Expression levels of atherosclerosis-associated miR-143 and miR-145 in the plasma of patients with hyperhomocysteinaemia. BMC Cardiovasc Disord 2017; 17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Markus B, Grote K, Worsch M, Parviz B, Boening A, Schieffer B, Parahuleva MS. Differential expression of microRNAs in endarterectomy specimens taken from patients with asymptomatic and symptomatic carotid plaques. Aikawa E, ed. PLoS One. 2016; 11:e0161632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kontaraki JE, Marketou ME, Zacharis EA, Parthenakis FI, Vardas PE. Differential expression of vascular smooth muscle-modulating microRNAs in human peripheral blood mononuclear cells: novel targets in essential hypertension. J Hum Hypertens 2014; 28:510–516. [DOI] [PubMed] [Google Scholar]
- 76. Santovito D, Mandolini C, Marcantonio P, De Nardis V, Bucci M, Paganelli C, Magnacca F, Ucchino S, Mastroiacovo D, Desideri G, Mezzetti A, Cipollone F. Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert Opin Ther Targets 2013; 17:217–223. [DOI] [PubMed] [Google Scholar]
- 77. de Gonzalo-Calvo D, Cenarro A, Civeira F, Llorente-Cortes V. microRNA expression profile in human coronary smooth muscle cell-derived microparticles is a source of biomarkers. Clin Investig Arterioscler 2016; 28:167–177. [DOI] [PubMed] [Google Scholar]
- 78. Sala F, Aranda JF, Rotllan N, Ramírez CM, Aryal B, Elia L, Condorelli G, Catapano AL, Fernández-Hernando C, Norata GD. MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr-/-mice. Thromb Haemost 2014; 112:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hergenreider E, Heydt S, Tréguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 2012; 14:249–256. [DOI] [PubMed] [Google Scholar]
- 80. Boettger T, Beetz N, Kostin S, Schneider J, Krüger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest 2009; 119:2634–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dahan D, Hien TT, Tannenberg P, Ekman M, Rippe C, Boettger T, Braun T, Tran-Lundmark K, Tran PK, Swärd K, Albinsson S. MicroRNA-dependent control of serotonin-induced pulmonary arterial contraction. J Vasc Res 2017; 54:246–256. [DOI] [PubMed] [Google Scholar]
- 82. Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 2005; 123:819–831. [DOI] [PubMed] [Google Scholar]
- 83. Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, Paulaitis ME, Piper MG, Marsh CB. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 2013; 121:984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Deiuliis JA, Syed R, Duggineni D, Rutsky J, Rengasamy P, Zhang J, Huang K, Needleman B, Mikami D, Perry K, Hazey J, Rajagopalan S. Visceral adipose microRNA 223 is upregulated in human and murine obesity and modulates the inflammatory phenotype of macrophages. Xu P, ed. PLoS One. 2016; 11:e0165962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen Y, Song Y, Huang J, Qu M, Zhang Y, Geng J, Zhang Z, Liu J, Yang G-Y. Increased circulating exosomal miRNA-223 is associated with acute ischemic stroke. Front Neurol. 2017; 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li Z, Yu F, Zhou X, Zeng S, Zhan Q, Yuan M, Yang Q, Liu Y, Xia J. Promoter hypomethylation of microRNA223 gene is associated with atherosclerotic cerebral infarction. Atherosclerosis 2017; 263:237–243. [DOI] [PubMed] [Google Scholar]
- 87. Wang Y, Zhang Y, Huang J, Chen X, Gu X, Wang Y, Zeng L, Yang GY. Increase of circulating miR-223 and insulin-like growth factor-1 is associated with the pathogenesis of acute ischemic stroke in patients. BMC Neurol 2014; 14:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS One 2012; 7:e42971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. A novel regulator of macrophage activation: miR-223 in obesity associated adipose tissue inflammation. Circulation 2012; 125:2892–2903. [DOI] [PubMed] [Google Scholar]
- 90. Vegter EL, Ovchinnikova ES, van Veldhuisen DJ, Jaarsma T, Berezikov E, van der Meer P, Voors AA. Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clin Res Cardiol 2017; 106:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Meder B, Katus HA, Rottbauer W. Right into the heart of microRNA-133a. Genes Dev 2008; 22:3227–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 2008; 118:1567–1576. [DOI] [PubMed] [Google Scholar]
- 94. miRBase accession for hsa-miR-1. <http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000651>. Accessed 25 September 2017.
- 95. Kontaraki JE, Marketou ME, Parthenakis FI, Maragkoudakis S, Zacharis EA, Petousis S, Kochiadakis GE, Vardas PE. Hypertrophic and antihypertrophic microRNA levels in peripheral blood mononuclear cells and their relationship to left ventricular hypertrophy in patients with essential hypertension. J Am Soc Hypertens 2015; 9:802–810. [DOI] [PubMed] [Google Scholar]
- 96. Dal-Pra S, Hodgkinson CP, Mirotsou M, Kirste I, Dzau VJ. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by miR ComboNovelty and significance. Circ Res 2017; 120:1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol 2011; 51:872–875. [DOI] [PubMed] [Google Scholar]
- 98. Patel SA, Winkel M, Ali MK, Narayan KM, Mehta NK. Cardiovascular mortality associated with 5 leading risk factors: national and state preventable fractions estimated from survey data. Ann Intern Med 2015; 163:245–253. [DOI] [PubMed] [Google Scholar]
- 99. van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med 2014; 6:851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res 2012; 110:496–507. [DOI] [PubMed] [Google Scholar]
- 101. Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, Benson MD, O’Sullivan JF, Keshishian H, Farrell LA, Fifer MA, Vasan RS, Sabatine MS, Larson MG, Carr SA, Wang TJ, Gerszten RE. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 2016; 134:270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, Polisky BA, Zinnen S. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 2005; 41:1349–1356. [DOI] [PubMed] [Google Scholar]
- 103. Sullenger BA, Nair S. From the RNA world to the clinic. Science 2016; 352:1417–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Beaucage SL, Caruthers MH. Synthetic strategies and parameters involved in the synthesis of oligodeoxyribonucleotides according to the phosphoramidite method. Curr Protoc Nucleic Acid Chem 2000; Chapter 3:00:3.3:3.3.1-3.3.20. [DOI] [PubMed] [Google Scholar]
- 105. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 106. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861–874. [DOI] [PubMed] [Google Scholar]
- 107. Murakami K. Non-coding RNAs and hypertension-unveiling unexpected mechanisms of hypertension by the dark matter of the genome. Curr Hypertens Rev 2015; 11:80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sharp PA. RNA interference–2001. Genes Dev 2001; 15:485–490. [DOI] [PubMed] [Google Scholar]
- 109. Hanna M. Novel drugs targeting transthyretin amyloidosis. Curr Heart Fail Rep 2014; 11:50–57. [DOI] [PubMed] [Google Scholar]
- 110. Dragon F, Lemay V, Trahan C. snoRNAs: biogenesis, structure and function. In: Maccarrone M (ed) Encyclopedia of Life Sciences. John Wiley & Sons: Chichester, UK, 2006. [Google Scholar]
- 111. Padgett RA, Padgett AR. mRNA splicing: role of snRNAs. In: Cooper DN (ed) eLS. John Wiley & Sons: Chichester, UK, 2015, pp 1–8. [Google Scholar]
- 112. Jarroux J, Morillon A, Pinskaya M.History, Discovery, and Classification of lncRNAs. Springer: Singapore, 2017, pp 1–46. [DOI] [PubMed] [Google Scholar]
- 113. Mishra PK, Tyagi N, Kundu S, Tyagi SC. MicroRNAs are involved in homocysteine-induced cardiac remodeling. Cell Biochem Biophys 2009; 55:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nandakumar P, Tin A, Grove ML, Ma J, Boerwinkle E, Coresh J, Chakravarti A. MicroRNAs in the miR-17 and miR-15 families are downregulated in chronic kidney disease with hypertension. Endlich N, ed. PLoS One. 2017; 12:e0176734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Huang Y, Huang C, Chen J, Li J, Feng Y. Plasma expression level of miRNA let-7 is positively correlated with carotid intima-media thickness in patients with essential hypertension. J Hum Hypertens 2017; 31:843–847. [DOI] [PubMed] [Google Scholar]
- 116. Qi H, Liu Z, Liu B, Cao H, Sun W, Yan Y, Zhang L. micro-RNA screening and prediction model construction for diagnosis of salt-sensitive essential hypertension. Medicine (Baltimore) 2017; 96:e6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nie X, Chen Y, Tan J, Dai Y, Mao W, Qin G, Ye S, Sun J, Yang Z, Chen J. MicroRNA-221-3p promotes pulmonary artery smooth muscle cells proliferation by targeting AXIN2 during pulmonary arterial hypertension. Vascul Pharmacol 2017. [DOI] [PubMed] [Google Scholar]
- 118. Deng H, Song Z, Xu H, Deng X, Zhang Q, Chen H, Wang Y, Qin Y, Li Y. MicroRNA-1185 promotes arterial stiffness though modulating VCAM-1 and E-selectin expression. Cell Physiol Biochem 2017; 41:2183–2193. [DOI] [PubMed] [Google Scholar]
- 119. Lin B, Feng DG, Wang F, Wang JX, Xu CG, Zhao H, Cheng ZY. MiR-365 participates in coronary atherosclerosis through regulating IL-6. Eur Rev Med Pharmacol Sci 2016; 20:5186–5192. [PubMed] [Google Scholar]
- 120. de Gonzalo-Calvo D, Dávalos A, Montero A, García-González Á, Tyshkovska I, González-Medina A, Soares SM, Martínez-Camblor P, Casas-Agustench P, Rabadán M, Díaz-Martínez ÁE, Úbeda N, Iglesias-Gutiérrez E. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol (1985) 2015; 119:124–134. [DOI] [PubMed] [Google Scholar]
- 121. Cengiz M, Yavuzer S, Kılıçkıran Avcı B, Yürüyen M, Yavuzer H, Dikici SA, Karataş ÖF, Özen M, Uzun H, Öngen Z. Circulating miR-21 and eNOS in subclinical atherosclerosis in patients with hypertension. Clin Exp Hypertens 2015; 37:643–649. [DOI] [PubMed] [Google Scholar]
- 122. Vogel B, Keller A, Frese KS, Leidinger P, Sedaghat-Hamedani F, Kayvanpour E, Kloos W, Backe C, Thanaraj A, Brefort T, Beier M, Hardt S, Meese E, Katus HA, Meder B. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J 2013; 34:2812–2822. [DOI] [PubMed] [Google Scholar]
- 123. Fu BC, Lang JL, Zhang DY, Sun L, Chen W, Liu W, Liu KY, Ma CY, Jiang SL, Li RK, Tian H. Suppression of miR-34a expression in the myocardium protects against ischemia-reperfusion injury through SIRT1 protective pathway. Stem Cells Dev 2017; 26:1270–1282. [DOI] [PubMed] [Google Scholar]
- 124. Yang F, Li H, Du Y, Shi Q, Zhao L. Downregulation of microRNA‑34b is responsible for the elevation of blood pressure in spontaneously hypertensive rats. Mol Med Rep 2017; 15:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang P, Xu J, Hou Z, Wang F, Song Y, Wang J, Zhu H, Jin H. miRNA-34a promotes proliferation of human pulmonary artery smooth muscle cells by targeting PDGFRA. Cell Prolif 2016; 49:484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Boström P, Che L, Zhang C, Spiegelman BM, Rosenzweig A. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab 2015; 21:584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kriegel AJ, Baker MA, Liu Y, Liu P, Cowley AW Jr, Liang M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 2015; 66:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med 2014; 20:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem 2009; 284:29514–29525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 2007; 100:1579–1588. [DOI] [PubMed] [Google Scholar]
- 131. Xu Y, Bei Y, Shen S, Zhang J, Lu Y, Xiao J, Li X. MicroRNA-222 promotes the proliferation of pulmonary arterial smooth muscle cells by targeting P27 and TIMP3. Cell Physiol Biochem 2017; 43:282–292. [DOI] [PubMed] [Google Scholar]
- 132. Qin B, Xiao B, Liang D, Xia J, Li Y, Yang H. MicroRNAs expression in ox-LDL treated HUVECs: MiR-365 modulates apoptosis and Bcl-2 expression. Biochem Biophys Res Commun 2011; 410:127–133. [DOI] [PubMed] [Google Scholar]
- 133. Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol 2011; 300:H1762–H1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, Yuan W, Rui YC. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens 2010; 28:1646–1654. [DOI] [PubMed] [Google Scholar]
- 135. Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci U S A 2010; 107:13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R, Federici M. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 2009; 120:1524–1532. [DOI] [PubMed] [Google Scholar]
- 137. Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006; 108:3068–3071. [DOI] [PubMed] [Google Scholar]