Abstract
We previously mapped loci for the genome-wide association studies (GWAS) and genome-wide gene-by-alcohol dependence interaction (GW-GxAD) analyses of risky sexual behaviors (RSB). This study extends those findings by analyzing the ancestry- and sex-specific AD-stratified effects on RSB. We examined the concordance of findings for the AD-stratified GWAS and the GW-GxAD analysis of RSB, with concordance defined as genome-wide significance in one analysis and at least nominal significance in the second analysis. 2,173 African-American (AA) and 1,751 European-American (EA) subjects were investigated. Information regarding RSB (lifetime experiences of unprotected sex and multiple sexual partners) and DSM-IV diagnosis of lifetime AD were derived from the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). In our ancestry- and sex-specific analyses, we identified four independent genome-wide significant (GWS) loci (p<5*10−8) and one suggestive locus (p<6*10−8). In men, we observed a GWS signal in FAM162A (rs2002594, p=4.96*10−8). In women, there was a suggestive locus in PLGRKT (rs3824435, p=5.52*10−8). In AAs, there was a GWS signal in GRK5 (rs1316543, p=1.25*10−9). In AA men, we observed an intergenic GWS signal (rs12898370, p=4.49*10−8) near LINGO1. In EA men, there was a GWS signal in CCSER1 (rs62313897; p=7.93*10−10). The loci identified in this GWAS implicate molecular mechanisms related to psychiatric illness and personality features, suggesting that the interplay between AD and RSB is mediated by alleles associated with behavioral traits.
Keywords: alcohol use disorders, GWAS, gene-by-environment interaction, unprotected sex, multiple sexual partners
Introduction
Heavy drinking is a leading cause of morbidity and mortality in industrialized countries [Spanagel, 2009]. There are multiple adverse effects of chronic exposure to alcohol, including the development of alcohol use disorders [Polimanti et al., 2017c]. Heavy drinking is associated with negative behavioral traits such as risky sexual behaviors (RSB).For example, heavy drinking is associated with an increased likelihood of sexual activity [Thompson et al., 2014], with heavy drinkers being more likely to have multiple sexual partners in a year [Thomson Ross et al., 2014], and an elevated blood alcohol concentration is associated with a greater likelihood of engaging in unprotected sex [Davis et al., 2009]. “Alcohol myopia theory” describes the alcohol-induced behavioral changes: alcohol ingestion leads to a state of short-sightedness by reducing cognitive processing capacity [Sevincer and Oettingen, 2014].
Genome-wide association studies (GWAS) and gene-by-environment genome-wide interaction study (GEWIS) are powerful tools to identify molecular mechanisms of complex traits [Polimanti et al., 2017a]. A recent genome-wide association study of sexual behaviors reported a genetic correlation between reproductive and behavioral traits [Day et al., 2016] and a phenome-wide scan indicated that alcohol risk alleles are associated with reproductive health traits [Polimanti et al., 2016]. Our trans-population genome-wide gene-by-alcohol dependence (GW-GxAD) analysis of RSB identified LHPP as a risk locus, linking genetic risk and AD to RSB and sexually transmitted disease [Polimanti et al., 2017b]. To provide a deeper understanding of these findings, we investigated ancestry, sex, and ancestry-by-sex, because both ancestry and sex could contribute to specific risk mechanisms [Polimanti et al., 2015]. In particular, the investigation of cohorts including individuals of differing ancestry can provide better coverage of genetic factors associated with alcohol use behaviors in the greater general population, reducing the vexing ethnic disparity in human genetic research [Chartier et al., 2017a; Chartier et al., 2017b; Scott, 2017].
In the present study, we performed ancestry- and sex-specific GW-GxAD analyses and AD-stratified GWAS of RSB in a sample of 3,924 subjects. To define genome-wide significant (GWS) loci, we considered regions that were concordant for both AD-stratified GWAS and GW-GxAD analysis of RSB with a p-value < 5*10−8 (GWS threshold) in one analysis and at least nominal significance in the other analysis (p-value < 0.05). Using this approach, we identified four independent GWS loci (p<5*10−8) and a suggestive locus (p<6*10−8) that are consistent with both our prior GW-GxAD analysis and GWAS.
Methods and Materials
Subjects and Diagnostic Procedures
All subjects were phenotyped using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA; [Pierucci-Lagha et al., 2005]), which yields DSM-IV diagnoses of lifetime alcohol and drug dependence and other major psychiatric traits, along with additional phenotypic information. A total of 2,173 AAs and 1,751 EAs, recruited in the Eastern United States [Gelernter et al., 2014b], were included in the present analyses. The institutional review board at each participating site approved the study and we obtained written informed consent from each participant. RSBs and AD were determined using items from the SSADDA. Detailed information regarding the phenotypic definitions are available in our previous report [Polimanti et al., 2017b]. Briefly, RSBs were defined as a score based on lifetime experiences of unprotected sex and multiple sexual partners using questions from the SSADDA section on antisocial personality: I35B (‘Have you ever had sex with 10 different people within a single year?’) and I37 (‘Have you more than once had unprotected sex (without a condom) with someone you believed could give you a disease, or when you had a disease that could be spread that way?’). On the basis of these two questions, we calculated an RSB score that ranged from 0 to 2 based on the number of affirmative responses. A diagnosis of lifetime AD was defined by DSM-IV criteria. We included only subjects who reported having ever consumed more than 3 drinks in a 24-hour period (i.e., more than minimally alcohol exposed) and had ever had sexual intercourse with least 10 sexual partners (i.e., sexually experienced). The characteristics of the study population are reported in our previous trans-population GW-GxAD analysis of RSB [Polimanti et al., 2017b] (Supplemental Table 1).
Genotyping and Imputation
The samples were genotyped using two different arrays: The Illumina HumanOmni1-Quad v1.0 microarray containing 988,306 autosomal SNPs to genotype 2,660 subjects (1,581 AAs and 1,079 EAs); and the Illumina HumanCoreExome array, which contains over 550,000 markers split between common tagging and low-frequency variants, to genotype 1,264 subjects (592 AAs and 672 EAs). Principal component (PC) analysis was conducted based on each genotyping array and for each ancestry group (AAs and EAs) using Eigensoft [Shriner, 2011] and SNPs that were common to the GWAS datasets and HapMap panel (after pruning the genome-wide SNPs for linkage disequilibrium (LD) (r2) > 80%) to characterize the underlying genetic architecture of the samples. Detailed information about the pre-imputation quality control pipeline is available in our published AD GWAS [Gelernter et al., 2014a]. Imputation was performed using Impute2 software [Howie et al., 2011] and the 1,000 Genomes Phase 1 reference panel. After imputation quality control, SNPs with minor allele frequency (MAF) > 5% and high imputation quality (certainty > 0.9, info > 0.8) were used in the analyses.
Data analysis methods
We performed AD-stratified GWAS and GW-GxAD analysis using the R package GWAF to fit a generalized estimating equations (GEE) model to adjust for correlations among related individuals [Chen and Yang, 2010]. In the AD-stratified GWAS, we tested the association of the imputed allele dosage with RSB score considered as the phenotype, after adjustment for DSM-IV cocaine dependence (CD), opioid dependence (OD), and nicotine dependence (ND) diagnoses, age, and the first three ancestry PCs. In the GW-GxAD analysis, we tested the interaction between the imputed minor allele dosage and DSM-IV AD diagnosis for the RSB score, after adjusting for DSM-IV CD, OD, and ND diagnoses, age, and the first three ancestry PCs. The results of the two analyses were combined by meta-analysis using the program METAL [Willer et al., 2010]. Details on the stratification and sample size for each meta-analysis are reported in Supplemental Table 2. In all meta-analyses, we applied a genomic control correction to all input files that showed negligible inflation of meta-analyzed p-values was observed (Supplemental Table 2; Supplemental Figures 1). To identify promising genome-wide significant (GWS) regions, we selected only regions that were concordant based on both the AD-stratified GWAS and the GW-GxAD analysis of RSB, such that there was a GWS p-value in one analysis, and at least nominal significance in the other. We performed multiple genome-wide analyses and used different stratification strategies and to ascertain whether this increased the risk of false positives, we performed a permutation analysis. This was accomplished by performing the same genome-wide investigation (i.e. AD-stratified GWAS and GW-GxAD analysis) considering each of 10 simulated phenotypes generated by random permutation of RSB score, and evaluating whether the actual observed GW hits were significantly different from the distributions of results from the permuted phenotypes.
Based on the results of the GW-GxAD analyses in AAs and EAs, we performed a gene-based association analysis in each ancestry group using VEGAS2 software [Mishra and Macgregor, 2015]. Reference panels of 1,000 Genomes Project European samples and African samples were used to correct for LD patterns in EAs and AAs, respectively. The results of gene-based association analysis were used to perform a protein network-based analysis using the R package dmGWAS [Jia et al., 2011]. We defined protein-protein interactions (PPI) using the Protein Interaction Network Analysis platform (PINA) v2.0 [Cowley et al., 2012] and subsequently used the R package dmGWAS to identify PPI modules enriched with small p-values. We used both AAs and EAs to search for PPI modules enriched for small p-values, using the “dual-evaluation” strategy of dmGWAS. Specifically, we applied a dense module search in the EAs and a follow-up analysis in AAs. Finally, we used DAVID 6.7 [Huang da et al., 2009] to perform functional annotation clustering on the protein interactive network identified. High classification stringency and Bonferroni correction for multiple comparisons were applied in the DAVID analyses.
Results
In our ancestry- and sex-specific analyses, we identified four independent GWS loci (p<5*10−8) and one suggestive locus (p<6*10−8) for the trait RSB. Table 1 reports the details of GWS signals observed in the meta-analyses.
Table 1.
Summary of GWS and suggestive GWS results.
| Meta-analysis (GW analysis) | rsID | Chr | BP | Gene | AF | N | P value | Direction |
|---|---|---|---|---|---|---|---|---|
| AAs (GWAS) | rs1316543 | 10 | 121204660 | GRK5 | 0.932 | 1541 | 1.25E-09 | + |
| Men (GW-GxAD) | rs2002594 | 3 | 122111561 | FAM162A | 0.399 | 2653 | 4.96E-08 | + |
| Women (GWAS) | rs3824435 | 9 | 5377115 | PLGRKT | 0.532 | 898 | 5.52E-08 | − |
| AA men (GW-GxAD) | rs12898370 | 15 | 77808598 | NA | 0.872 | 1488 | 4.49E-08 | − |
| EA men (GW-GxAD) | rs62313897 | 4 | 91136741 | CCSER1 | 0.942 | 1165 | 7.93E-10 | − |
| rs78694949 | 4 | 91119554 | CCSER1 | 0.941 | 1165 | 8.90E-10 | − |
AA-specific meta-analysis
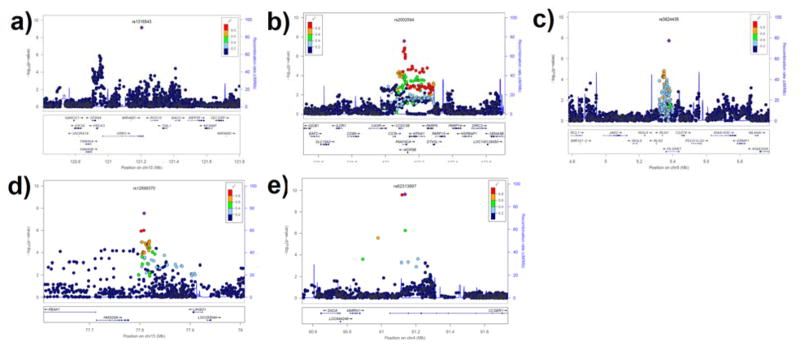
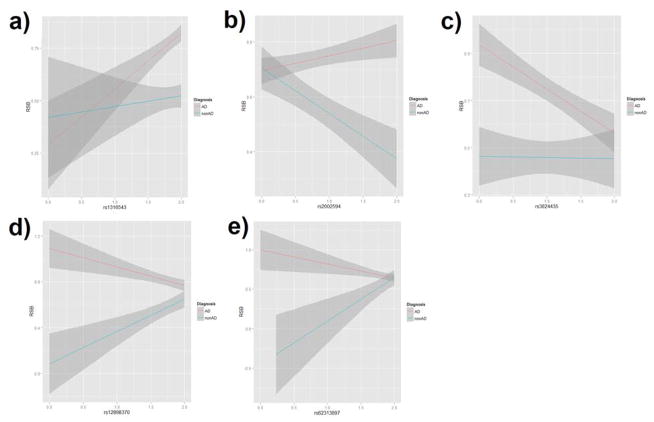
In the meta-analysis in AA subjects (including both males and females), there were three GWS signals in the AD-stratified GWAS of RSB (Supplemental Table 3). Rs1316543, located within GRK5, which was GWS in the AD-stratified GWAS of RSB (p=1.25* 10−9, Figure 1a), was nominally significant in the GW-GxAD analysis of RSB (p=0.042), with the rs1316543 imputed allele dosage showing greater association with RSB in AD subjects than non-AD subjects (Figure 2a). In EA-specific meta-analysis, no locus met the GWS criteria.
Figure 1.
Regional Manhattan Plots of the GWS and suggestive hits: a) GRK5 rs1316543 in AA meta-analysis of AD-stratified GWAS of RSB; b) FAM162A rs2002594 in male meta-analysis of GW-GxAD analyses of RSB; c) PLGRKT rs3824435 in female meta-analysis of AD-stratified GWAS of RSB; d); rs12898370 in AA-male meta-analysis of GW-GxAD analyses of RSB; and e) CCSER1 rs62313897 and rs78694949 in EA-male meta-analysis of GW-GxAD analyses of RSB.
Figure 2.
Relationship between imputed allele dosage of GWS hits and RSB in AD and nonAD subjects: a) GRK5 rs1316543 in AA samples; b) FAM162A rs2002594 in male samples; c) PLGRKT rs3824435 in female samples; d) rs12898370 in AA-male samples; and e) CCSER1 rs62313897 and rs78694949 in EA-male samples.
Male-specific meta-analysis
In the meta-analyses in men (including both EAs and AAs), we observed a GWS signal in the GW-GxAD analysis of RSB for rs2002594 in FAM162A (p=4.96*10−8, Figure 1b). This SNP was nominally significant in the AD-stratified GWAS (p=0.009), such that the imputed rs2002594 allele dosage was negatively associated with RSB in non-AD subjects and positively associated in AD subjects (Figure 2b).
Female-specific meta-analysis
In the AD-stratified GWAS of RSB in women, we observed a suggestive signal for rs3824435 in PLGRKT (p=5.52*10−8, Figure 1c), such that the imputed allele dosage for the SNP was negatively associated with RSB in AD women (including both AAs and EAs) (Figure 2c). This variant also showed nominal significance in the GW-GxAD analysis of RSB (p=0.017).
AA Male-specific meta-analysis
In the meta-analysis in AA men, rs12898370, located near LINGO1, was GWS in the GW-GxAD analysis (p=4.49*10−8; Figure 1d) and nominally significant in the AD-stratified GWAS (p=8.29*10−4). That is, the rs12898370 imputed allele dosage was positively associated with RSB in non-AD subjects and negatively associated in AD subjects (Figure 2d).
EA Male-specific meta-analysis
In the meta-analysis in EA men, there were several linked GWS signals in the GW-GxAD analysis (Supplemental Table 4). Two SNPs (rs62313897 and rs78694949) in high LD (r2>0.8) located in CCSER1 were GWS in the GW-GxAD analysis (p=7.93*10−10 and p=8.90*10−10; Figure 1e) and nominally significant in the AD-stratified GWAS (p=0.009). Specifically, the imputed allele dosages for both SNPs were positively associated with RSB in non-AD subjects and negatively associated in AD samples (Figure 2e).
Permutation analysis
The permuted analyses, performed to evaluate the possibility of false positives due to the multiple genome-wide investigations and stratification analyses, confirmed that our GW hits are significantly different from the result distributions of the phenotypes generated by random permutations (Supplemental Table 5).
Protein interactive network analysis
We used the results of the meta-analyses of GW-GxAD analysis of RSB in AAs and EAs to perform a gene-based association analysis (Supplemental Table 6, respectively). These gene-based associations were then used to implement a gene interactive network analysis. We used the dual evaluation approach in the R package dmGWAS, in which we considered EAs as the discovery dataset and AAs as the evaluation dataset. We tested the hypothesis that, although different SNPs and genes might be implicated in different populations, the underlying biological mechanisms are similar. Thirty-six PPI modules were significant after dual evaluation analysis (Supplemental Figure 2). Seventy-seven genes were included in this protein interactive network associated with the interplay between AD and RSB. Performing a term enrichment analysis, we observed two functional annotation clusters associated with terms related to nucleotide binding and organelle location (Table 2).
Table 2.
Gene ontology analysis of protein network associated with the interplay of AD and RSB
| Enrichment score | Term | Genes | Fold Enrichment | P-value | PBonferroni |
|---|---|---|---|---|---|
| 5.04 | GO:0070013~intracellular organelle lumen | RBL2, SMAD5, TP53, PRKDC, YTHDC1, CDC73, CDC27, PPARGC1B, CXXC1, AKT1, STAT4, PAPOLA, PTRF, ATG4C, SMARCB1, MAPK14, SMARCA5, UBC, TEP1, YAP1, NCOR1, CHD3, HSPA9 | 2.75 | 6.37E-06 | 1.04E-03 |
| GO:0043233~organelle lumen | 2.69 | 9.27E-06 | 1.51E-03 | ||
| GO:0031974~membrane-enclosed lumen | 2.64 | 1.28E-05 | 2.08E-03 | ||
| 4.51 | GO:0005524~ATP binding | EGFR, ITK, COASY, NLRP4, HSP90AA1, PFKFB2, TP53, PRKDC, PRKCE, AKT1, PDIK1L, PAPOLA, MAPK14, MAP3K8, PI4K2A, SMARCA5, TEP1, CHTF18, CDK14, CHD3, HSPA9 | 3.03 | 5.44E-06 | 1.28E-03 |
| GO:0032559~adenyl ribonucleotide binding | 2.99 | 6.68E-06 | 1.58E-03 | ||
| GO:0030554~adenyl nucleotide binding | 2.83 | 1.47E-05 | 3.46E-03 | ||
| GO:0001883~purine nucleoside binding | 2.79 | 1.84E-05 | 4.33E-03 | ||
| GO:0001882~nucleoside binding | 2.77 | 2.04E-05 | 4.79E-03 | ||
| GO:0032555~purine ribonucleotide binding | 2.43 | 1.33E-04 | 3.09E-02 | ||
| GO:0032553~ribonucleotide binding | 2.43 | 1.33E-04 | 3.09E-02 | ||
| GO:0017076~purine nucleotide binding | 2.33 | 2.44E-04 | 5.59E-02 |
Discussion
Meta-analyses based on sex, ancestry, and ancestry-by-sex stratifications yielded evidence for the effects of numerous genetic loci and AD, as well as the interaction of the two, on an ordinal measure of RSB. These findings contribute to our understanding of the contribution of ancestry and sex differences to the molecular mechanisms that underlie sexual behavior and its interplay with AD. Indeed, because RSB differs greatly by sex, it would be expected that different mechanisms and different risk loci could be involved in men and women. Because different population groups also have different risk loci and alleles for many genetically complex traits, the same may the case for RSB.
In meta-analyses in AA males and females, GRK5 rs1316543 showed GWS association with RSB in AA subjects with AD and nominal significance for the interaction with AD in relation to RSB. This intronic variant is involved in distal transcriptional regulation (ENCODE assay: Chromatin Interaction Paired-End Tags). GRK5 encodes a serine/threonine kinase that phosphorylates the activated forms of a variety of G-protein-coupled receptors [Baameur et al., 2010]. G protein-coupled receptor dysfunction has been thoroughly investigated with respect to neuropsychiatric disorders, and current pharmacological therapies target these proteins [Moreno et al., 2013]. A recent study that examined the activation systems of these proteins showed an increased level of GRK5 expression in schizophrenia [Funk et al., 2014].
In meta-analyses in males of both populations, the strongest evidence for association was at rs2002594, a SNP that showed a GWS interaction with AD in relation to RSB and a nominally significant association with RSB in AD men. This intronic variant is located at the FAM162A locus, and is potentially involved in distal transcriptional regulation (ENCODE assay: Chromatin Interaction Paired-End Tags) and RNA binding protein-mediated regulation (ENCODE assay: RNA IP Sequencing). FAM162A encodes a transmembrane protein that may be involved in hypoxia-induced neuronal cell death [Lee et al., 2004]. A previous mega-analysis of GWAS performed by the Psychiatric Genomics Consortium investigators showed a suggestive association (p=6*10−6) of this locus with major depressive disorder [Major Depressive Disorder Working Group of the Psychiatric et al., 2013]. PARP15 rs2173763 is 217 Kb from FAM162A rs2002594. Although the close mapping of PARP15 rs2173763 and FAM162A rs2002594 may be due to chance, we observed two GWS loci (i.e., LHPP rs34997829 [Polimanti et al., 2017b] and FAM162A rs2002594) that may link the mechanisms by which AD may result in both RSB and major depression disorder.
In meta-analyses in females of both populations, the strongest finding was a possible association (p<6*10−8) to RSB in AD women that was also nominally significant in the GxAD analysis. This finding was for rs3824435, an intronic variant located within the PLGRKT locus. ENCODE data support the regulatory function of this variant (RNA binding protein mediated regulation; ENCODE assay: RNA IP Sequencing). PLGRKT encodes a receptor for plasminogen that regulates several neuroendocrine processes [Lighvani et al., 2011]. Investigating previous GWAS findings in this chromosomal region, we found a GWS finding that was 2.5 Mb from the PLGRKT SNP rs3824435, related to “openness to experience” (assessed using the Korean short version of the original NEO-PI-R, a 90-item measure of the five factors of personality [Costa et al., 1992]) in young women [Kim et al., 2013].
In meta-analyses in AA men, we observed a GWS finding for rs12898370, an intergenic variant involved in distal transcriptional regulation (ENCODE assay: Chromatin Interaction Paired-End Tags) in the GW-GxAD analysis that was also nominally significant in the AD-stratified GWAS. Variants located in LINGO1 (97 Kb from rs12898370) are in LD with rs12898370 (r2>0.2). LINGO1 encodes a functional component of the Nogo receptor signaling complex in RhoA activation responsible for inhibition of axonal regeneration by myelin-associated factors [Kwon et al., 2014]. A recent study observed altered Lingo-1 signaling in schizophrenia brain [Fernandez-Enright et al., 2014]. Both of the findings related to meta-analyses in AAs of both sexes and AA men highlight loci also reportedly involved in the pathogenesis of schizophrenia. Furthermore, although those alleles are present in both AAs and EAs (MAF > 5%), we observed significant associations in the African-descent cohorts only. As also reported in other GWAS of substance use disorders [Smith et al., 2017], this may be due to the presence of epistatic interactions specific to African genomic background [Iorio et al., 2017; Karaca et al., 2016; Polimanti et al., 2015].
In the meta-analysis of EA males, a strong GWS finding was present in the GW-GxAD analysis that was also nominally significant in the AD-stratified GWAS: SNPs rs62313897 and rs78694949 are intronic variants mapped to the CCSER1 gene. The strongest signal, rs62313897, is involved in distal transcriptional regulation (ENCODE assay: Chromatin Interaction Paired-End Tags). No functional mechanism is apparent for rs78694949. CCSER1 encodes a protein with unknown function. However, multiple GWAS of brain-related phenotypes have identified this locus as a suggestive candidate for multiple traits. A GWAS and a replication study supported the association of CCSER1 variants with attention deficit hyperactivity disorder, and verified its expression in cerebellum [Lantieri et al., 2010; Neale et al., 2008]. Two independent GWAS identified CCSER1 variants as suggestive loci associated to d-amphetamine response and monoamine metabolite levels in human cerebrospinal fluid [Hart et al., 2012; Luykx et al., 2014].
Some of the identified loci are involved in biological processes potentially related to RSB. LHPP [Polimanti et al., 2017b], FAM162A (males), GRK5 (AAs), and LINGO1 (AA males) were previously implicated in major depressive disorder and schizophrenia. There is a consistent literature showing that RSBs are more frequent among psychiatric patients than in the general population [Ramrakha et al., 2000; Tull and Gratz, 2013]. Accordingly, our findings suggest that AD shares with other psychiatric disorders some of the molecular processes responsible for RSB. In females, we observed a possible genetic convergence between RSB and “openness to experience,” which are correlated traits: “openness to experience” has been positively associated with sexual experience, liberal attitudes toward sex, and sexual drive [Brown et al., 1996]. This evidence and our results indicate that the identified female-specific locus may contribute to both of these traits.
To investigate further the genetics of the interplay between RSB and AD, we considered protein network information in relation to the GW-GxAD analysis. This approach can be useful to identify molecular mechanisms related to loci associated to alcohol drinking behaviors [Polimanti and Gelernter, 2017]. Because different ancestry groups can show different risk alleles for a given trait but often nevertheless present the same genetic architecture, we used the GW-GxAD meta-analysis in EAs as the discovery sample and the GW-GxAD meta-analysis in AAs for replication. Our investigation identified a protein interactive network comprised of 77 genes. Enrichment analysis indicated that two large functional annotation clusters related to intra-cellular lumen and ATP/purine-nucleotide binding are included in the protein network. Although these features are shared by numerous mechanisms, the purinergic regulation of hypothalamo-pituitary functions is an intriguing possibility in relation to the AD-RSB interplay. The hypothalamo-pituitary system is strongly involved in the hormonal regulation of several behaviors, including sexual behavior, and ATP and its metabolic products are involved in its regulation by activating adenosine and/or purinergic receptors [Stojilkovic, 2009]. Alcohol-related disinhibition has also been related to hypothalamic neurons [Spanagel, 2009].
In conclusion, we performed ancestry- and sex-specific genome-wide analyses of RSB and the interplay of RSB with AD, which identified several loci. Our findings indicate that different mechanisms related to mental illness, personality features and other brain mechanisms may underlie the interplay. Further ancestry-specific and sex-specific genome-wide studies in larger samples are needed to replicate our findings and identify other loci. This may require new sample collection, as it is fairly uncommon for both of these kinds of data to be available simultaneously. Additionally, our results support that future studies should be specifically designed to take ancestry-specific and sex-specific genetic associations into account. The use of newer genotyping arrays with better representation of different population groups and updated imputation reference panels can permit us to improve the coverage in ethnically diverse samples significantly, increasing the statistical power of future multi-ancestry analysis [Bien et al., 2016]. The inclusion of sex chromosomes in GWAS is highly desirable but not straightforward; it requires the application of specific analytic approaches to overcome complications in genotype calling, imputation, and selection of test statistics. Addressing these issues can result in a more consistent amount of information for those traits with sex-specific features [Konig et al., 2014; Powers et al., 2017; Wise et al., 2013]. The current findings may provide the first insight regarding how ancestry and sex differences modulate the link between psychiatric disorders and the predisposition to RSB and their consequences, such as sexually transmitted disease [Polimanti et al., 2017b; Wang et al., 2016]. if confirmed, they may have implications for designing personalized treatments to minimize the harm associated with RSB in AD subjects.
Supplementary Material
Q-Q plots of meta-analyses with GW or suggestive GWS results.
Protein network associated with the interplay between AD and RSB. The grey color gradient of a node is proportional to its p-value.
Acknowledgments
This study was supported by National Institutes of Health grants R01 DA12690, R01 AA017535, P50 AA012870, the Connecticut and Crescenz VA MIRECCs, and a NARSAD Young Investigator Award (to RP) from the Brain & Behavior Research Foundation. We appreciate the work in recruitment and assessment provided at Yale University School of Medicine and the APT Foundation by James Poling, PhD; at McLean Hospital by Roger Weiss, M.D., at the Medical University of South Carolina by Kathleen Brady, MD, PhD, and at the University of Pennsylvania by David Oslin, MD. Genotyping services for a part of our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University Center for Genome Analysis. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Catherine Aldi and Christa Robinson for their excellent technical assistance, to the SSADDA interviewers, led by Yaira Nunez and Michelle Slivinsky, who devoted substantial time and effort to phenotype the study sample and to John Farrell and Alexan Mardigan for database management assistance. Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Indivior and Lundbeck. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, and XenoPort. The other authors reported no biomedical financial interests or potential conflicts of interest.
References
- Baameur F, Morgan DH, Yao H, Tran TM, Hammitt RA, Sabui S, McMurray JS, Lichtarge O, Clark RB. Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol Pharmacol. 2010;77(3):405–415. doi: 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien SA, Wojcik GL, Zubair N, Gignoux CR, Martin AR, Kocarnik JM, Martin LW, Buyske S, Haessler J, Walker RW, Cheng I, Graff M, Xia L, Franceschini N, Matise T, James R, Hindorff L, Le Marchand L, North KE, Haiman CA, Peters U, Loos RJ, Kooperberg CL, Bustamante CD, Kenny EE, Carlson CS, Study P. Strategies for Enriching Variant Coverage in Candidate Disease Loci on a Multiethnic Genotyping Array. PLoS One. 2016;11(12):e0167758. doi: 10.1371/journal.pone.0167758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GR, Wise TN, Costa PT, Jr, Herbst JH, Fagan PJ, Schmidt CW., Jr Personality characteristics and sexual functioning of 188 cross-dressing men. J Nerv Ment Dis. 1996;184(5):265–273. doi: 10.1097/00005053-199605000-00001. [DOI] [PubMed] [Google Scholar]
- Chartier KG, Hesselbrock MN, Hesselbrock VM. Conclusion: Special issue on genetic and alcohol use disorder research with diverse racial/ethnic groups: Key findings and potential next steps. Am J Addict. 2017a;26(5):532–537. doi: 10.1111/ajad.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier KG, Hesselbrock MN, Hesselbrock VM. Introduction: Special issue on genetic research of alcohol use disorder in diverse racial/ethnic populations. Am J Addict. 2017b;26(5):422–423. doi: 10.1111/ajad.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26(4):580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, McCrae RR Psychological Assessment Resources I. Revised NEO Personality Inventory (NEO PI-R) and NEO Five-Factor Inventory (NEO-FFI) Psychological Assessment Resources; 1992. [Google Scholar]
- Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40(Database issue):D862–865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KC, George WH, Norris J, Schacht RL, Stoner SA, Hendershot CS, Kajumulo KF. Effects of alcohol and blood alcohol concentration limb on sexual risk-taking intentions. J Stud Alcohol Drugs. 2009;70(4):499–507. doi: 10.15288/jsad.2009.70.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Helgason H, Chasman DI, Rose LM, Loh PR, Scott RA, Helgason A, Kong A, Masson G, Magnusson OT, Gudbjartsson D, Thorsteinsdottir U, Buring JE, Ridker PM, Sulem P, Stefansson K, Ong KK, Perry JR. Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet. 2016 doi: 10.1038/ng.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Enright F, Andrews JL, Newell KA, Pantelis C, Huang XF. Novel implications of Lingo-1 and its signaling partners in schizophrenia. Transl Psychiatry. 2014;4:e348. doi: 10.1038/tp.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk AJ, Haroutunian V, Meador-Woodruff JH, McCullumsmith RE. Increased G protein-coupled receptor kinase (GRK) expression in the anterior cingulate cortex in schizophrenia. Schizophr Res. 2014;159(1):130–135. doi: 10.1016/j.schres.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014a;19(1):41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014b;19(6):717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, Engelhardt BE, Wardle MC, Sokoloff G, Stephens M, de Wit H, Palmer AA. Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CDH13) PLoS One. 2012;7(8):e42646. doi: 10.1371/journal.pone.0042646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio A, De Angelis F, Di Girolamo M, Luigetti M, Pradotto LG, Mazzeo A, Frusconi S, My F, Manfellotto D, Fuciarelli M, Polimanti R. Population diversity of the genetically determined TTR expression in human tissues and its implications in TTR amyloidosis. BMC Genomics. 2017;18(1):254. doi: 10.1186/s12864-017-3646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27(1):95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca S, Karaca M, Civelek E, Ozgul RK, Sekerel BE, Polimanti R. Haplotype analysis of non-HLA immunogenetic loci in Turkish and worldwide populations. Gene. 2016;587(2):132–136. doi: 10.1016/j.gene.2016.04.050. [DOI] [PubMed] [Google Scholar]
- Kim HN, Roh SJ, Sung YA, Chung HW, Lee JY, Cho J, Shin H, Kim HL. Genome-wide association study of the five-factor model of personality in young Korean women. J Hum Genet. 2013;58(10):667–674. doi: 10.1038/jhg.2013.75. [DOI] [PubMed] [Google Scholar]
- Konig IR, Loley C, Erdmann J, Ziegler A. How to include chromosome X in your genome-wide association study. Genet Epidemiol. 2014;38(2):97–103. doi: 10.1002/gepi.21782. [DOI] [PubMed] [Google Scholar]
- Kwon HS, Nakaya N, Abu-Asab M, Kim HS, Tomarev SI. Myocilin is involved in NgR1/Lingo-1-mediated oligodendrocyte differentiation and myelination of the optic nerve. J Neurosci. 2014;34(16):5539–5551. doi: 10.1523/JNEUROSCI.4731-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantieri F, Glessner JT, Hakonarson H, Elia J, Devoto M. Analysis of GWAS top hits in ADHD suggests association to two polymorphisms located in genes expressed in the cerebellum. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(6):1127–1133. doi: 10.1002/ajmg.b.31110. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Kim JY, Suk K, Park JH. Identification of the hypoxia-inducible factor 1 alpha-responsive HGTD-P gene as a mediator in the mitochondrial apoptotic pathway. Mol Cell Biol. 2004;24(9):3918–3927. doi: 10.1128/MCB.24.9.3918-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani S, Baik N, Diggs JE, Khaldoyanidi S, Parmer RJ, Miles LA. Regulation of macrophage migration by a novel plasminogen receptor Plg-R KT. Blood. 2011;118(20):5622–5630. doi: 10.1182/blood-2011-03-344242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luykx JJ, Bakker SC, Lentjes E, Neeleman M, Strengman E, Mentink L, DeYoung J, de Jong S, Sul JH, Eskin E, van Eijk K, van Setten J, Buizer-Voskamp JE, Cantor RM, Lu A, van Amerongen M, van Dongen EP, Keijzers P, Kappen T, Borgdorff P, Bruins P, Derks EM, Kahn RS, Ophoff RA. Genome-wide association study of monoamine metabolite levels in human cerebrospinal fluid. Mol Psychiatry. 2014;19(2):228–234. doi: 10.1038/mp.2012.183. [DOI] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GC. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Muller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O’Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Volzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18(1):86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- Moreno JL, Holloway T, Gonzalez-Maeso J. G protein-coupled receptor heterocomplexes in neuropsychiatric disorders. Prog Mol Biol Transl Sci. 2013;117:187–205. doi: 10.1016/B978-0-12-386931-9.00008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, Vasquez AA, Asherson P, Chen W, Banaschewski T, Buitelaar J, Ebstein R, Gill M, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Sonuga-Barke E, Mulas F, Taylor E, Laird N, Lange C, Daly M, Faraone SV. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80(3):303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Gelernter J. ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am J Med Genet B Neuropsychiatr Genet. 2017 doi: 10.1002/ajmg.b.32523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Kaufman J, Zhao H, Kranzler HR, Ursano RJ, Kessler RC, Gelernter J, Stein MB. A genome-wide gene-by-trauma interaction study of alcohol misuse in two independent cohorts identifies PRKG1 as a risk locus. Mol Psychiatry. 2017a doi: 10.1038/mp.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Kranzler HR, Gelernter J. Phenome-wide association study for alcohol and nicotine risk alleles in 26,394 Women. Neuropsychopharmacology. 2016;41(11):2688–2696. doi: 10.1038/npp.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Wang Q, Meda SA, Patel KT, Pearlson GD, Zhao H, Farrer LA, Kranzler HR, Gelernter J. The Interplay Between Risky Sexual Behaviors and Alcohol Dependence: Genome-Wide Association and Neuroimaging Support for LHPP as a Risk Gene. Neuropsychopharmacology. 2017b;42(3):598–605. doi: 10.1038/npp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Yang C, Zhao H, Gelernter J. Dissecting ancestry genomic background in substance dependence genome-wide association studies. Pharmacogenomics. 2015;16(13):1487–1498. doi: 10.2217/pgs.15.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Zhang H, Smith AH, Zhao H, Farrer LA, Kranzler HR, Gelernter J. Genome-wide association study of body mass index in subjects with alcohol dependence. Addict Biol. 2017c;22(2):535–549. doi: 10.1111/adb.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MS, Smith PH, McKee SA, Ehringer MA. From sexless to sexy: Why it is time for human genetics to consider and report analyses of sex. Biol Sex Differ. 2017;8:15. doi: 10.1186/s13293-017-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrakha S, Caspi A, Dickson N, Moffitt TE, Paul C. Psychiatric disorders and risky sexual behaviour in young adulthood: cross sectional study in birth cohort. BMJ. 2000;321(7256):263–266. doi: 10.1136/bmj.321.7256.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS. Commentary: Perspectives on alcohol-related gene and environment interplay in diverse populations. Am J Addict. 2017;26(5):526–531. doi: 10.1111/ajad.12584. [DOI] [PubMed] [Google Scholar]
- Sevincer AT, Oettingen G. Alcohol myopia and goal commitment. Front Psychol. 2014;5:169. doi: 10.3389/fpsyg.2014.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D. Investigating population stratification and admixture using eigenanalysis of dense genotypes. Heredity (Edinb) 2011;107(5):413–420. doi: 10.1038/hdy.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Jensen KP, Li J, Nunez Y, Farrer LA, Hakonarson H, Cook-Sather SD, Kranzler HR, Gelernter J. Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Mol Psychiatry. 2017;22(3):346–352. doi: 10.1038/mp.2016.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89(2):649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Stojilkovic SS. Purinergic regulation of hypothalamopituitary functions. Trends Endocrinol Metab. 2009;20(9):460–468. doi: 10.1016/j.tem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RG, Jr, Eaton NR, Hu MC, Grant BF, Hasin DS. Regularly drinking alcohol before sex in the United States: effects of relationship status and alcohol use disorders. Drug Alcohol Depend. 2014;141:167–170. doi: 10.1016/j.drugalcdep.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson Ross L, Zeigler S, Kolak AM, Epstein D. Sexual Hookups and Alcohol Consumption Among African American and Caucasian College Students: A Pilot Study. J Psychol. 2014:1–19. doi: 10.1080/00223980.2014.946461. [DOI] [PubMed] [Google Scholar]
- Tull MT, Gratz KL. Major Depression and Risky Sexual Behavior among Substance Dependent Patients: The Moderating Roles of Distress Tolerance and Gender. Cognit Ther Res. 2013;37(3):483–497. doi: 10.1007/s10608-012-9490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Polimanti R, Kranzler HR, Farrer LA, Zhao H, Gelernter J. Genetic factor common to schizophrenia and HIV infection is associated with risky sexual behavior: antagonistic vs. synergistic pleiotropic SNPs enriched for distinctly different biological functions. Hum Genet. 2016 doi: 10.1007/s00439-016-1737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AL, Gyi L, Manolio TA. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am J Hum Genet. 2013;92(5):643–647. doi: 10.1016/j.ajhg.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Q-Q plots of meta-analyses with GW or suggestive GWS results.
Protein network associated with the interplay between AD and RSB. The grey color gradient of a node is proportional to its p-value.