Abstract
BACKGROUND
Understanding the interactions between genetics, sodium (Na+) intake, and blood pressure (BP) will help overcome the lack of individual specificity in our current treatment of hypertension. This study had 3 goals: expand on the relationship between striatin gene (STRN) status and salt-sensitivity of BP (SSBP); evaluate the status of Na+ and volume regulating systems by striatin risk allele status; evaluate potential SSBP mechanisms.
METHODS
We assessed the relationship between STRN status in humans (HyperPATH cohort) and SSBP and on volume regulated systems in humans and a striatin knockout mouse (STRN+/−).
RESULTS
The previously identified association between a striatin risk allele and systolic SSBP was demonstrated in a new cohort (P = 0.01). The STRN–SSBP association was significant for the combined cohort (P = 0.003; β = +5.35 mm Hg systolic BP/risk allele) and in the following subgroups: normotensives, hypertensives, men, and older subjects. Additionally, we observed a lower epinephrine level in risk allele carriers (P = 0.014) and decreased adrenal medulla phenylethanolamine N-methyltransferase (PNMT) in STRN+/− mice. No significant associations were observed with other volume regulated systems.
CONCLUSIONS
These results support the association between a variant of striatin and SSBP and extend the findings to normotensive individuals and other subsets. In contrast to most salt-sensitive hypertensives, striatin-associated SSBP is associated with normal plasma renin activity and reduced epinephrine levels. These data provide clues to the underlying cause and a potential pathway to achieve, specific, personalized treatment, and prevention.
Keywords: blood pressure, epinephrine, genetics, hypertension, salt sensitivity, striatin
It is estimated that 1.3 billion individuals in the world have hypertension (HTN) and is now the leading condition associated with death, disability, and health care costs worldwide.1 Typically, treatment of HTN is empirical, with little or no insight into underlying etiology for any given patient. Because the blood pressure (BP) lowering is usually not directed at the underlying cause, it may not result in the maximum improvement in cardiovascular health. For example, 1 mechanistic HTN subgroup is primary aldosteronism, the underlying disease in 5–15% of hypertensives. In this disease, there is dissociation between the BP level and the degree of cardiovascular and renal damage.2 Thus, treating the BP, and not the underlying cause (dysregulated aldosterone (ALDO)), results in suboptimal outcomes.
With the recent focus on precision medicine applied to both prevention and treatment strategies, there has been a renewed interest in understanding the mechanisms of the likely processes underlying HTN. Gene–environment interaction underscores progression to HTN, with BP sensitivity to salt in animals and humans as the major example.3–5 We assembled a large set of carefully phenotyped hypertensive and normotensive subjects on liberal (LIB) and restricted (RES) salt diets that allow us to test the hypothesis that some hypertensives have increased BP secondary to dysfunction(s) in one or more variants in genes related to Na+/volume homeostasis, with confirmatory mechanistic studies in comparable, usually genetically manipulated, animal models.
We recently identified one such hypertensive subset with the candidate gene being striatin. A polymorphic variant in this gene is associated with SSBP in humans and in STRN+/− mice.6 The knockout mice also have increased vasoconstriction in response to phenylephrine or potassium and decreased vasodilation in response to acetyl choline, all while on LIB diets.7 Striatin, also known as calmodulin-binding protein, was identified by Castets and colleagues using rat brain synaptosome.8 It is widely expressed in the heart, kidney, various digestive organs, and the adrenal.9 Striatin also was shown to interact with the estrogen receptor-α (ER-α) and to play an obligatory role in ER-α’s nongenomic pathway related to estrogen’s cardiovascular protective effects.10,11 We reported that striatin could likewise interact with the mineralocorticoid receptor and regulate the rapid/nongenomic responses to ALDO in mouse and human vascular endothelial cells.12,13
The goals of the present study were 3-fold: first, to expand on the association between striatin risk allele (rs254093) and SSBP in a different group of subjects using a similar protocol as that used by Garza et al.6; second, in the combined population, to assess whether this association was limited to one sex, race, age, or disease state; and third, in exploratory analyses, we evaluated potential mechanisms underlying the SSBP. A better definition of the breadth of the association will more precisely guide potential treatment/prevention strategies.
METHODS
Population
Subjects came from the approximately 2,000 subject HyperPATH cohort. Although results from the HyperPATH cohort have been previously reported, the present studies have only been reported in part.6 Specifically, the data from the new subcohort has not been described before. In brief, HyperPATH is an on-going, multi-institutional, international program to study the genetic underpinnings of hormonal mechanisms of HTN and cardiovascular disease. Normotensive and stage I hypertensive subjects, with or without diabetes, were recruited and withdrawn from all antihypertensive medications for 1–3 months prior to study.5,6,12,14–22 Subjects were studied under 2 dietary conditions, at least 1 week apart: LIB (200 mmol/day Na+) and RES (10 mmol/day Na+), with all other components similar in the 2 diets, for 5–7 days followed by a 24-hour urine collection. Subjects were admitted to the Clinical Research Unit and kept in a supine posture overnight. The following morning, blood was obtained to measure a variety of hormones, as noted below, and para-aminohippuric acid was infused to assess renal blood flow. All study procedures were approved by the corresponding Institutional Review Board, and written informed consent was obtained from participants prior to inclusion in the study.
Genotyping
The Partners Center for Personalized Genetic Medicine (PCPGM) performed the genotyping by techniques as we have previously reported.14–19,21,22 This study used the same tagging single-nucleotide polymorphism, rs2540923, a nonfunctional, nonsense variant located within the noncoding region of the STRN. It was identified in a previous publication and used for analysis purposes for the reasons defined in that publication.6
Generation of striatin heterozygous mouse and experimental protocol
The trans-NIH Knock-Out Mouse Project (KOMP) was used to generate the STRN+/− strain (CSD26933) as we have previously reported.6,7 All study procedures were approved by the Animal Review Board (IACUC) prior to study.
Analyte measurements
Hormones and electrolytes in serum and urine were measured in the Brigham and Women’s Hospital’s Research Assay Core Laboratory as previously reported.5,6,14–22 ALDO levels were measured using Coat-A-Count Radioimmunometric Assay (RIA) kit (SIEMENS, Los Angeles, CA); plasma renin activity by RIA assay (DiaSorin, Stillwater, MN); cortisol by Roche Access and catecholamines and para-aminohippuric acid by LC Mass Spec.
Western blot analysis
The mouse adrenal medulla was isolated from the adrenal cortex by mechanical dissection. Protein analysis was performed as previously described.6,17,18,22,23 Protein was extracted from the medulla by homogenizing the tissue; cell lysates were prepared; samples were size fractionated by electrophoresis on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE); proteins transferred to nitrocellulose membranes by electroblotting; blots were incubated with primary antibody overnight at 4 °C (BD biosciences) and then incubated with conjugated secondary antibody with horseradish-peroxidase for 1 hour at room temperature. The samples were analyzed using Enhanced Chemiluminescence (ECL) (Perkin-Elmer Life Sciences, Waltham, MA). Blots were reprobed for β-actin (Sigma) and results were normalized to correct for loading.
Statistics
All statistical analyses were performed with the Stata/SE 14.1 (StataCorp). Data and results are presented as mean ± SD and percentages for categorical variables. Univariate analyses were performed using a chi-square test and/or an unpaired Student’s t-test. Observed and expected values for allele frequencies to evaluate Hardy–Weinberg Equilibrium were compared with a chi-square test. We applied a multiple linear regression model to test striatin risk alleles associations with the primary phenotype (i.e., SSBP). All association analyses between striatin risk allele and the primary endpoint, systolic SSBP, and secondary endpoints were performed assuming an additive model and adjusted for age, gender, race, body mass index, and disease state (HTN vs. normal and diabetes vs. HTN and normal) in the subjects in the HyperPATH Cohort that had the appropriate variables. The P value threshold for significance was set at 0.05 for the primary endpoint. Where multiple comparisons were made, the alpha was 0.05/3 = 0.017.
RESULTS
New cohort
Demographics.
The new cohort consisted of 331 individuals. Approximately 50% were female, 27% were Blacks, 10% were diabetic, and 55% were normotensives without diabetes. The distribution of these characteristics did not differ significantly by rs2540923 genotype except for race (Table 1). Blacks carried the minor (risk) allele more frequently than Caucasians (P < 0.0001).
Table 1.
Tagging SNP rs2540923 analysis for new cohort
| Major allele homozygotes (GG), (n = 290) | Minor allele carriers (AG + AA), (n = 41) | P value | |
|---|---|---|---|
| Sex | |||
| Females (%) | 143 (49.3) | 23 (56.1) | |
| Males (%) | 147 (50.7) | 18 (43.9) | 0.42 |
| Race (%) | |||
| Caucasians | 204 (77.9) | 8 (19.5) | |
| Blacks | 58 (22.1) | 33 (80.5) | <0.0001 |
| Disease status (%) | |||
| Hypertensive with diabetes mellitus | 26 (9.3) | 5 (12.5) | |
| Hypertensive | 80 (28.4) | 15 (37.5) | |
| Normotensive | 175 (62.3) | 20 (50.0) | 0.14 |
| Age, years | 43.2 ± 11.6 | 41.9 ± 10.2 | 0.49 |
| BMI, kg/m2 | 26.5 ± 4.4 | 28.7 ± 4.8 | 0.003 |
| SBP on liberal salt, mm Hg | 123.3 ± 22.4 | 137.4 ± 27.4 | 0.001 |
| Mean aldosterone, ng/dl | 4.4 ± 3.4 | 4.4 ± 3.7 | 0.98 |
| Upright PRA, ng/(ml * hour) | 8.4 ± 6.3 | 6.6 ± 5.0 | 0.08 |
Data for continuous variables represent means ± SD. Abbreviations: BMI, body mass index; PRA, plasma renin activity; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
SSBP.
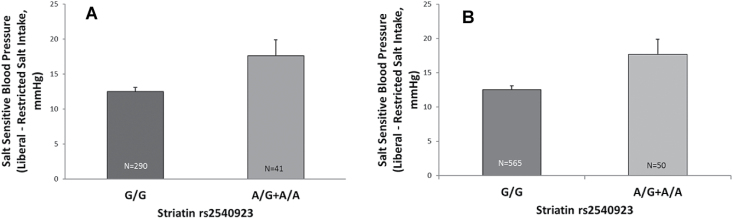
As in our original cohort, there was a statistically significant difference between the minor allele (AA and A/G) (risk) carriers and major allele homozygote (GG) noncarriers for SSBP (change in systolic BP between RES and LIB diets) (P = 0.001; β = +6.75 mm Hg per risk allele, 95% confidence interval: 2.7–11.1) (Table 2). This difference remained significant even after adjusting for age, body mass index, race, gender, and disease state (HTN vs. normal and diabetes vs. normal and HTN) (P = 0.017; β = +4.84 mm Hg per risk allele, 95% confidence interval: 0.84–8.8) (Table 2). The trend analysis by genotype was also significant (P = 0.005) (Figure 1a). With increasing number of risk alleles, the carriers had a greater SSBP.
Table 2.
Systolic salt sensitivity of blood pressure (SSBP) by univariate and multivariate analysis for tagging SNP rs2540923
| Cohort | Sample size | Univariate analysis | Multivariate analysisa | ||
|---|---|---|---|---|---|
| β (95% CI) | P value | β (95% CI) | P value | ||
| New | 270 | 6.75 (2.70, 10.81) | 0.001 | 4.84 (0.84, 8.83) | 0.01 |
| Combined | 615 | 5.25 (1.67, 8.84) | 0.004 | 5.35 (1.86, 8.83) | 0.003 |
Abbreviations: BMI, body mass index; CI, confidence interval; SNP, single-nucleotide polymorphism.
aAdjustment made for age, sex, race, BMI, disease state (hypertensive vs. normal and diabetic vs. hypertensive and normal). The SNP rs2540923 was stratified by comparing the GG group to the AG + AA group.
Figure 1.
New cohort, systolic SSBP according to striatin genotype (a). Data represent mean ± SEM by t-test, there was a significant (P = 0.005) association between systolic SSBP and genotype, with greater systolic SSBP noted in risk allele carrier group (A/G = 38, A/A = 3). Combined population, systolic SSBP according to striatin genotype (b). Data represents mean ± SEM. Based on t-test, there is again a significant (P = 0.005) association between systolic SSBP and genotype, with minor allele carriers experiencing greater SSBP than noncarriers, an effect more pronounced with increasing numbers of risk alleles (A/G = 46, A/A = 4).
Combined cohort
Demographics.
The data from the new and previously publish cohort were then combined. The combined cohort consisted of 696 individuals of whom 8.5% were carriers of the risk (minor) allele. Approximately 45% were females, 14% were Blacks, 5% were diabetics, and 27% were normotensives without diabetes. The distribution of the several characteristics did not differ significantly by rs2540923 genotype except for race (Table 3). Blacks carried the risk (minor) allele more frequently than Caucasians (P < 0.0001).
Table 3.
Tagging SNP rs2540923 analysis for combined study population
| Major allele homozygotes (GG), (n = 637) | Minor allele carriers (AG + AA), (n = 59) | P value | |
|---|---|---|---|
| Sex | |||
| Females (%) | 284 (44.6) | 32 (54.2) | |
| Males (%) | 353 (55.4) | 27 (45.8) | 0.15 |
| Race (%) | |||
| Caucasians | 551 (90.5) | 26 (44.1) | |
| Blacks | 58 (9.5) | 33 (55.9) | <0.0001 |
| Disease status (%) | |||
| Hypertensive with diabetes mellitus | 26 (4.1) | 5 (8.6) | |
| Hypertensive | 427 (68.0) | 33 (56.8) | |
| Normotensive | 175 (27.9) | 20 (34.6) | 0.29 |
| Age, years | 45.9 ± 10.5 | 45.3 ± 10.6 | 0.66 |
| BMI, kg/m2 | 27.3 ± 4.2 | 28.9 ± 4.7 | 0.005 |
| SBP on liberal salt, mm Hg | 137.0 ± 23.4 | 142.2 ± 25.6 | 0.12 |
| Mean aldosterone, ng/dl | 5.1 ± 3.8 | 4.7 ± 3.8 | 0.55 |
| Upright PRA, ng/(ml * hour) | 7.8 ± 6.5 | 6.3 ± 4.9 | 0.11 |
Data for continuous variables represent means ± SD. Abbreviations: BMI, body mass index; PRA, plasma renin activity; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
SSBP.
As in the new cohort, there was a statistically significant difference between the minor (risk) allele carriers and major allele homozygote noncarriers for systolic SSBP (P = 0.004; β = +5.25 mm Hg per risk allele, 95% confidence interval: 1.7–8.8) (Table 2). This difference remained significant even after adjusting for age, body mass index, race, sex, and the 2 disease states (HTN and diabetes) (P = 0.003; β = +5.35 mm Hg per risk allele, 95% confidence interval: 1.9–8.8) (Table 2). The trend analysis by genotype was also significant (P = 0.005) (Figure 1b). With increasing number of risk alleles the carriers had a greater SSBP.
Relation of STRN status and SSBP in demographic subgroups.
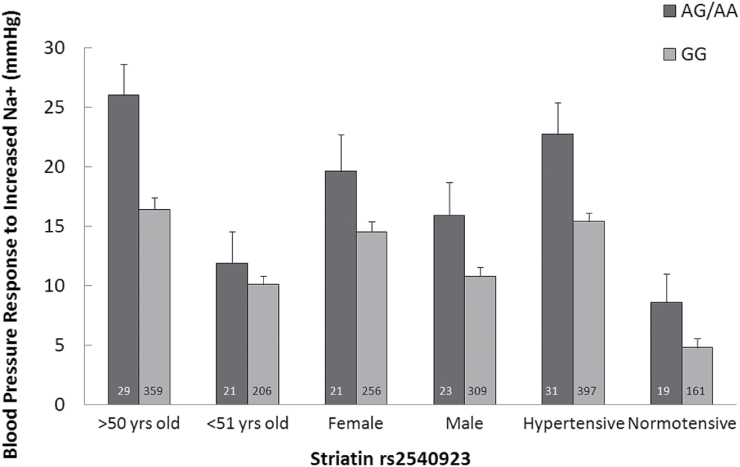
Because of the independent association of SSBP, using trend analyses, we assessed the effect of risk allele status (0, 1, 2 alleles) on SSBP in several demographic groups. In these analyses, risk allele carriers demonstrated greater systolic SSBP than noncarriers, and this difference was significant across several subgroups for age, sex, and HTN status (Figure 2). As was reported in the previous publications for Caucasian hypertensives, Caucasian risk allele carrier had significantly higher systolic SSBP (P = 0.001) than noncarriers. Despite the greater frequency of the risk allele in Blacks, this did not result in a significant association between risk allele number and SSBP. There were not enough diabetics to assess SSBP by allele carrier status.
Figure 2.
Combined population, systolic SSBP stratified by age, gender, and hypertensive state. Risk allele carriers had greater systolic SSBP than noncarriers if they were normotensive (P = 0.03), hypertensive (P = 0.004), male (P = 0.02), female (P = 0.09), or older than 50 years of age (0.006). Because of multiple comparisons only those P values <0.017 reached statistical significance. The rest are considered trends. Data represents mean ± SEM.
Exploratory analyses
Given the results of the previously reported studies in mice,6,7 we examined several factors that were available only in some but not all subjects. These factors were divided into 2 groups: hormones (renin–angiotensin–ALDO system, cortisol, and catecholamines); and renal function (renal blood flow in response to Na+ intake and angiotensin II infusion). On a population basis, there were no differences between carriers and noncarriers for changes in renal blood flow in response to angiotensin II infusion or change in salt intake, creatinine clearance, cortisol, ALDO, or norepinephrine levels whether unadjusted or fully adjusted in linear regressions models. Plasma renin activity was mildly greater in nonrisk allele carriers (7.7 ng/ml/hr) than risk allele carriers (6.3 ng/ml/hr) (P = 0.049). However, these differences became nonsignificant in the fully adjusted model. Furthermore, the frequency of low renin HTN (plasma renin activity <2.4 ng/ml/hr upright on a RES salt diet) did not differ by genotype.
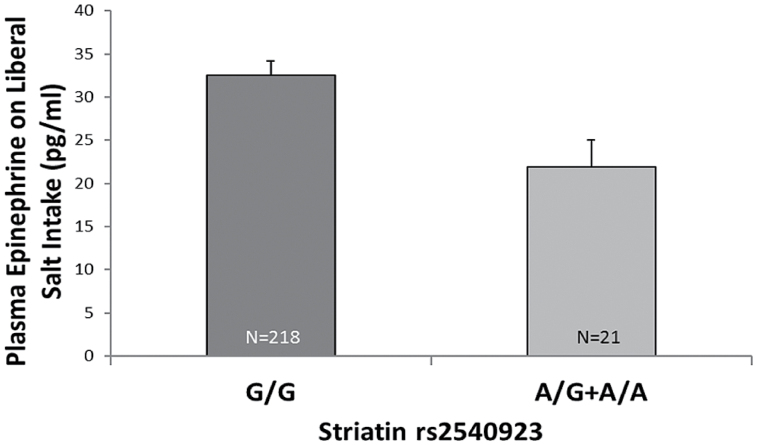
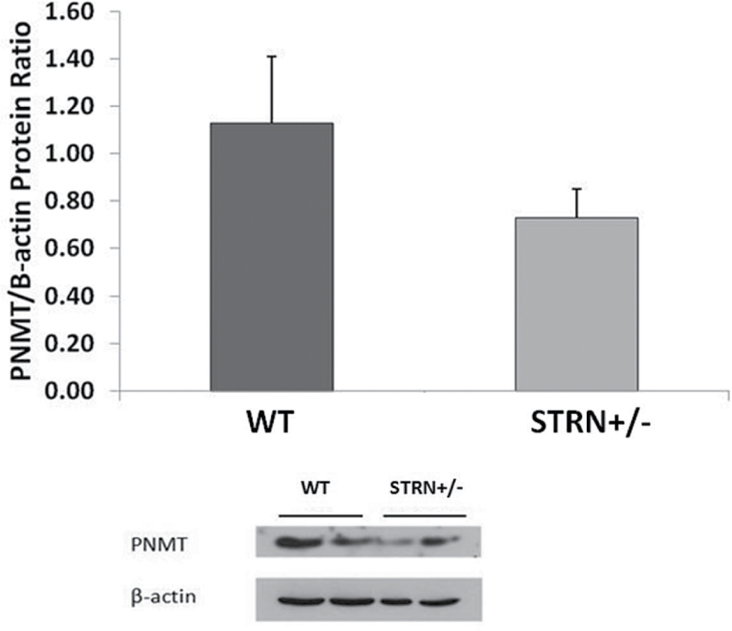
Of interest, risk allele carriers of rs2540923 had statistically lower concentration of plasma epinephrine than noncarriers (P = 0.014) (Figure 3) (trend analyses for number of risk alleles). Given the role of phenylethanolamine N-methyltransferase (PNMT) in converting norepinephrine to epinephrine in the adrenal medulla, we assessed the level of PNMT protein in wild type (WT) and STRN+/− littermates mouse adrenal medulla tissue. The STRN+/− mice had lower protein levels of PNMT than WT (Figure 4), suggesting that the decreased epinephrine in the rs2540923 carriers could be secondary to decreased levels of PNMT.
Figure 3.
Plasma epinephrine levels on LIB diet according to striatin genotype. Based on trend analysis, risk allele carriers of rs2540923 had statistically lower concentration of plasma epinephrine than noncarriers (P = 0.02). Data represents mean ± SEM. Abbreviation: LIB, liberal.
Figure 4.
PNMT protein levels in adrenal medulla from Wild type (WT) and STRN+/− mice. The STRN+/− mice had lower protein levels of PNMT than WT. Data represents mean ± SD, n = 2.
DISCUSSION
The purpose of this study was three fold: 1) to expand on the findings from our prior publication6 that rs2540923 genotype is associated with SSBP by analyzing data from a second, cohort studied on a similar protocol; 2) by combining data from both cohorts, assess the relationship between striatin genotype and SSBP in different demographic subgroups; and 3) to explore potential mechanisms that underlie the association of striatin genotype and SSBP. Our results document a similar association between SSBP and striatin risk allele in a second cohort, thereby confirming the original report.6 There was a moderate effect size with an estimated increase in systolic SSBP of +5.35 mm Hg for those subjects who carrier one risk allele and nearly +11 mm Hg for those few individuals who carry 2 risk alleles. Importantly, the combined cohort contained data from a general population. Even though the risk allele carriers comprised less than 10% of the cohort, their effect size on systolic SSBP was substantial. The data from the combined cohorts suggest that this positive relationship exists in normotensives, hypertensives, males, Caucasians, and older individuals with a trend toward significance in females. In exploratory studies, lower plasma epinephrine levels were associated with the striatin risk allele, and in STRN+/− mice, adrenal medulla PMNT levels were lower than in WT mice. Finally, the risk allele in this cohort was not associated with HTN, per se, but systolic SSBP.
There has been increased interest in assessing the genetic underpinnings of HTN with limited success. Despite extensive analyses of several cohorts by a variety of techniques including candidate gene and genome-wide association studies, consistent findings have been elusive. In contrast, several rare, monogenic causes of HTN have been described,24–29 and mutations in WNK1 and WNK4 cause pseudo-hypoaldosteronism Type II.30–32 Recently, more common polymorphic variants in genes associated with SSBP have been reported. These associations include, for example, angiotensinogen,5,16,21 caveolin-1,18 α-adducin,14,33 β2-adrenergic receptor,17,34,35 lysine-specific demethylase 1 (LSD1),22 the P450 arachidonic acid monooxygenase (ω/ω-1 hydroxylase CYP4A11),36 and common variants in ENaC.37 From the present study, striatin can be added to this list. Of interest, each of these genotype/phenotype associations was determined by a candidate gene approach, and all have been confirmed in a second cohort and/or in a genetically altered rodent model. Currently, individuals who fall into each of these subsets comprise approximately 30% of the hypertensive population and about half of the SSBP group.
The mechanisms underlying these genetic associations with SSBP generally fall into 2 patterns: low renin HTN (which is often directly or indirectly mediated by hormonal changes) and normal/high renin HTN (typically mediated by renal mechanisms). The SSBP/striatin subgroup belongs to the normal renin group. Studies in genetically engineered mice provide a proposed mechanism(s) for the SSBP. The complete striatin knockout mouse is embryologically lethal. However, the STRN+/− mouse has SSBP similar to humans that carry the striatin risk allele. The mice have defective vasodilatory capacity secondary to decreased cGMP generation/activation on a LIB diet. It has been proposed that this defect leads to reduced renal vasodilation and SSBP.7 However, further mechanistic studies are needed to elucidate this.
For some of these genotype associations with SSBP, the relationships apply to only some, but not all demographic subgroups. For example, the angiotensinogen genotype subset which can lead to “nonmodulation”5,16,21 only is present in hypertensive Caucasians, while the LSD1 risk allele is only associated with SSBP in Blacks and not hypertensive Caucasians.22 For striatin risk allele carriers, SSBP is present in normotensives, hypertensives, men, and older individuals regardless of their race, sex, and disease state. Thus, compared to the other genotypes, the striatin risk allele seems to confer a risk of SSBP on most subjects regardless of their demographic characteristics. In those subgroups where it did not confer such a risk, the sample size may have been too small to detect.
Of note, though there have been numerous genome-wide association studies on HTN, none have yet reported on the STRN with BP. This is not altogether surprising, considering that, even within our own data set, no association was seen with HTN alone, but rather with SSBP, suggestive of how heterogeneous the hypertensive population can be.
Intriguingly, in our exploratory analyses, a reduction in epinephrine levels on the LIB diet was found in the striatin risk allele carriers. Since no significant difference in norepinephrine levels between carriers and noncarriers was seen, one potential mechanism underlying the observed SSBP might involve the conversion of norepinephrine to epinephrine by PNMT. This hypothesis is supported by our in vitro studies, in which STRN+/− mice had lower levels of PNMT. Epinephrine is a mediator of nitric oxide cGMP generation.38 Thus, reduced epinephrine would lead to reduced cGMP, as observed in Strn+/ mice.7
The results of this study have important implications for the treatment of human disease. Understanding the connection between genotype and phenotype in the setting of SSBP will better allow us to target subgroups with the appropriate therapy. Although our study was not designed specifically to assess cGMP levels, prior studies have shown that striatin knockout mice have reduced cGMP levels in response to increased Na+ intake.7 Thus, these individuals could potentially benefit from the agonists being developed to generate cGMP directly.39 It also suggests that the striatin risk allele might be a precursor or predictor for the development of HTN, and that normotensive carriers might be responsive to a specific, preventive intervention, such as a reduction in salt intake.
There are several limitations of our study. First, additional mechanistic studies in both humans and mice will be necessary to explore further the pathophysiology of the SSBP. Second, the currently available data suggest that we are not using a functional single-nucleotide polymorphism. Further, we performed analyses using GTEx, ENCODE, and Ensembl and found no results indicating this particular single-nucleotide polymorphism as an eQTL, regardless of tissue. Third, our sample size may have hindered us from observing a change in plasma renin activity, ALDO, or renal blood flow. However, the percentage of individuals with the striatin risk allele does not exceed 7–8% of the general population, so the relative sample size in our study is not unreasonable. We believe that this subset of salt-sensitivity has not yet been observed in larger genetic studies because of the uncontrolled heterogeneity that exists in most hypertensive cohorts. Since our study was tightly controlled and we used an intermediate phenotype (SSBP), we were better able to detect the physiological response to Na+ intake and to therefore identify this novel subset of BP sensitivity to salt intake.
In conclusion, the current study expands on our previous findings that a striatin risk allele at rs254093 is associated with SSBP.6 Furthermore, when the data from the 2 cohorts are combined, this SSBP risk is present in several demographic groups, including normotensives. Finally, in an exploratory analysis, epinephrine levels were lower in human subjects with the striatin risk allele, and STRN+/− mice had decreased PNMT levels that may explain the decreased cGMP generation previously reported.7 Whether these contribute to altered SSBP will require additional mechanistic studies. These data suggest that a genetic variant in striatin may be a useful molecular biomarker to identify individuals for specific mechanistically driven therapy to treat and/or prevent SSBP—personalized medicine.
DISCLOSURE
The authors declared no conflict of interest.
ACKNOWLEDGMENTS
We would like to thank the Center for Clinical Investigation and Brigham and Women’s Hospital’s staff as well as other investigators and staff of the HyperPath Protocol and participants at each protocol site, including the Clinical Investigation Centre, INSERM CIC 9201, Hôpital Européen Georges Pompidou (Paris, France), University of Utah Medical Center (Salt Lake City, UT), Vanderbilt University (Nashville, TN), and University of La Sapiena (Rome, Italy). We are particularly grateful for the many research subjects who gave their time and effort as participants in the HyperPATH program and the research fellows that cared for them and performed the research protocols that generated the data used in this study. With sincere gratitude we acknowledge financial assistance, in partial support for these studies, from the following sources: the Brigham and Women’s Hospital, Division of Endocrinology, Diabetes, Hypertension and Harvard Medical School; National Institute of Health Grants P50HL055000 (HyperPATH Cohort); UL1RR025758 Harvard Clinical and Translational Science Center, from the National Center for Research Resources; M01-RR02635, Brigham and Women’s Hospital, General Clinical Research Center, from the National Center for Research Resources; T32 training grant T32HL007609; and National Institutes of Health research grants R01HL086907, R01HL11476, R01HL69208, R01HL4765, R01HL59424, R01HL69208, R01HL096518, R01HL104032, and T32HL 007609 from the National Heart, Lung and Blood Institute and a grant from the National Center for Advanced Translational Science RM-07-2002. G.H.W., J.S.W., A.G., J.R., and PH designed the research studies and/or oversaw the HyperPATH protocols in Boston, MA, and Salt Lake City, Utah. M.C., N.A., T.G. coordinated the human subjects and acquired data. J.W.T. and W.M. performed the data analyses. P.Y.T. performed the cell-based study. T.G., M.C., and N.A. wrote the manuscript with the oversight of J.S.W. and G.H.W. A.G. and J.R.R. also provided revisions for the manuscript.
REFERENCES
- 1. Mills KT, Bundy JD, Kelly TN, Reed JE, Kearney PM, Reynolds K, Chen J, He J. Global disparities of hypertension prevalence and control: a systematic analysis of population-based studies from 90 countries. Circulation 2016; 134:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrosino P, Lupoli R, Tortora A, Cacciapuoti M, Lupoli GA, Tarantino P, Nasto A, Di Minno MN. Cardiovascular risk markers in patients with primary aldosteronism: a systematic review and meta-analysis of literature studies. Int J Cardiol 2016; 208:46–55. [DOI] [PubMed] [Google Scholar]
- 3. Hurwitz S, Fisher ND, Ferri C, Hopkins PN, Williams GH, Hollenberg NK. Controlled analysis of blood pressure sensitivity to sodium intake: interactions with hypertension type. J Hypertens 2003; 21:951–959. [DOI] [PubMed] [Google Scholar]
- 4. Luzardo L, Noboa O, Boggia J. Mechanisms of salt-sensitive hypertension. Curr Hypertens Rev 2015; 11:14–21. [DOI] [PubMed] [Google Scholar]
- 5. Williams GH, Hollenberg NK. Non-modulating hypertension. A subset of sodium-sensitive hypertension. Hypertension 1991; 17:I81–I85. [DOI] [PubMed] [Google Scholar]
- 6. Garza AE, Rariy CM, Sun B, Williams J, Lasky-Su J, Baudrand R, Yao T, Moize B, Hafiz WM, Romero JR, Adler GK, Ferri C, Hopkins PN, Pojoga LH, Williams GH. Variants in striatin gene are associated with salt-sensitive blood pressure in mice and humans. Hypertension 2015; 65:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garza AE, Pojoga LH, Moize B, Hafiz WM, Opsasnick LA, Siddiqui WT, Horenstein M, Adler GK, Williams GH, Khalil RA. Critical role of striatin in blood pressure and vascular responses to dietary sodium intake. Hypertension 2015; 66:674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castets F, Bartoli M, Barnier JV, Baillat G, Salin P, Moqrich A, Bourgeois JP, Denizot F, Rougon G, Calothy G, Monneron A. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J Cell Biol 1996; 134:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breitman M, Zilberberg A, Caspi M, Rosin-Arbesfeld R. The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. Biochim Biophys Acta 2008; 1783:1792–1802. [DOI] [PubMed] [Google Scholar]
- 10. Bernelot Moens SJ, Schnitzler GR, Nickerson M, Guo H, Ueda K, Lu Q, Aronovitz MJ, Nickerson H, Baur WE, Hansen U, Iyer LK, Karas RH. Rapid estrogen receptor signaling is essential for the protective effects of estrogen against vascular injury. Circulation 2012; 126:1993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu Q, Pallas DC, Surks HK, Baur WE, Mendelsohn ME, Karas RH. Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor alpha. Proc Natl Acad Sci U S A 2004; 101:17126–17131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coutinho P, Vega C, Pojoga LH, Rivera A, Prado GN, Yao TM, Adler G, Torres-Grajales M, Maldonado ER, Ramos-Rivera A, Williams JS, Williams G, Romero JR. Aldosterone’s rapid, nongenomic effects are mediated by striatin: a modulator of aldosterone’s effect on estrogen action. Endocrinology 2014; 155:2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pojoga LH, Coutinho P, Rivera A, Yao TM, Maldonado ER, Youte R, Adler GK, Williams J, Turchin A, Williams GH, Romero JR. Activation of the mineralocorticoid receptor increases striatin levels. Am J Hypertens 2012; 25:243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grant FD, Romero JR, Jeunemaitre X, Hunt SC, Hopkins PN, Hollenberg NH, Williams GH. Low-renin hypertension, altered sodium homeostasis, and an alpha-adducin polymorphism. Hypertension 2002; 39:191–196. [DOI] [PubMed] [Google Scholar]
- 15. Hopkins PN, Hunt SC, Jeunemaitre X, Smith B, Solorio D, Fisher ND, Hollenberg NK, Williams GH. Angiotensinogen genotype affects renal and adrenal responses to angiotensin II in essential hypertension. Circulation 2002; 105:1921–1927. [DOI] [PubMed] [Google Scholar]
- 16. Kosachunhanun N, Hunt SC, Hopkins PN, Williams RR, Jeunemaitre X, Corvol P, Ferri C, Mortensen RM, Hollenberg NK, Williams GH. Genetic determinants of nonmodulating hypertension. Hypertension 2003; 42:901–908. [DOI] [PubMed] [Google Scholar]
- 17. Pojoga L, Kolatkar NS, Williams JS, Perlstein TS, Jeunemaitre X, Brown NJ, Hopkins PN, Raby BA, Williams GH. Beta-2 adrenergic receptor diplotype defines a subset of salt-sensitive hypertension. Hypertension 2006; 48:892–900. [DOI] [PubMed] [Google Scholar]
- 18. Pojoga LH, Underwood PC, Goodarzi MO, Williams JS, Adler GK, Jeunemaitre X, Hopkins PN, Raby BA, Lasky-Su J, Sun B, Cui J, Guo X, Taylor KD, Chen YD, Xiang A, Raffel LJ, Buchanan TA, Rotter JI, Williams GH. Variants of the caveolin-1 gene: a translational investigation linking insulin resistance and hypertension. J Clin Endocrinol Metab 2011; 96:E1288–E1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Underwood PC, Sun B, Williams JS, Pojoga LH, Chamarthi B, Lasky-Su J, Raby BA, Hopkins PN, Jeunemaitre X, Brown NJ, Adler GK, Williams GH. The relationship between peroxisome proliferator-activated receptor-gamma and renin: a human genetics study. J Clin Endocrinol Metab 2010; 95:E75–E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watkins WS, Hunt SC, Williams GH, Tolpinrud W, Jeunemaitre X, Lalouel JM, Jorde LB. Genotype-phenotype analysis of angiotensinogen polymorphisms and essential hypertension: the importance of haplotypes. J Hypertens 2010; 28:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams GH, Dluhy RG, Lifton RP, Moore TJ, Gleason R, Williams R, Hunt SC, Hopkins PN, Hollenberg NK. Non-modulation as an intermediate phenotype in essential hypertension. Hypertension 1992; 20:788–796. [DOI] [PubMed] [Google Scholar]
- 22. Williams JS, Chamarthi B, Goodarzi MO, Pojoga LH, Sun B, Garza AE, Raby BA, Adler GK, Hopkins PN, Brown NJ, Jeunemaitre X, Ferri C, Fang R, Leonor T, Cui J, Guo X, Taylor KD, Ida Chen YD, Xiang A, Raffel LJ, Buchanan TA, Rotter JI, Williams GH, Shi Y. Lysine-specific demethylase 1: an epigenetic regulator of salt-sensitive hypertension. Am J Hypertens 2012; 25:812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo C, Ricchiuti V, Lian BQ, Yao TM, Coutinho P, Romero JR, Li J, Williams GH, Adler GK. Mineralocorticoid receptor blockade reverses obesity-related changes in expression of adiponectin, peroxisome proliferator-activated receptor-gamma, and proinflammatory adipokines. Circulation 2008; 117:2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Botero-Velez M, Curtis JJ, Warnock DG. Brief report: Liddle’s syndrome revisited–a disorder of sodium reabsorption in the distal tubule. N Engl J Med 1994; 330:178–181. [DOI] [PubMed] [Google Scholar]
- 25. Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, Lifton RP. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science 2000; 289:119–123. [DOI] [PubMed] [Google Scholar]
- 26. Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet 1995; 11:76–82. [DOI] [PubMed] [Google Scholar]
- 27. Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel JM. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature 1992; 355:262–265. [DOI] [PubMed] [Google Scholar]
- 28. Mune T, Rogerson FM, Nikkilä H, Agarwal AK, White PC. Human hypertension caused by mutations in the kidney isozyme of 11 beta-hydroxysteroid dehydrogenase. Nat Genet 1995; 10:394–399. [DOI] [PubMed] [Google Scholar]
- 29. Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR Jr, Ulick S, Milora RV, Findling JW. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell 1994; 79:407–414. [DOI] [PubMed] [Google Scholar]
- 30. Wilson FH, Disse-Nicodème S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science 2001; 293:1107–1112. [DOI] [PubMed] [Google Scholar]
- 31. Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci U S A 2003; 100:680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest 2003; 111:1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cusi D, Bianchi G. Renal mechanisms of genetic hypertension: from the molecular level to the intact organism. Kidney Int 1996; 49:1754–1759. [DOI] [PubMed] [Google Scholar]
- 34. Sun B, Williams JS, Svetkey LP, Kolatkar NS, Conlin PR. Beta2-adrenergic receptor genotype affects the renin-angiotensin-aldosterone system response to the Dietary Approaches to Stop Hypertension (DASH) dietary pattern. Am J Clin Nutr 2010; 92:444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svetkey LP, Harris EL, Martin E, Vollmer WM, Meltesen GT, Ricchiuti V, Williams G, Appel LJ, Bray GA, Moore TJ, Winn MP, Conlin PR. Modulation of the BP response to diet by genes in the renin-angiotensin system and the adrenergic nervous system. Am J Hypertens 2011; 24:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams JS, Hopkins PN, Jeunemaitre X, Brown NJ. CYP4A11 T8590C polymorphism, salt-sensitive hypertension, and renal blood flow. J Hypertens 2011; 29:1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao Q, Gu D, Hixson JE, Liu DP, Rao DC, Jaquish CE, Kelly TN, Lu F, Ma J, Mu J, Shimmin LC, Chen J, Mei H, Hamm LL, He J; Genetic Epidemiology Network of Salt Sensitivity Collaborative Research Group . Common variants in epithelial sodium channel genes contribute to salt sensitivity of blood pressure: The GenSalt study. Circ Cardiovasc Genet 2011; 4:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qi R, Ozaki Y, Satoh K, Yang LB, Asazuma N, Yatomi Y, Kume S. Intracellular levels of cyclic AMP and cyclic GMP differentially modify platelet aggregate size in human platelets activated with epinephrine or ADP. J Cardiovasc Pharmacol 1996; 28:215–222. [DOI] [PubMed] [Google Scholar]
- 39. Oettrich JM, Dao VT, Frijhoff J, Kleikers P, Casas AI, Hobbs AJ, Schmidt HH. Clinical relevance of cyclic GMP modulators: A translational success story of network pharmacology. Clin Pharmacol Ther 2016; 99:360–362. [DOI] [PubMed] [Google Scholar]