Abstract
Background
Giardia spp. are flagellated protozoan parasites that infect humans and many other vertebrates worldwide. Currently seven species of Giardia are considered valid.
Results
Here, we report a new species, Giardia cricetidarum n. sp. in hamsters. Trophozoites of G. cricetidarum n. sp. are pear-shaped with four pairs of flagella and measure on average 14 μm (range 12–18 μm) in length and 10 μm (range 8–12 μm) in width. The trophozoites of the new species are generally larger and stouter than those of most of the other Giardia spp. and exhibit the lowest length/width ratio (c.1.40) of all recognized Giardia species. Cysts of G. cricetidarum n. sp. are ovoid and measure on average 11 μm (range 9–12 μm) in length and 10 μm (range 8–10 μm) in width and are indistinguishable from the cysts of other Giardia species. Molecular phylogenetic analyses based on beta-giardin, small subunit rRNA, and elongation factor-1 alpha loci all demonstrated that G. cricetidarum n. sp. is genetically distinct from all currently accepted Giardia spp. Investigation of the host range indicated that the new species was only found in hamsters (including Phodopus sungorus, P. campbelli and Mesocricetus auratus), while all the other described mammal-parasitizing species (G. muris, G. microti and G. intestinalis) each infect multiple hosts. Cross-transmission studies further demonstrated the apparent host specificity of G. cricetidarum n. sp. as it only infected hamsters. Trophozoites were found in high numbers in hamster intestines (5 × 105 – 5 × 106) and was rarely detected co-infecting with other Giardia spp. in the common hamster, suggesting it has some advantages in parasitizing hamsters.
Conclusions
This study has identified a new species of Giardia, which appears to be specific to hamsters, and together with the three other mammal-parasitizing Giardia species with different host ranges, may be able to be used as a model system for the study of evolutionary divergence of host parasitism strategies in Giardia.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2786-8) contains supplementary material, which is available to authorized users.
Keywords: Giardia cricetidarum n. sp., Host range and specificity, Morphology, Phylogenetic analysis
Background
Since Antony van Leeuwenhoek first observed and recorded Diplomonadida in 1681, the flagellated protozoan intestinal parasite, Giardia Künstler, 1882, has attracted the attention of scientists for many reasons in the past 300 years [1]. Giardia spp. are the most common intestinal protozoan parasites of humans and many other vertebrates worldwide; infection can cause giardiasis, the symptoms of which include acute or chronic diarrhea, nausea and weight loss. Previously up to 51 species of Giardia were incorrectly described on the basis of assumed host specificity [2]. Currently seven species of Giardia are considered valid: G. agilis Künstler, 1882 in amphibians, G. ardeae Filice, 1952 and G. psittaci Filice, 1952 in birds, G. microti Filice, 1952 and G. muris Filice, 1952 in rodents, G. intestinalis Lambl, 1859 (syns. G. duodenalis and G. lamblia) in most vertebrates including human beings [3], and the most recently described species, G. peramelis Hillman, 2016, in Australian bandicoots (Isoodon obesulus Driessen) [4].
The taxonomy of Giardia spp. is usually based on morphological characters of the trophozoites and cysts [3]. The shape and size of the trophozoites and cysts are the most common characters used for describing and identifying Giardia spp. Today, sequence analysis at multiple loci is essential for the identification and classification of Giardia, and the following genes are most widely used: the glutamate dehydrogenase, beta-giardin, elongation factor-1 alpha, triose phosphate isomerase and small subunit rRNA (SSU rRNA) [5]. Within G. intestinalis, genetic analysis has identified eight assemblages, A-H: assemblages A and B in humans and other mammals; assemblages C and D in dogs and other canids; assemblage E mainly in ungulates, assemblage F mainly in cats; assemblage G in rats and mice; and assemblage H in marine mammals [6, 7].
In the present study, morphological and molecular characterisation and experimental cross-transmission studies were used to describe a new species, G. cricetidarum n. sp. in hamsters.
Methods
Collection and purification of trophozoites and cysts
Giardia trophozoites were collected from hamsters (Phodopus sungorus, Kunming, China) which were bought from pet markets. The hamsters were anesthetized to death with chloroform and then their intestines were removed and cut into 0.5 cm segments. Segments of intestines were collected in 2 ml centrifuge tubes with normal saline. The centrifuge tubes were chilled in ice for 20 min. The suspension was briefly centrifuged at 1,000× g for 1 s to remove the large fragments. The supernatant was removed into new centrifuge tubes and then was centrifuged at 500× g for 5 min to concentrate the trophozoites. The pellet was resuspended with TYI-S-33 medium and cultivated at 37 °C for 3 h [8]. Then the medium was replaced with fresh medium when the majority of G. cricetidarum cells had adhered to the wall of the centrifuge tube [9]. The medium was replaced with phosphate buffered saline (PBS) buffer and the trophozoites were cultivated at 37 °C for 2 h. The centrifuge tubes were chilled in ice for 20 min and centrifuged at 10,000× g for 2 min, and the supernatant was removed to collect the trophozoites, which were frozen at -20 °C for later use. Cysts were obtained from the feces of infected hamsters and prepared for morphometric and host range studies by re-suspending in water and filtering the suspension through a 60 mesh sieve and centrifugation at 1500× g for 10 min. A 33% zinc sulfate solution with the same amount of sediment was added to resuspend the sediment, and centrifuged at 1200× g for 15 min. The supernatant was diluted with 4 volumes of water and centrifuged again at 1500× g for 10 min. Then the last two steps were repeated twice to further purify the cysts.
Host range and experimental cross-transmission studies
The host range was examined by screening feces and intestinal contents from a range of vertebrate animals including frogs (wild), parrots (domestic), mice (domestic and wild), small oriental voles (wild) and hamsters (domestic), from different areas or markets, with at least 10 individuals examined for each species. These animals were anesthetized to death with chloroform and their intestines were removed and cut into 0.5 cm segments. The segments of intestines were collected in 2 ml centrifuge tubes with normal saline and were chilled in ice for 20 min. The suspension was briefly centrifuged at 1000× g for 1 s to remove the large fragments. The supernatant was removed into new centrifuge tubes and centrifuged at 500× g for 5 min to concentrate Giardia. The sediment was resuspended with normal saline and then examined under a bright field microscope. Cross-transmission studies were performed by adding ~106 purified cysts of G. cricetidarum n. sp. to 80 ml of the drinking water for Rattus norvegicus, Mus musculus and Oryctolagus cuniculus f. domesticus (n = 10 for each species), after first confirming that their feces was negative for Giardia spp. by microscopy. After two weeks, the animals were examined for infection by visual identification of intestines and feces according to Farthing et al. [10]. Ten Giardia-uninfected hamsters were also infected in the same way and used as controls. All the animals were housed in different cages.
Microscopy
For haematoxylin and eosin staining, PBS solution containing G. cricetidarum trophozoites was dried on a glass slide; the glass slide was placed into Carnoy's fluid for 15 min and washed with PBS buffer for 5 min. Then the glass slide was processed through the following steps: stained with haematoxylin for 30 min and then rinsed with tap water; dipped in hydrochloric-alcohol solution (saturated hydrochloric acid diluted in 70% alcohol, with a final concentration of 1% hydrochloric acid) for 7 s and then rinsed with tap water; dipped in 2% ammonium hydroxide for 10 s and then rinsed with tap water; stained with 0.5% eosin (acetic acid in eosin solution with a final concentration of 2% acetic acid) for 10 min and washed by water; dehydrated through an ascending ethanol series (70%, 80%, 90%, 95% and 100%); cleared in dimethylbenzene and mounted in neutral balsam.
Slides of trophozoites and cysts were prepared as described above. All slides were examined under oil immersion by a 100× HCX PL APO objective on a Leica DM2500 microscope (Leica, Wetzlar, Germany). The images were captured by a Leica DFC450 C digital camera. The length and width of the trophozoites (n = 20 per sample) and cysts (n = 10 per sample) from 87 samples were measured. Sample handling and photomicrographs of scanning electron microscope were processed at the Kunming Medical University (Kunming, China) using HITACHI 3700N (Tokyo, Japan).
DNA extraction, PCR amplification and sequencing
Genomic DNA of the new species was extracted from purified trophozoites using the Qiagen QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Total fecal DNA was extracted from the host animals using the TIANamp Stool DNA Kit (Tiangen, Beijing, China). The primers specific to both ends of the sequences of the three genes, SSU rRNA, β-giardin and elongation factor-1 alpha, were designed according to the conserved sequences of these genes of G. intestinalis (50586 isolate) using PRIMER PREMIER program version 5.00 (Biosoft International) [11], and the expected product lengths of the SSU rRNA, β-giardin and elongation actor-1 alpha were 950 bp, 500 bp and 630 bp, respectively (see primer sequences in Additional file 1). The primers were used to amplify the sequences of the three genes from all Giardia samples by PCR. The PCR reactions were set up in 25 μl containing 1× GC buffer, 0.08 mM dNTP, 1 μM of each primer, 0.25 μl (0.5U) LA-Taq DNA polymerase (Takara, Tokyo, Japan) and 0.5–1 μl of cell sample or DNA. Thermocycling conditions were as follows: 94 °C for 2 min followed by 32 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 30 s, followed by 72 °C for 10 min.
The PCR products were purified using the Wizard SV Gel and PCR clean-up system kit (Qiagen), and cloned into pMD-19T vectors using TaKaRa pMD-19T VectorCloning Kit (TaKaRa, Tokyo, Japan). The ligation products were transformed into DH5α chemically competent E. coli. Colony PCR was carried out with the vector-specific primers provided in the kit, and colonies were selected and Sanger-sequenced using vector-specific forward and reverse primers by Sangon Biotech (Shanghai, China).
Molecular phylogenetic analysis
In order to perform the phylogentic analysis, we sequenced, identified and retrieved the elongation factor-1 alpha of G. cricetidarum n. sp. (GenBank: MG733773), G. intestinalis strain ATCC 50803 (GenBank: KX131163), G. intestinalis strain Ad-23 (GenBank: AF069572), G. intestinalis strain P-15 (GenBank: AF069571), G. psittaci (GenBank: AB714979), G. ardeae (GenBank: AF069567), G. muris (GenBank: AF069566), Spironucleus vortens (GenBank: U94406); SSU rRNA of G. cricetidarum n. sp. (GenBank: MF185957), G. muris (GenBank: MF185956), G. microti (GenBank: MF185958), Spironucleus sp. (GenBank: FM897198), S. barkhanu (GenBank: DQ186590), G. psittaci (GenBank: AF473853.1), G. muris (GenBank: X65063 S53320), G. ardeae (GenBank: Z17210 S53313), G. microti (GenBank: AF006677), G. intestinalis(dog) (GenBank: AF199449), G. intestinalis Assemblage A isolate WB GL50803 r0019 (GenBank: M54878 M19); and β-giardin sequences of G. cricetidarum n. sp. (GenBank: MF185953), G. agilis (GenBank: MF185954), G. microti (GenBank:MF185955), G. intestinalis Assemblage_A (GenBank: KJ363393), G. intestinalis Assemblage_B (GenBank: KJ363389), G. intestinalis Assemblage_D (GenBank: KJ027418), G. intestinalis Assemblage_E (GenBank: KJ363399), G. intestinalis Assemblage_F (GenBank: KJ027424), G. muris (GenBank: EF455599), G. muris (GenBank: AY258618), G. psittaci (GenBank: AB714977).
The number of isolates of G. cricetidarum n. sp. for each locus sequenced is as follows: 8 for SSU rRNA; 12 for β-giardin; and 9 for elongation factor-1 alpha. The other sequences used in this work were all retrieved from the GenBank database (see accession numbers in Additional file 2: Table S1).
The maximum likelihood phylogenetic trees based on SSU rRNA, β-giardin, and elongation factor-1 alpha DNA sequences were reconstructed by using the PhyML software [12]. The sequence data of the three loci are from 7, 12 and 7 Giardia species or isolates; and two SSU rRNA sequences for Spironucleus spp. and one elongation factor-1 alpha sequence for Spironucleus vortens were used as outgroups. Multiple sequence alignments were performed with ClustalW 2.0 program [13], and the alignments were visually inspected to eliminate poorly aligned positions. The best-fit DNA model used for reconstructing the maximum likelihood phylogeny was selected by the JModelTest software [14, 15]. Trees were constructed using the PhyML 3.0 based on 9 taxa with 437 nt positions for SSU rRNA gene, 12 taxa with 284 nt positions for β-giardin gene, and 8 taxa with 550 nt positions for elongation factor-1 alpha gene. Tree reliability was determined by using bootstrap analyses with 1000 replicates.
Results
Order Diplomonadida Wenyon, 1926
Family Hexamitidae Kent, 1881
Genus Giardia Künstler, 1882
Giardia cricetidarum n. sp.
Type-host: Phodopus sungorus Pallas (Rodentia: Cricetidae), Dzhunfarian hamster.
Other natural hosts: Phodopus campbelli and Mesocricetus auratus
Type-locality: Kunming, Yunnan, China.
Type-specimens: Frozen G. cricetidarum trophozoites were deposited at the Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences, Kunming, Yunnan, China. Hapantotype permanent mounts and photomicrographs were deposited in the Kunming Natural History Museum of Zoology and Kunming Institute of Zoology under accession numbers KIZ170814 and KIZ170925.
Site of infection: Intestine.
Prepatent period: 7–14 days.
Patent period: Unknown.
Representative DNA sequences: Partial sequences of SSU rRNA, beta-giardin and elongation factor-1 alpha genes were submitted to the GenBank database under the accession numbers MF185957, MF185953 and MG733773, respectively.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [16], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid: zoobank.org:pub:F2CBD145-DE59-4C68-A333-8DE022F86892. The LSID for the new name Giardia cricetidarum is urn:lsid:zoobank.org:act:C3001577-97B2-4E05-A277-302A39CC6EAC.
Etymology: The new species is named the Cricetinae Fischer (hamsters), a subfamily of the Cricetidae Fischer.
Description
Trophozoite. [Based on scanning electron microscope micrographs from 36 samples; Fig. 1d and e.] Trophozoites stout, pyriform, 12–18 (14 ± 1.37) μm long (n = 720), 8–12 (10 ± 1.02) μm wide (n = 720). Ventral disc 7–8 (8 ± 0.53) μm long (n = 50), 6–8 (8 ± 0.67) μm wide (n = 50). Two nuclei, 2–3 (3) μm long (n = 10), 1–3 (2) μm wide (n = 10). Flagella 4 pairs, equal in length, 12–16 (15) μm give range for flagellum length.
Fig. 1.
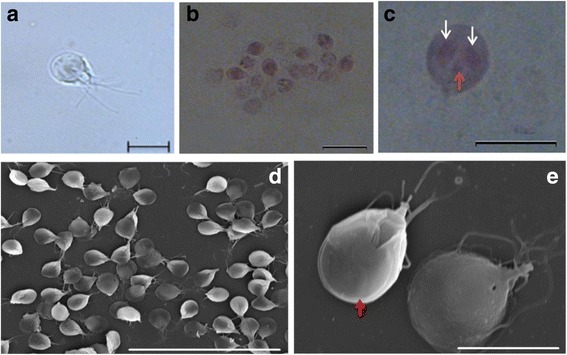
Trophozoites of Giardia cricetidarum n. sp. a Photomicrographs under bright-field microscope. b Stained trophozoites. c A stained trophozoite of G. cricetidarum (white arrows point to the nucleus and red arrow points to the median body). d Trophozoites under scanning electron microscope. e Trophozoites under scanning electron microscope, at high magnification (red arrow points the ventral disk). Scale-bars: a, c, e, 10 μm; b, 20 μm; d, 70 μm
Cyst. [Based on photomicrographs of bright field microscope from 36 samples; Fig. 2.] Cysts ovoidal, 9–12 (11 ± 0.96) μm long (n = 360), 8–10 (10 ± 0.60) μm wide (n = 360).
Fig. 2.
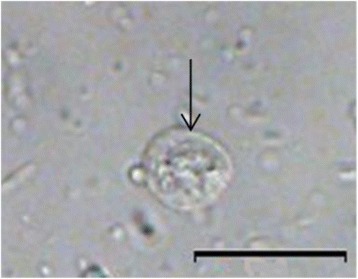
Cyst of Giardia cricetidarum (arrowed). Scale-bar: 20 μm
Remarks
Giardia cricetidarum n. sp. is most similar to G. muris on the basis of the morphology of the trophozoites shape and molecular data. The new species differs from the other Giardia spp. in: (i) its trophozoites are generally larger and stouter than most of the other Giardia species (Fig. 1) and exhibit the lowest length/width ratio (~1.40) among all the described Giardia species (Table 1); (ii) it is usually detected in high numbers with high density and all individuals were infected in one infected population; (iii) it was only found in hamsters yet. However, the cysts of G. cricetidarum were ovoidal and measured 9–12 (11 ± 0.96) μm long and 8–10 (10 ± 0.60) μm wide (Fig. 2), which are similar to other Giardia spp. such as G. intestinalis, which are also ovoidal and measure 8–12 μm in length and 7–10 μm in width [17]. Thus, the cysts of G. cricetidarum are morphologically indistinguishable from those of other Giardia species.
Table 1.
Trophozoite size for Giardia cricetidarum and other Giardia spp
| Species | Length (μm) | Width (μm) | Length/width ratio |
|---|---|---|---|
| G. cricetidarum n. sp. | 12–18 | 8–12 | 1.40 |
| G. intestinalis | 12–15 | 6–8 | 1.79 |
| G. muris | 9–12 | 5–7 | 1.75 |
| G. microti | 12–15 | 6–8 | 1.79 |
| G. agilis | 20–30 | 4–5 | 5.56 |
| G. ardeae | c.10 | c.6.5 | 1.54 |
| G. psittaci | c.14 | c.6 | 2.33 |
Note: The data for Giardia spp. are from Lujan et al. [3]
The host range of G. cricetidarum
During our investigation of the host range of G. cricetidarum, we detected G. intestinalis in Rattus norvegicus (Berkenhout) and Phodopus sungorus (Pallas), G. microti in Eothenomys melanogaster (Milne-Edwards), G. muris in R. norvegicus, P. sungorus, and G. agilis in Babina pleuraden (Boulenger), but G. cricetidarum n. sp. was only found in hamsters, i.e. P. sungorus, P. campbelli (Thomas) and Mesocricetus auratus (Waterhouse). The detailed results are shown in Table 2. Our cross-transmission studies showed that the rodents including R. norvegicus and M. musculus and lagomorphs including Oryctolagus cuniculus f. domesticus did not produce detectable infections with G. cricetidarum n. sp. while all of the control hamsters were easily infected. This suggests that G. cricetidarum n. sp. may specifically parasitize hamsters and has the narrowest host range within all the known mammal-parasitizing Giardia spp. However, further research is required to confirm this. No significant loss in weight and no intestinal lesions of hamsters infected with G. cricetidarum n. sp. were found.
Table 2.
Giardia spp. identified in the host range investigation of G. cricetidarum n. sp
| Host species | No. examined | Giardia species | Prevalence (%) n infected/n examined |
|---|---|---|---|
| Babina pleuraden | 62 | G. agilis | 100 (62/62) |
| Rana chaochiaoensis | 34 | G. agilis | 65 (22/34) |
| Rattus norvegicus | 23 | G. muris | 17 (4/23) |
| G. intestinalis | 4 (1/23) | ||
| Eothenomys melanogaster | 7 | G. microti | 100 (7/7) |
| Phodopus sungorus | 87 | G. cricetidarum n. sp. | 41 (36/87) |
| G. intestinalis | 7 (6/87) | ||
| G. muris | 3 (3/87) | ||
| Phodopus campbelli | 9 | G. cricetidarum n. sp. | 100 (9/9) |
| Mesocricetus auratus | 11 | G. cricetidarum n. sp. | 100 (11/11) |
Comparison of the prevalence features between G. cricetidarum and the other mammal-parasitizing Giardia
Our investigation revealed an interesting distribution feature of G. cricetidarum n. sp. in feeding populations of hamsters (bought from pet markets); for a certain feeding population, either all of the individuals or none of them were infected by G. cricetidarum, exhibiting an “all or none” infection pattern. Two of the ten hamster populations examined were infected, thus the infection rate per population was 20% [overall prevalence of 52% (56/107)]. Interestingly the numbers of G. cricetidarum trophozoites in all infected hamsters examined were high (5 × 105–5 × 106). This “all or none” prevalence has seldom been documented for the three other mammal-parasitizing Giardia species in their corresponding hosts [3]. In the present study, some hamsters co-infected with G. intestinalis and G. muris were identified but the infection rates of G. intestinalis and G. muris in all of the hamsters examined were only 6% (6 out of 107) and 3% (3 out of 107), respectively. In addition, hamsters infected with G. intestinalis and G. muris shed much lower numbers of trophozoites (less than 104) and of the 36 G. cricetidarum-infected hamsters, only one and two individuals were co-infected with G. intestinalis and G. muris, respectively. This suggests that hamsters are the most suitable hosts for G. cricetidarum n. sp. rather than other mammals.
Genetic distinction between G. cricetidarum n. sp. and other known Giardia species
Genetic distances between Giardia species at the three loci (Tables 3, 4 and 5) indicate that the genetic distances between G. cricetidarum n. sp. and all other Giardia species are equal or greater than the differences between currently accepted Giardia species. Although G. cricetidarum n. sp. is most genetically close to G. muris, it has a significant genetic distance with all of the other Giardia spp., including those that can parasitize the same host hamster.
Table 3.
Genetic distances between Giardia spp. for the SSU rRNA locus
| Species | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| 1 | G. intestinalis | ||||||
| 2 | G. psittaci | 0.546 | |||||
| 3 | G. ardeae | 0.107 | 0.575 | ||||
| 4 | G. microti | 0.017 | 0.556 | 0.117 | |||
| 5 | G. muris | 0.183 | 0.559 | 0.158 | 0.196 | ||
| 6 | G. peramelis | 0.581 | 0.592 | 0.582 | 0.574 | 0.603 | |
| 7 | G. cricetidarum n. sp. | 0.197 | 0.575 | 0.152 | 0.210 | 0.098 | 0.595 |
Table 4.
Genetic distances between Giardia spp. for the β-giardin locus
| Species | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 1 | G. intestinalis | |||||
| 2 | G. psittaci | 0.139 | ||||
| 3 | G. agilis | 0.087 | 0.128 | |||
| 4 | G. microti | 0.100 | 0.129 | 0.012 | ||
| 5 | G. muris | 0.108 | 0.170 | 0.099 | 0.113 | |
| 6 | G. cricetidarum n. sp. | 0.104 | 0.152 | 0.105 | 0.113 | 0.033 |
Table 5.
Genetic distances between Giardia spp. for the elongation factor-1 alpha locus
| Species | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 1 | G. intestinalis | ||||
| 2 | G. ardeae | 0.089 | |||
| 3 | G. muris | 0.121 | 0.086 | ||
| 4 | G. psittaci | 0.110 | 0.124 | 0.151 | |
| 5 | G. cricetidarum n. sp. | 0.111 | 0.076 | 0.072 | 0.137 |
Phylogenetic analysis of a fragment of the SSU rRNA (Fig. 3), the β-giardin (Fig. 4) and the elongation factor-1 alpha (Fig. 5) loci showed that although G. cricetidarum is genetically most closely related to G. muris, it is genetically distinct from all other Giardia species, including those that can parasitize hamsters.
Fig. 3.
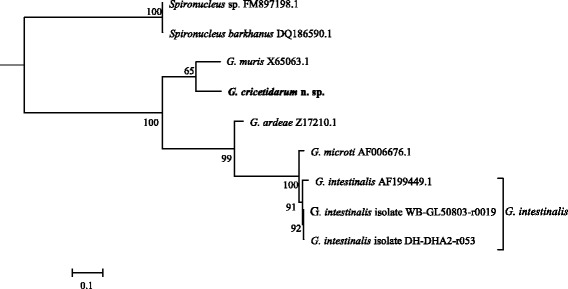
Phylogenetic tree from ML based on SSU rRNA sequences indicating that Giardia cricetidarum n. sp. is distinct from all Giardia spp. currently considered valid. Spironucleus spp. were used as the outgroup
Fig. 4.
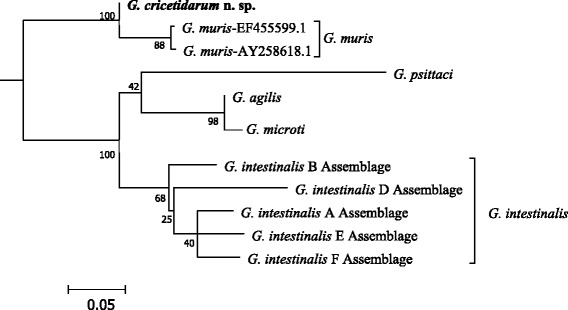
Phylogenetic tree from ML based on β-giardin sequences indicating that Giardia cricetidarum n. sp. is distinct from all Giardia spp. currently considered valid
Fig. 5.
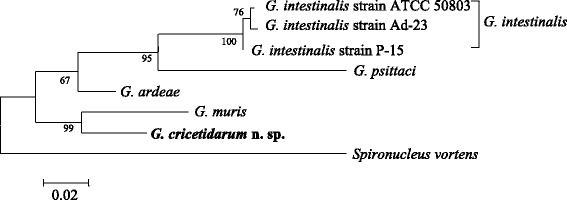
Phylogenetic tree from ML based on elongation factor-1 alpha sequences indicating that G. cricetidarum n. sp. is distinct from all Giardia spp. currently considered valid. Spironucleus vortens was used as the outgroup
Discussion
In the present study, a new species of Giardia, G. cricetidarum n. sp. was described from hamsters. Cysts of G. cricetidarum are difficult to distinguish morphologically from the other described Giardia species, but the trophozoites of the new species are distinctly larger and stouter than those in all the other described Giardia spp., including the three mammal-parasitizing Giardia, i.e. G. muris, G. microti and G. intestinalis. The phylogenetic analyses based on partial sequences of the three loci (SSU rRNA, β-giardin, and elongation factor-1 alpha) all support the genetic distinctness of G. cricetidarum n. sp. Our investigations of the host range and cross-transmission studies both suggest that G. cricetidarum is a hamster-specific parasite; however further evidence is required to confirm this. Therefore, the combination of evidence based on biological and molecular data strongly supports that G. cricetidarum n. sp. is not only a new species of Giardia, but also a mammal-parasitizing Giardia with the narrowest host range found so far. Furthermore, we found several special prevalence features of this new species: (i) “all or none” prevalence pattern in hamster populations; (ii) heavy infections in the intestines of infected hamsters without observed clinical signs; and (iii) rare co-infections with other Giardia spp. in the common hamster. These features imply that G. cricetidarum n. sp. obviously has some advantages in parasitizing hamsters over other mammal-parasitizing Giardia. Therefore, the new species, together with the three other mammal-parasitizing Giardia spp. with wider host ranges, may be able to be used as a model system for the study of evolutionary differences in the host parasitism strategy of Giardia species.
Conclusion
A new species of Giardia, G. cricetidarum n. sp. is described. The identification of G. cricetidarum may not just add a new member to the ancient genus Giardia, but may benefit studies on parasitic adaptation and evolutionary differences in the host parasitism strategies of Giardia spp.
Additional files
The primer pairs designed for amplification of SSU rRNA, β-giardian and elongation factor-1 alpha. (PDF 16 kb)
Table S1. The SSU rRNA, β-giardian and elongation factor-1 alpha sequences retrieved from GenBank and used for molecular phylogenetic analyses. (PDF 71 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC), Grant No. 31401972 to JRS, Grant No. 31572256 and 31772452 to JFW, and Grant No. 31401973 to BC; the grant from Yunnan Province No. 2015FB181 to JRS; the grant from State Key Laboratory of Genetic Resources and Evolution, KIZ, CAS No. GREKF14-11 to YQ. The funding bodies played no role in the design of the study, collection, analysis, interpretation of the data and writing the manuscript.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files. The newly generated sequences are submitted in the GenBank database under the accession numbers MG733773, MF185956, MF185957, MF185958, MF185953, MF185954 and MF185955. Hapantotypes were submitted to Kunming Natural History Museum of Zoology and Kunming Institute of Zoology under the accession numbers KIZ170814 and KIZ170925.
Abbreviations
- SSU rRNA
small subunit ribosomal RNA
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
Authors' contributions
ZL carried out the experiments, collected and analyzed the data, and wrote the manuscript. JS carried out the experiments and collected the data. MX, QY, BC and YQ took part in the investigation of host range and partial data analysis. JW made a contribution to conception, design, and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval
We followed the guidelines of The Animal Care and Use Committee of the American Society of Mammalogists for the use of wildlife in our research [18]. All the experimental procedures and animal care were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences (PAOKIZ131215, 12/2013).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2786-8) contains supplementary material, which is available to authorized users.
Contributor Information
Zhangxia Lyu, Email: zhangxialv@outlook.com.
Jingru Shao, Email: shaojr@mail.kiz.ac.cn.
Min Xue, Email: xuemin@mail.kiz.ac.cn.
Qingqing Ye, Email: yeqingqing@mail.kiz.ac.cn.
Bing Chen, Email: chenbing@mail.kiz.ac.cn.
Yan Qin, Email: qinyan1024@163.com.
Jianfan Wen, Email: wenjf@mail.kiz.ac.cn.
References
- 1.Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14(3):447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filice FP. Studies on the cytology and life-history of a Giardia from a laboratory rat. Oakland: University of California Press; 1952. [Google Scholar]
- 3.Lujan HD, Svard S. (Editors) Giardia. A model organism. Wien: Springer-Verlag; 2011. [Google Scholar]
- 4.Hillman A, Ash A, Elliot A, Lymbery A, Perez C, Thompson RCA. Confirmation of a unique species of Giardia parasitic in the quenda (Isoodon obesulus) Int J Parasitol Parasites Wildl. 2016;5(1):110–115. doi: 10.1016/j.ijppaw.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol BiochemParasit. 2008;160(2):75. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Jerlström-Hultqvist J, Franzén O, Ankarklev J, Xu F, Nohýnková E, Andersson JO, Svärd SG, Andersson B. Genome analysis and comparative genomics of a Giardia intestinalis assemblage E isolate. BMC Genomics. 2010;11(1):543. doi: 10.1186/1471-2164-11-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasek-Nesselquist E, Welch DM, Sogin ML. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int J Parasitol. 2010;40(9):1063–1074. doi: 10.1016/j.ijpara.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keister DB. Axenic culture of Giardia lamblia, in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77(4):487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S, Frisardi M, Rogers R, Samuelson J. How Giardia swim and divide. Infect Immun. 2001;69(12):7866–7872. doi: 10.1128/IAI.69.12.7866-7872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farthing MJ, Mata L, Urrutia JJ. Natural history of Giardia infection of infants and children in rural Guatemala and its impact on physical growth. Am J Clin Nutr. 1986;43(3):395–405. doi: 10.1093/ajcn/43.3.395. [DOI] [PubMed] [Google Scholar]
- 11.Lalitha S. Primer Premier 5. Biot Soft Int Rep. 2004;1(6):270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 12.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W. Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Posada D. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 2009;537:93. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- 15.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–169. [Google Scholar]
- 17.Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol. 2010;8(6):413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- 18.Sikes RS. The Animal Care and Use Committee of the American Society of Mammalogists. 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal. 2016;97:663–688. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The primer pairs designed for amplification of SSU rRNA, β-giardian and elongation factor-1 alpha. (PDF 16 kb)
Table S1. The SSU rRNA, β-giardian and elongation factor-1 alpha sequences retrieved from GenBank and used for molecular phylogenetic analyses. (PDF 71 kb)
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional files. The newly generated sequences are submitted in the GenBank database under the accession numbers MG733773, MF185956, MF185957, MF185958, MF185953, MF185954 and MF185955. Hapantotypes were submitted to Kunming Natural History Museum of Zoology and Kunming Institute of Zoology under the accession numbers KIZ170814 and KIZ170925.


