ABSTRACT
Recent declines in bumble bee populations are of great concern and have prompted critical evaluations of the role of pathogen introductions and host resistance in bee health. One factor that may influence host resilience when facing infection is the gut microbiota. Previous experiments with Bombus terrestris, a European bumble bee, showed that the gut microbiota can protect against Crithidia bombi, a widespread trypanosomatid parasite of bumble bees. However, the particular characteristics of the microbiome responsible for this protective effect have thus far eluded identification. Using wild and commercially sourced Bombus impatiens, an important North American pollinator, we conducted cross-wise microbiota transplants to naive hosts of both backgrounds and challenged them with a Crithidia parasite. As with B. terrestris, we find that microbiota-dependent protection against Crithidia operates in B. impatiens. Lower Crithidia infection loads were experimentally associated with high microbiome diversity, large gut bacterial populations, and the presence of Apibacter, Lactobacillus Firm-5, and Gilliamella spp. in the gut community. These results indicate that even subtle differences between gut community structures can have a significant impact on a microbiome's ability to defend against parasite infections.
IMPORTANCE Many wild bumble bee populations are under threat due to human activity, including through the introduction of pathogens via commercially raised bees. Recently, it was found that the bumble bee gut microbiota can help defend against a common parasite, Crithidia bombi, but the particular factors contributing to this protection are unknown. Using both wild and commercially raised bees, we conducted microbiota transplants to show that microbiome diversity, total gut bacterial load, and the presence of certain core members of the microbiota may all impact bee susceptibility to Crithidia infection. Bee origin (genetic background) was also a factor. Finally, by examining this phenomenon in a previously uninvestigated bee species, our study demonstrates that microbiome-mediated resistance to Crithidia is conserved across multiple bumble bee species. These findings highlight how intricate interactions between hosts, microbiomes, and parasites can have wide-ranging consequences for the health of ecologically important species.
KEYWORDS: Bombus impatiens, trypanosomatid, gut microbiota, host-microbe interaction, symbiosis
INTRODUCTION
As one of the most common insect pollinators of flowering plants, bumble bees fulfill vital roles in both natural ecosystems and agricultural croplands. However, bumble bee populations have experienced recent decline, most likely due to land use change and the spread of pathogens (1, 2). One protozoan parasite, Crithidia bombi, infects multiple bumble bee species (3). Crithidia bombi increases bumble bee mortality under stressful conditions (4) and reduces colony-founding success of infected queens by 40%, while also leading to significant reductions in mass among infected queens, colony size, male production, and overall fitness (5).
Recently, it has been found that the presence of the bumble bee gut microbiota can protect against C. bombi infection (6) and that variation in this protective capability is driven at least as much by the microbiota as by host genetics (7). Neither the precise aspects of the microbiome that confer this defensive benefit nor its mechanism of action are known (8). Unlike honey bees, lone bumble bee queens establish new colonies annually; thus, the microbiome of a bumble bee colony reflects that of its founding queen, as well as bacteria acquired during foraging and contact with nestmates (9–11). The single-queen generational bottleneck and the relatively small colony size (typically <100 individuals, depending on species) are hypothesized to give rise to microbiome heterogeneity between bumble bee colonies (7, 12).
In this study, we conducted controlled microbiota transplants from different queens into sterile worker bees to determine how C. bombi infection loads are affected by microbiome composition via measurements of diversity, bacterial load, and abundances of bacterial phylotypes (closely related clusters corresponding to species or species groups). Furthermore, we compared bees of wild and commercial (captive-bred) origins. Bumble bees have been bred for use as commercial pollinators since 1988 and have been used to supplement or even replace other pollination methods (13). Commercial bumble bees often have higher pathogen infection rates than wild bumble bees and can spread those diseases to wild bumble bee populations (14). Such transmission routes may be a contributing factor in the decline of wild bees (14, 15–19). Both host genetics and the microbiomes of wild versus commercial bees could differ in their ability to confer resistance to C. bombi, and we tested these factors using a crosswise experimental design.
Previous studies showing the protective effect of the microbiota against C. bombi were conducted with Bombus terrestris, a common European bumble bee (6–8). Here, we found that this effect also extends to a species endemic to North America, Bombus impatiens. B. terrestris and B. impatiens are the major commercial species bred in their respective regions (13). We found that bees of commercial origin have reduced susceptibility to the tested C. bombi strain and that parasite infection load is negatively correlated with microbiome diversity and bacterial load. Finally, we identified several bacterial phylotypes that are strongly associated with C. bombi resistance; these represent potentially beneficial microbes that warrant further investigation.
RESULTS
The following is a brief outline of the experiment: newly eclosed bees (adult bees that emerge from pupation), which lack gut bacteria (6), were inoculated with 5 different microbiota treatments (harvested from the guts of queen bees of different origins). These were then challenged with the Crithidia parasite. After 7 days, guts were removed, the parasites were enumerated, the bacterial community compositions were determined by 16S rRNA gene sequencing, and absolute bacterial loads were quantified by quantitative PCR (qPCR). A detailed description of the experiment and treatments can be found in Materials and Methods.
The five different microbiota treatments administered prior to C. bombi infection were given to bees from both wild and commercial origins. Four of the treatments involved inoculation with gut microbiota derived from a single wild queen, a pooled combination of four wild queens, a single commercial queen, or a pooled combination of four commercial queens. The fifth inoculum consisted of pooled material from the other four inocula, filtered to remove most cells, while retaining acellular factors that may influence host immunity or parasite resistance. Our rationale behind using several separate microbiome treatment inocula was that different source bees can have different microbiome compositions. Different compositions may give rise to different interactions with the parasite (7). Having this variation is necessary for separation of the factors contributing to parasite resistance. We tested four different inocula (from single bees as well as combined inocula from several bees) with the expectation that there would be differences between them. We used combination inocula in case inocula from single individuals were insufficient to capture the diversity of the bee gut microbiota. The inocula were sequenced along with samples from treatment bees at the end of the experiment to determine their community compositions.
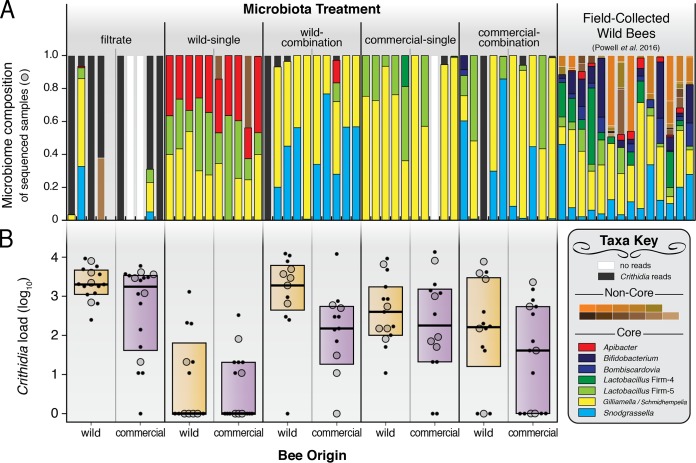
All bees were infected with a common strain of C. bombi administered 7 days after microbiota treatment. The gut microbiome compositions of five randomly chosen samples from each origin-treatment combination were assessed 7 days after C. bombi exposure by PCR amplification and high-throughput sequencing of the bacterial 16S rRNA gene (Fig. 1A). Following quality filtering, a total of 2,190,342 sequences were retrieved across 49 samples. These formed 30 operational taxonomic units of 97% or greater sequence identity (OTUs97). For 13 of these clusters, the top BLASTN hits corresponded to sequences of phylotypes previously sampled from bees. Seven typical bee-associated phylotypes were represented by single OTU97 (Apibacter, Bifidobacterium, Bombiscardovia, Lactobacillus Firm-4, Lactobacillus Firm-5, Saccharibacter, or Snodgrassella), while another typical bee-associated phylotype was represented by six OTUs97 (Gilliamella). Gilliamella OTUs may also comprise “Candidatus Schmidhempelia” (20), a closely related bacterium which was not distinguishable by our short-read data set.
FIG 1.
Microbiome composition and C. bombi infection load in the bumble bee B. impatiens. (A) Gut communities, based on bacterial 16S rRNA gene profiling, of randomly selected bees from five experimental treatments and from wild field-caught bees from a previous study (69) for comparison. Crithidia-specific reads were removed prior to downstream analyses. The y axis indicates relative abundances (sums to 1 for each sample). (B) C. bombi infection loads of individual bees across experimental treatments and host backgrounds (wild or commercial bees). Large markers, microbiota-profiled samples; small markers, unprofiled samples. Boxes show quartiles and medians. C. bombi load expressed as number of cells per 10 μl of gut homogenate. Noncore taxa corresponding to colored labels from top left to bottom right in key: Asaia, Enterobacteriaceae, Fructobacillus, Gluconobacter, Lactobacillus kunkeei, Leuconostoc, Microbacterium, Parasaccharibacter, Saccharibacter, Staphylococcus, and Zymobacter.
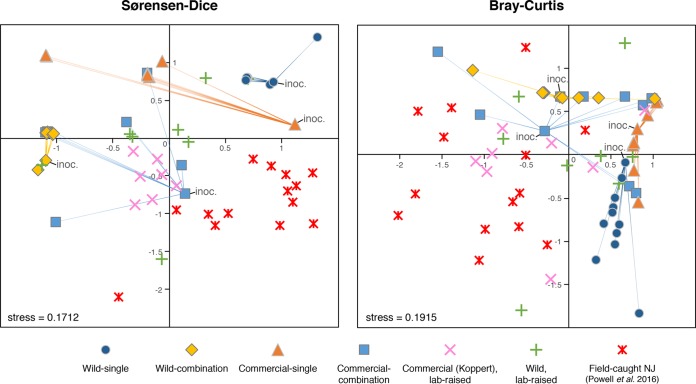
Each treatment resulted in bees with distinct microbiome profiles (Fig. 1A), as assessed by community dissimilarity metrics (Bray-Curtis analysis of similarity [ANOSIM] R = 0.55, P < 0.001, PERMDISP F4 = 2.5, P = 0.068; Sørensen-Dice ANOSIM R = 0.57, P < 0.001, PERMDISP F4 = 0.57, P = 0.69). Community compositions were not statistically different between wild or commercial bees (Bray-Curtis ANOSIM R = 0.00, P = 0.94, PERMDISP F1 = 0.060, P = 0.81; Sørensen-Dice ANOSIM R = 0.00, P = 0.90, PERMDISP F1 = 0.19, P = 0.68), or between source colonies (Bray-Curtis ANOSIM R = 0.06, P = 0.16, PERMDISP F9 = 0.51, P = 0.85; Sørensen-Dice ANOSIM R = 0.02, P = 0.37, PERMDISP F9 = 0.42, P = 0.91) in our experiment. The wild-single treatment produced microbiomes with considerably greater alpha diversity than the other treatments, as measured by OTUs97 (average, 4.0 OTU97; Bonferroni-adjusted t tests, P ≤ 0.01) and Shannon's H (average, 1.58; Bonferroni-adjusted t tests, P ≤ 0.001). However, all treatments had less diverse microbiomes than those of wild field-caught B. impatiens from a previous study (average, 10.3 OTU97, 2.65 Shannon's H; Fig. 1A). The microbiomes of untreated bees in lab-reared B. impatiens colonies (both newly established from the wild and from commercial origins) were also found to differ from those of field-caught bees and treatment bees (Fig. 2; see also Fig. S1 in the supplemental material).
FIG 2.
Nonmetric multidimensional scaling (NMDS) plots of community dissimilarities based on Sørensen-Dice and Bray-Curtis distances. Input samples used for inoculation of experimental microbiota treatments are denoted by “inoc.” and by lines connecting to the resulting output microbiomes. Communities were compared at a depth of 5,000 reads per sample; samples with <600 reads and samples from the filtrate treatment were excluded.
Crithidia bombi infection levels were measured for 138 samples (13 to 18 samples from each origin-treatment combination; Fig. 1B). Infection loads varied significantly according to microbiome treatment [two-way analysis of variance (ANOVA), F(4,138) = 19.0, P < 0.0001] and bee origin [two-way ANOVA, F(1,138) = 10.8, P = 0.0013], but no interaction between bee origin and microbiome treatment was detected [two-way ANOVA, F(4,138) = 0.598, P = 0.66]. The filtrate treatment produced, on average, bees with the greatest C. bombi infection loads (2,495 cells per 10 μl), while the wild-single treatment resulted in the lowest infection loads (78 cells per 10 μl).
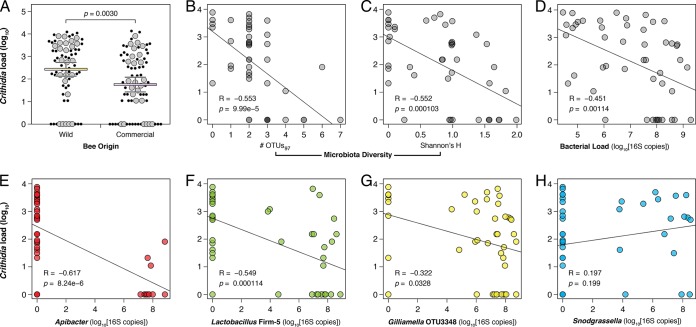
Both bee origin and microbiome diversity were significant correlates of C. bombi infection load. Commercial captive-raised bees had lower C. bombi loads than did wild bees (Fig. 3A). Bees with more diverse microbiomes also had fewer C. bombi parasites (Fig. 3B and C). To investigate the relationship between bacterial abundance and C. bombi abundance, we performed quantitative PCR (qPCR) targeting the bacterial 16S rRNA gene as a proxy to quantify absolute loads of the gut microbiota. A strong negative correlation was found between C. bombi load and bacterial load (Fig. 3D), suggesting that the strength of the bumble bee microbiota's protective effect scales with the number of gut bacteria present.
FIG 3.
Correlations between C. bombi infection load and host and microbiome characteristics. (A) Lower C. bombi susceptibility in bees of commercial origin (two-tailed t test). Large markers, microbiota-profiled samples; small markers, unprofiled samples. Means and 95% confidence intervals shown. (B and C) Lower C. bombi loads in bees with high microbiome diversity. (D) Lower C. bombi loads in bees with larger gut bacterial populations. (E to G) Lower C. bombi loads associated with greater abundance of Apibacter, Lactobacillus Firm-5, and Gilliamella in the gut. (H) No impact of Snodgrassella abundance on C. bombi infection load. Pearson correlation statistics shown. C. bombi load expressed as number of cells per 10 μl of gut homogenate.
However, this trend might also be driven by specific bacterial phylotypes. To assess this possibility, we examined the relationship between C. bombi load and the absolute abundances of each OTU. Four OTUs were significantly correlated with lower parasite loads: Apibacter (Fig. 3E), Lactobacillus Firm-5 (Fig. 3F), and two Gilliamella OTUs (Fig. 3G and S2). Apibacter had the largest effect, and clustering of reads at 100% identity showed that a single Apibacter strain was predominant in the wild-single treatment (see Data Set S2 in the supplemental material). Snodgrassella alvi, another common bumble bee-associated bacterial phylotype, showed no relationship with C. bombi infection load (Fig. 3H).
The observed correlations were not solely driven by the wild-single microbiota treatment (the treatment with the most pronounced outcome in our experiment). Diminished C. bombi load with increasing microbiota diversity, bacterial load, and abundances of Lactobacillus Firm-5 and Gilliamella, as well as the neutral effect of Snodgrassella spp., were all trends that persisted following the removal of wild-single samples from our analyses (Table S1). The effect size and statistical significance of the associations were reduced in these cases, however. Since our experimental microbiota inoculations preceded C. bombi infection, the results presented here point toward causative relationships, not mere correlations. The microbiomes of posttreatment bees closely resembled those of their respective input inocula (Fig. 2 and S1), suggesting that exposure to C. bombi is not responsible for the observed gut community differences but rather that the microbiota induces changes to C. bombi susceptibility.
DISCUSSION
Of the five microbiome treatment groups, the wild-single microbiome had the greatest protective effect against C. bombi, followed by the commercial-combination treatment (Fig. 1B). The microbiome treatments explained 33.3% of the observed variation in parasite load, whereas bee origin (likely reflecting host genetic differences) explained only 4.7% of the observed variation (Fig. 3A). This extends previous findings for B. terrestris that the gut microbiome is a much more important determinant of C. bombi resistance than host background (7). In our experiment, B. impatiens bees of commercial origin were less susceptible to C. bombi than wild B. impatiens bees. This finding is consistent with the hypothesis that commercial captive-bred bees (where C. bombi infection prevalence is generally higher) are better able to tolerate parasites, leaving wild bees vulnerable to pathogen spillovers from commercial operations (19). However, our experiments do not allow us to claim a general difference between commercial and wild bees, since we only sampled from limited sources for either category. Also, different parasite-host combinations can result in diverse infection outcomes (7), and we only tested a single C. bombi strain; further experiments are needed to validate the resilience of commercial bees.
The more important factor in infection resistance, microbiome composition, displayed substantial variation between our treatments (Fig. 1A). This allowed us to tease apart the components of gut community structure that are most influential in promoting C. bombi resistance. A potentially important component in host health is gut microbiome diversity; for instance, low microbiome diversity in humans is associated with inflammatory bowel disorder and Clostridium difficile infections (21). We found that greater microbiome diversity was associated with lower infection loads (Fig. 3B and C). Our experimental treatments generally had lower diversity than field-collected bees (Fig. 1A), which is consistent with observations that bees acquire “noncore” environmental bacteria (from foraging on flowers, etc.) when not confined to the indoors (22, 23). In contrast, our experimental bees had microbiomes almost exclusively consisting of the “core” gut bacteria. While it appears that a higher diversity of the core microbiota can protect against C. bombi, the same may not be true of noncore bacteria: previous surveys of wild bumble bees showed increased C. bombi infection with increasing noncore diversity (24, 25). Surveys of gut microbiota of Bombus species in China and North America indicate that all species possess the core phylotypes, including Gilliamella, Snodgrassella, and Lactobacillus Firm-5, but that some individual bees of each species are dominated by noncore environmental bacteria (25, 26). Potentially, retention of the core phylotypes is more important than diversity per se in resisting pathogen colonization.
Correlation of absolute abundances of microbial phylotypes with C. bombi load revealed three bacterial taxa as promising candidates for inhibiting C. bombi: Apibacter, Lactobacillus Firm-5, and Gilliamella (Fig. 3 and S2). Lactobacillus Firm-5 and Gilliamella are well-known core members of the honey bee and bumble bee gut microbiota (27–31), and they participate in the digestion of complex polysaccharides in the bee hindgut (32–35). Gut colonization by Gilliamella spp. has been associated with decreased Crithidia infection levels in field-caught bees, but the effect was small (25) or not significant (6). Another core microbiota member, Snodgrassella alvi (36), was found to be negatively associated with C. bombi in one survey of field-caught bees (6) but not in another (25). In our experimental transplant study, we found no correlation between Snodgrassella and C. bombi levels in the gut (Fig. 3H). A recent study in honey bees also showed no benefit of Snodgrassella spp. in suppressing infection by a related protozoan parasite, Lotmaria passim (37).
Apibacter spp. exhibited the strongest negative correlation with C. bombi infection (Fig. 3E). Previously classified as an unidentified Bacteroidetes/Flavobacteriales bacterium (7, 25, 30, 38), representatives of the Apibacter clade have now been cultivated from both honey bees and bumble bees (39, 40). Their biological role within the gut remains uncharacterized, although our results suggest that Apibacter spp. can colonize to high numbers in a given individual (>30% relative abundance). In field-caught bees, Apibacter spp. constituted a much smaller proportion of the gut community (average, 2.6%; Fig. 1A). Interestingly, Apibacter spp. were only present in the wild-single treatment, which resulted in significantly lower levels of Crithidia infection in both wild and commercial bees. The wild-combination treatment, which failed to reduce infection loads, lacked Apibacter spp. (save for a single sample, which had <500 reads and was removed from the analysis). This suggests substantial heterogeneity in the microbiomes of overwintered queens, which may result in bee colonies with differing abilities to resist various pathogens and parasites (7).
A previous experimental microbiome transplant study by Koch and Schmid-Hempel (7) in the European bumble bee B. terrestris offers evidence consistent with our results. Although not noted by the authors at the time, their bees with the highest Crithidia infection loads were found in a treatment group lacking Lactobacillus Firm-5 (as “Lactobacillus sp.”), and the lowest infection loads were recorded in a group harboring Apibacter spp. (as “Bacteroidetes”) (see Fig. 4 and S2 in reference 7). That both studies point to these two bacteria as key players in the interaction between microbiome, host, and parasite is compelling evidence of a conserved interaction network present across bumble bee species.
Several possibilities for the mechanism behind microbiota-mediated resistance should be explored going forward. Since both C. bombi and the microbiota colonize the bumble bee hindgut (41), it is possible that the microbiota directly inhibits C. bombi through the secretion of antimicrobial compounds, competition for resources, or spatial interference. Indirect interaction via the host may also play a role: the bee gut microbiota can stimulate the immune system, with different combinations of microbes provoking different immune responses (8, 42, 43). Host innate immunity is likely a key component in combatting C. bombi (44–46), but working out the mechanisms, especially as applied to wild bee populations, requires further study of the complex interplay between environmental influences, host genotype, parasite genotype, and the microbiota (47).
There is considerable evidence of the gut microbiota promoting host health in other insects and affecting the growth of eukaryotic parasites. In mosquitoes, gut bacteria are essential for host development (48) and can affect the infection cycle of Plasmodium falciparum, the causative agent of malaria (49). In sand flies, the diversity of the gut microbiome decreases over the course of Leishmania infection but is also essential for the parasite's survival, as shown by Leishmania growth suppression in antibiotic-treated flies (50). In bumble bees, Crithidia can be transmitted between members of a colony by fecal-oral contact and can spread across multiple colonies via contact at shared floral resources (51). Infection prevalence increases (up to ∼80%) and then decreases over the course of a year, reflecting the annual growth and decline of bumble bee colonies (52). Crithidia survival in overwintering queens seeds the cycle in the following year (47).
Our results indicate a potential for coevolutionary interactions between gut microbiomes and parasites in overwintering queens. While the effect of infection intensity on the spread of Crithidia spp. has not been explicitly tested, higher Crithidia loads are associated with the higher transmission rates typically seen early in the colony cycle (52). By lowering Crithidia abundance at the individual level, particular microbiomes might decrease overall infection rates across a colony. If so, why have “protective” microbiomes not become the norm? Perhaps any given bacterial strain can only inhibit specific Crithidia strains (7) and Crithidia genotype frequencies constantly shift to escape inhibition. There might also be fitness tradeoffs for possessing certain microbiome compositions, independent of Crithidia. Furthermore, it has been hypothesized that Crithidia infection alters bee-foraging activities, potentially leading to self-medication (51, 53). How such behavioral changes induced by Crithidia spp. affect the microbiome is unknown. Microbiome changes after Crithidia infection also may impact fitness; however, in this study, we only tested the effect of the microbiome prior to parasite exposure.
Understanding the factors affecting bumble bee health is vital to agriculture and to the sustainability of natural ecosystems. Wild bumble bees are among the most common and important nonmanaged insect pollinators (54), and B. impatiens in particular is the most valuable species in its range across eastern North America (55). It is also currently the primary species sold commercially across North America and provides highly efficient pollination services for various greenhouse crops, including tomatoes (56), muskmelons (57), and sweet peppers (58), as well as for a number of field crops (54, 59, 60). As commercial reliance on captive-bred bees grows (13, 61), the impact of pathogen exposure on wild bee populations, and the corresponding methods to alleviate such stressors, will require increasingly urgent and critical evaluation.
MATERIALS AND METHODS
Bumble bee colonies.
Wild B. impatiens queens were collected in Polk County, TX (30.7449, −94.6218) in March 2015, coinciding with their emergence from winter hibernation. The wild queens were kept in individual cages in a laboratory incubator at 28°C and 60% humidity, fed a diet of sterile irradiated pollen and sucrose water (1:1 [wt/vol]), and allowed to start a colony. Commercial bumble bees were ordered from Biobest (Westerlo, Belgium; Leamington, Ontario, Canada, for North America) and kept in the lab on a diet of sterile irradiated pollen (Betterbee, Greenwich, NY) and sucrose water.
Preparation of treatment groups.
Worker B. impatiens from both wild and commercial colonies were inoculated with one of five microbiome treatments derived from wild-caught or commercially produced queens: wild-single, wild-combination, commercial-single, commercial-combination, and filtrate. In order to generate the wild-single microbiome treatment, one whole gut was removed from a wild queen and homogenized in 250 μl of 10 mM phosphate-buffered saline (PBS). Of this, 100 μl was set aside for the filtrate treatment. Glycerol was added to the remaining 150 μl, resulting in a total volume of 300 μl at 15% (vol/vol) glycerol. This mixture was then separated into 5-μl aliquots and frozen at −80°C.
In order to generate the wild-combination treatment, whole guts were removed from four wild queens and homogenized in 1,000 μl PBS. Of this, 400 μl was set aside for the filtrate treatment. The remaining 600 μl was diluted in glycerol to yield 1,200 μl at 15% (vol/vol) glycerol. The mixture was then separated into 5-μl aliquots and frozen at −80°C.
The above-mentioned process was followed to generate the commercial-single and commercial-combination microbiome treatments from commercial queens. The filtrate treatment was generated from 75 μl of homogenized guts set aside from each treatment. The guts were filtered through 4 layers of cheese cloth and then brought to a 15% (vol/vol) glycerol concentration. This method of filtration was done to remove intact bacterial cells while retaining other nonbacterial gut particulates that may affect pathogen infectivity or host immune response. We attempted separation with 0.2-μm-pore-size filters. However, these filters were easily clogged, and we were able to obtain only a few microliters of liquid from ∼200 μl of bee guts. This volume was insufficient for administering our treatments, so we used folded cheese cloth, which yielded sufficient material while removing the vast majority of bacterial cells. The final filtrates were combined and then divided into 5-μl aliquots and frozen at −80°C. This treatment was a control for potential unintended effects of feeding bees homogenized gut material (e.g., if the gut material itself induced protection against the parasite or if there was an acellular agent present that made the bees sick). While not all filtrate-fed bees were completely bacteria-free by the end of the experiment (Fig. 1A), a comparison of absolute bacterial loads (see Data Set S3 in the supplemental material) shows that filtrate-fed bees had a much lower load of bacteria, often at or below the detection limit of our method.
In order to generate germfree bumble bees, B. impatiens worker cocoons were removed from the wild and commercial colonies. Four wild colonies and six commercial colonies were used for the experiment. The pupae were removed from the cocoon and allowed to mature under sterile conditions at 28°C and 60% humidity. This method has been previously used and validated to generate microbiota-free bees (6, 62). Upon eclosion, they were transferred to individual sterile plastic cages and inoculated with one of the five treatments. Bees from each colony were assigned randomly to treatment groups. The prepared 5-μl treatment aliquots were thawed on ice and then combined with 10 μl of sterile sugar water for a final 15-μl inoculum. To encourage feeding, bees were starved for 3 to 5 h prior to inoculation. The bees were monitored to ensure that they consumed the inoculum. Following the inoculation, workers were fed filter-sterilized sucrose water and gamma-irradiated pollen ad libitum and kept at room temperature in individual plastic cages. To produce sterile pollen, pollen purchased from Brushy Mountain Bee Farm (NC, USA) was irradiated at 30 kGy with gamma radiation (Sadex Corporation, IA, USA). Sterilization was validated by mixing ∼100 mg irradiated pollen in 1 ml PBS, plating 100 μl on lysogeny broth agar, and observing for growth of microbial colonies.
Crithidia infection.
After allowing 7 days for the administered microbiota to establish in the gut, bees were removed from their cages and starved for 3 to 5 h in preparation for Crithidia infection. The C. bombi strain used for infection was isolated from a B. impatiens worker bee collected in West Haven, CT, USA, in September 2012 (C. bombi strain 12.6 from reference 63). Crithidia was grown in Insectagro DS2 medium (Corning, Inc.) supplemented with 5% fetal bovine serum at 28°C and 3% CO2. The concentration of the Crithidia culture was determined using a hemocytometer (Neubauer improved cell counting chamber), and a 2:1 solution of sucrose water to culture medium was prepared so that the bees were fed a total of 15,000 Crithidia cells each. The bees were monitored to ensure that they consumed the 15 μl of infection mixture.
After another 7 days, shown to be within the peak period of Crithidia infection (64), the bees were placed on ice and dissected. The gut was removed and homogenized in 200 μl PBS. Crithidia infection load was recorded by counting the number of Crithidia cells present in 10 μl of homogenized gut using a hemocytometer (Data Set S3). The remaining homogenized gut and bee carcass were stored at −80°C. Crithidia cells actively infected and multiplied within B. impatiens, as indicated by some bees having higher counts at the end of the experiment than what was expected from an initial infection inoculum of 15,000 cells.
Microbiome analysis.
Five individual guts from each of the 10 treatment groups were randomly selected for DNA extraction according to Engel et al. (65). The microbiome inocula for each treatment group were also similarly processed to assess whether the transplant of the original microbiota to naive bees was successful (Fig. 2 and S1). DNA concentrations were determined using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Inc.). Samples were diluted to 10 ng/μl and sent to the University of Texas at Austin Genomic Sequencing and Analysis Facility, where the V4 region of the bacterial 16S rRNA gene was amplified by PCR using universal bacterial primers 515F and 806R. Amplifications were carried out in triplicate and the products pooled to minimize PCR jackpot effects. Amplicon libraries were sequenced on the Illumina MiSeq platform with a 2 × 250-bp read design. The sequenced samples are listed in Data Set S3.
Sequences were processed and analyzed using QIIME version 1.9.1 (66). Primer sequences were trimmed from the reads, and forward and reverse reads were joined with SeqPrep. Quality filtering using split_libraries_fastq.py was performed using the settings Phred q ≥ 30, maximum N = 0, and read length fraction minimum of 0.8. Reads of <230 bp or >270 bp were removed (expected read length, ∼250 bp). Sequences were then clustered into operational taxonomic units (OTUs) at 97% sequence similarity using the pick_de_novo_otus pipeline. Any OTUs accounting for less than 0.5% of the reads for that sample were excluded from the analysis, since they could result from multiplexing barcode assignment errors. Taxonomic assignment of representative OTUs was manually determined using BLASTN against the GenBank nr database. The recovered OTUs were further filtered to remove reads derived from plastid, mitochondrial, and eukaryotic (e.g., Crithidia or host) sources. Samples with fewer than 500 total reads were also removed from the analysis. Alpha- and beta-diversity analyses were performed at a read depth of 550 reads per sample; the OTU corresponding to Crithidia was retained for the beta-diversity analysis to permit ordination of samples lacking any bacterial reads. Read/rarefaction thresholds were chosen based on sequencing depth and by maximizing the number of samples retained. There were no more than 12 OTUs in any sample. Visual inspection of rarefaction curves showed that gut community diversity was adequately captured. The OTU tables and representative sequences generated in this study are presented in Data Set S4.
Additional analysis of Apibacter reads was performed using CD-HIT-EST (67). Reads from the wild-single treatment were clustered at 100% identity. Clusters with fewer than 5 reads were excluded. Apibacter-specific clusters were identified by BLAST using the Apibacter representative sequence from the 97% OTU clustering as a query. The cluster sizes and sequences are listed in Data Set S2.
Quantitative PCR (qPCR) was performed according to Cariveau et al. (25) to determine the absolute number of 16S rRNA gene copies present per gut sample, as a proxy for the number of bacteria present in each gut. Total copy numbers were adjusted by the relative abundance output from OTU clustering to obtain absolute abundances of bacterial OTUs in each sample. Statistical analyses were conducted in R 3.2.3 (68) and Prism 6 (GraphPad Software, Inc.) and used log10(x + 1)-transformed C. bombi and bacterial load values. We acknowledge the potential for error in both the amplicon sequencing and the qPCR (due to PCR primer bias and different 16S rRNA gene copy numbers for each bacterial genome), and thus assessments of OTU abundance, particularly in a comparison between OTUs, should be interpreted cautiously.
Lab-reared B. impatiens microbiomes.
We additionally surveyed the microbiomes of lab-reared B. impatiens colonies on which we did not conduct experimental microbiome manipulations. Two adult workers each from each colony were sampled and processed as described above. Four colonies were established from wild queens collected in Polk County, TX, in March 2015, and another four colonies were purchased from a commercial bumble bee supplier (Koppert Biological Systems, The Netherlands; bumble bee production in Howell, MI, USA). The microbiomes were compared to that of wild field-caught wild B. impatiens bees collected in New Jersey, USA, from a previous study (69).
Accession number(s).
Parasite counts and qPCR data are placed in Data Sets S1 and S3 in the supplemental material. The 16S rRNA gene sequence reads are deposited under NCBI BioProject no. PRJNA371284.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eva Frederick for assistance in the qPCR assays, Antonio Castilla for help in bumble bee queen collecting, and Dylan Dey for assistance in bumble bee maintenance. The authors' contributions are as follows: conceptualization, B.K.M. and H.K.; investigation, methodology, and validation, B.K.M., W.K.K., and H.K.; funding acquisition, B.K.M. and N.A.M.; resources and supervision, N.A.M.; data curation, formal analysis, visualization, and writing of the original draft, B.K.M. and W.K.K.; and review and editing of the writing, B.K.M., W.K.K., N.A.M., and H.K.
We declare no competing interests.
B.K.M. received a University of Texas Undergraduate Research Fellowship. N.A.M. received a National Institutes of Health award 1R01-GM108477-01. H.K. received Swiss National Science Foundation Postdoctoral Fellowship 147881.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02335-17.
REFERENCES
- 1.Cameron SA, Lozier JD, Strange JP, Koch JB, Cordes N, Solter LF, Griswold TL. 2011. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci U S A 108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulson D, Nicholls E, Botias C, Rotheray EL. 2015. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- 3.Salathé RM, Schmid-Hempel P. 2011. The genotypic structure of a multi-host bumblebee parasite suggests a role for ecological niche overlap. PLoS One 6:e22054. doi: 10.1371/journal.pone.0022054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MJF, Loosli R, Schmid-Hempel P. 2000. Condition-dependent expression of virulence in a trypanosome infecting bumble bees. Oikos 91:421–427. doi: 10.1034/j.1600-0706.2000.910302.x. [DOI] [Google Scholar]
- 5.Brown MJF, Schmid-Hempel R, Schmid-Hempel P. 2003. Strong context-dependent virulence in a host-parasite system: reconciling genetic evidence with theory. J Anim Ecol 72:994–1002. doi: 10.1046/j.1365-2656.2003.00770.x. [DOI] [Google Scholar]
- 6.Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A 108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett 15:1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- 8.Näpflin K, Schmid-Hempel P. 2016. Immune response and gut microbial community structure in bumblebees after microbiota transplants. Proc Biol Sci 283:20160312. doi: 10.1098/rspb.2016.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch H, Abrol DP, Li J, Schmid-Hempel P. 2013. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol 22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 10.Newbold LK, Oliver AE, Cuthbertson L, Walkington SE, Gweon HS, Heard MS, van der Gast CJ. 2015. Rearing and foraging affects bumblebee (Bombus terrestris) gut microbiota. Environ Microbiol Rep 7:634–641. doi: 10.1111/1758-2229.12299. [DOI] [PubMed] [Google Scholar]
- 11.Billiet A, Meeus I, Van Nieuwerburgh F, Deforce D, Wäckers F, Smagghe G. 2017. Colony contact contributes to the diversity of gut bacteria in bumblebees (Bombus terrestris). Insect Sci 24:270–277. doi: 10.1111/1744-7917.12284. [DOI] [PubMed] [Google Scholar]
- 12.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Jaffé R, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci Adv 3:e1600513. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velthuis HHW, van Doorn A. 2006. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 37:421–451. doi: 10.1051/apido:2006019. [DOI] [Google Scholar]
- 14.Graystock P, Yates K, Evison SEF, Darvill B, Goulson D, Hughes WOH. 2013. The Trojan hives: pollinator pathogens, imported and distributed in bumblebee colonies. J Appl Ecol 50:1207–1215. doi: 10.1111/1365-2664.12134. [DOI] [Google Scholar]
- 15.Colla SR, Otterstatter MC, Gegear RJ, Thomson JD. 2006. Plight of the bumble bee: pathogen spillover from commercial to wild populations. Biol Conserv 129:461–467. doi: 10.1016/j.biocon.2005.11.013. [DOI] [Google Scholar]
- 16.Otterstatter MC, Thomson JD. 2008. Does pathogen spillover from commercially reared bumble bees threaten wild pollinators? PLoS One 3:e2771. doi: 10.1371/journal.pone.0002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachman-Ruiz B, Narváez-Padilla V, Reynaud E. 2015. Commercial Bombus impatiens as reservoirs of emerging infectious diseases in central México. Biol Invasions 17:2043–2053. doi: 10.1007/s10530-015-0859-6. [DOI] [Google Scholar]
- 18.Manley R, Boots M, Wilfert L. 2016. Emerging viral disease risk to pollinating insects: ecological, evolutionary and anthropogenic factors. J Appl Ecol 52:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graystock P, Blane EJ, McFrederick QS, Goulson D, Hughes WOH. Do managed bees drive parasite spread and emergence in wild bees? Int J Parasitol Parasites Wildl 5:64–75. doi: 10.1016/j.ijppaw.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinson VG, Magoc T, Koch H, Salzberg SL, Moran NA. 2014. Genomic features of a bumble bee symbiont reflect its host environment. Appl Environ Microbiol 80:3793–3803. doi: 10.1128/AEM.00322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meeus I, Parmentier L, Billiet A, Maebe K, Van Nieuwerburgh F, Deforce D, Wäckers F, Vandamme P, Smagghe G. 2015. 16S rRNA amplicon sequencing demonstrates that indoor-reared bumblebees (Bombus terrestris) harbor a core subset of bacteria normally associated with the wild host. PLoS One 10:e0125152. doi: 10.1371/journal.pone.0125152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parmentier L, Meeus I, Mosallanejad H, de Graaf DC, Smagghe G. 2015. Plasticity in the gut microbial community and uptake of Enterobacteriaceae (Gammaproteobacteria) in Bombus terrestris bumblebees' nests when reared indoors and moved to an outdoor environment. Apidologie 47:237–250. doi: 10.1007/s13592-015-0393-7. [DOI] [Google Scholar]
- 24.Koch H, Cisarovsky G, Schmid-Hempel P. 2012. Ecological effects on gut bacterial communities in wild bumblebee colonies. J Anim Ecol 81:1202–1210. doi: 10.1111/j.1365-2656.2012.02004.x. [DOI] [PubMed] [Google Scholar]
- 25.Cariveau DP, Elijah Powell J, Koch H, Winfree R, Moran NA. 2014. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J 8:2369–2379. doi: 10.1038/ismej.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Powell JE, Guo J, Evans JD, Wu J, Williams P, Lin Q, Moran NA, Zhang Z. 2015. Two gut community enterotypes recur in diverse bumblebee species. Curr Biol 25:R652–R653. doi: 10.1016/j.cub.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 28.Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corby-Harris V, Maes P, Anderson KE. 2014. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS One 9:e95056. doi: 10.1371/journal.pone.0095056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HC, Chu C-C, Seufferheld MJ, Cameron SA. 2015. Deep sequencing and ecological characterization of gut microbial communities of diverse bumble bee species. PLoS One 10:e0118566. doi: 10.1371/journal.pone.0118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Jaffé R, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci Adv 3:e1600515. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton ILG. 2015. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 34.Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SG, Vásquez A. 2015. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16:284. doi: 10.1186/s12864-015-1476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong WK, Moran NA. 2013. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol 63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 37.Schwarz RS, Moran NA, Evans JD. 2016. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci U S A 113:9345–9350. doi: 10.1073/pnas.1606631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch H, Schmid-Hempel P. 2011. Bacterial communities in central European bumble bees: low diversity and high specificity. Microb Ecol 62:121–133. doi: 10.1007/s00248-011-9854-3. [DOI] [PubMed] [Google Scholar]
- 39.Kwong WK, Moran NA. 2016. Apibacter adventoris gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from honey bees. Int J Syst Evol Microbiol 66:1323–1329. doi: 10.1099/ijsem.0.000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praet J, Aerts M, De Brandt E, Meeus I, Smagghe G, Vandamme P. 2016. Apibacter mensalis sp. nov.: a rare member of the bumble bee gut microbiota. Int J Syst Evol Microbiol 66:1645–1651. doi: 10.1099/ijsem.0.000921. [DOI] [PubMed] [Google Scholar]
- 41.Lipa JJ, Triggiani O. 1988. Crithidia bombi sp. n. A flagellated parasite of a bumblebee Bombus terrestris L. (Hymenoptera, Apidae). Acta Protozool 27:287–290. [Google Scholar]
- 42.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci 4:170003. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emery O, Schmidt K, Engel P. 2017. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol Ecol 26:2576–2590. doi: 10.1111/mec.14058. [DOI] [PubMed] [Google Scholar]
- 44.Brown MJ, Moret Y, Schmid-Hempel P. 2003. Activation of host constitutive immune defence by an intestinal trypanosome parasite of bumble bees. Parasitology 126:253–260. doi: 10.1017/S0031182002002755. [DOI] [PubMed] [Google Scholar]
- 45.Riddell CE, Lobaton Garces JD, Adams S, Barribeau SM, Twell D, Mallon EB. 2014. Differential gene expression and alternative splicing in insect immune specificity. BMC Genomics 15:1031. doi: 10.1186/1471-2164-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marxer M, Vollenweider V, Schmid-Hempel P. 2016. Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Philos Trans R Soc Lond B Biol Sci 371:20150302. doi: 10.1098/rstb.2015.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadd BM, Barribeau SM. 2013. Heterogeneity in infection outcome: lessons from a bumblebee-trypanosome system. Parasite Immunol 35:339–349. doi: 10.1111/pim.12043. [DOI] [PubMed] [Google Scholar]
- 48.Coon KL, Vogel KJ, Brown MR, Strand MR. 2014. Mosquitoes rely on their gut microbiota for development. Mol Ecol 23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahia AC, Dong Y, Blumberg BJ, Mlambo G, Tripathi A, BenMarzouk-Hidalgo OJ, Chandra R, Dimopoulos G. 2014. Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol 16:2980–2994. doi: 10.1111/1462-2920.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelly PH, Bahr SM, Serafim TD, Ajami NJ, Petrosino JF, Meneses C, Kirby JR, Valenzuela JG, Kamhawi S, Wilson ME. 2017. The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of Leishmania infantum. mBio 8:e01121-16. doi: 10.1128/mBio.01121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch H, Brown MJ, Stevenson PC. 2017. The role of disease in bee foraging ecology. Curr Opin Insect Sci 21:60–67. doi: 10.1016/j.cois.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Popp M, Erler S, Lattorff HM. 2012. Seasonal variability of prevalence and occurrence of multiple infections shape the population structure of Crithidia bombi, an intestinal parasite of bumblebees (Bombus spp.). Microbiologyopen 1:362–372. doi: 10.1002/mbo3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson LL, Bowers MD, Irwin RE. 2016. Nectar chemistry mediates the behavior of parasitized bees: consequences for plant fitness. Ecology 97:325–337. doi: 10.1890/15-0263.1. [DOI] [PubMed] [Google Scholar]
- 54.Garibaldi LA, Steffan-Dewenter I, Winfree R, Aizen MA, Bommarco R, Cunningham SA, Kremen C, Carvalheiro LG, Harder LD, Afik O, Bartomeus I, Benjamin F, Boreux V, Cariveau D, Chacoff NP, Dudenhöffer JH, Freitas BM, Ghazoul J, Greenleaf S, Hipólito J, Holzschuh A, Howlett B, Isaacs R, Javorek SK, Kennedy CM, Krewenka KM, Krishnan S, Mandelik Y, Mayfield MM, Motzke I, Munyuli T, Nault BA, Otieno M, Petersen J, Pisanty G, Potts SG, Rader R, Ricketts TH, Rundlöf M, Seymour CL, Schüepp C, Szentgyörgyi H, Taki H, Tscharntke T, Vergara CH, Viana BF, Wanger TC, Westphal C, Williams N, Klein AM. 2013. Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 339:1608–1611. doi: 10.1126/science.1230200. [DOI] [PubMed] [Google Scholar]
- 55.Kleijn D, Winfree R, Bartomeus I, Carvalheiro LG, Henry M, Isaacs R, Klein AM, Kremen C, M'Gonigle LK, Rader R, Ricketts TH, Williams NM, Lee Adamson N, Ascher JS, Báldi A, Batáry P, Benjamin F, Biesmeijer JC, Blitzer EJ, Bommarco R, Brand MR, Bretagnolle V, Button L, Cariveau DP, Chifflet R, Colville JF, Danforth BN, Elle E, Garratt MP, Herzog F, Holzschuh A, Howlett BG, Jauker F, Jha S, Knop E, Krewenka KM, Le Féon V, Mandelik Y, May EA, Park MG, Pisanty G, Reemer M, Riedinger V, Rollin O, Rundlöf M, Sardiñas HS, Scheper J, Sciligo AR, Smith HG, Steffan-Dewenter I, et al. 2015. Delivery of crop pollination services is an insufficient argument for wild pollinator conservation. Nat Commun 6:7414. doi: 10.1038/ncomms8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kevan PG, Clark EA, Thomas VG. 1990. Insect pollinators and sustainable agriculture. Am J Altern Agric 5:13–22. doi: 10.1017/S0889189300003179. [DOI] [Google Scholar]
- 57.Fisher RM, Pomeroy N. 1989. Pollination of green-house muskmelons by bumble bees (Hymenoptera: Apidae). Ann Entomol Soc Am 82:1061–1066. [Google Scholar]
- 58.Shipp JL, Whitefield GH, Papadopoulos AP. 1994. Effectiveness of the bumble bee, Bombus impatiens Cr. (Hymenoptera: Apidae), as a pollinator of greenhouse sweet pepper. Sci Hortic 57:29–39. doi: 10.1016/0304-4238(94)90032-9. [DOI] [Google Scholar]
- 59.Stanghellini MS, Ambrose JT, Schultheis JR. 1997. The effects of A. mellifera and B. impatiens pollination on fruit set and abortion of cucumber and watermelon. Am Bee J 137:386–391. [Google Scholar]
- 60.Stubbs CS, Drummond FA. 2001. Bombus impatiens (Hymenoptera: Apidae): an alternative to Apis mellifera (Hymenoptera: Apidae) for lowbush blueberry pollination. J Econ Entomol 94:609–616. doi: 10.1603/0022-0493-94.3.609. [DOI] [PubMed] [Google Scholar]
- 61.Inari N, Nagamitsu T, Kenta T, Goka K, Hiura T. 2005. Spatial and temporal pattern of introduced Bombus terrestris abundance in Hokkaido, Japan, and its potential impact on native bumblebees. Popul Ecol 47:77–82. doi: 10.1007/s10144-004-0205-9. [DOI] [Google Scholar]
- 62.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer-Young EC, Sadd BM, Stevenson PC, Irwin RE, Adler LS. 2016. Bumble bee parasite strains vary in resistance to phytochemicals. Sci Rep 6:37087. doi: 10.1038/srep37087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmid-Hempel P, Schmid-Hempel R. 1993. Transmission of a pathogen in Bombus terrestris, with a note on division of labour in social insects. Behav Ecol Sociobiol 33:319–327. doi: 10.1007/BF00172930. [DOI] [Google Scholar]
- 65.Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA. 2013. Standard methods for research on Apis mellifera gut symbionts. J Apic Res 52:1–24. doi: 10.3896/IBRA.1.52.4.07. [DOI] [Google Scholar]
- 66.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 69.Powell E, Ratnayeke N, Moran NA. 2016. Strain diversity and host specificity in a specialized gut symbiont of honey bees and bumble bees. Mol Ecol 25:4461–4471. doi: 10.1111/mec.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.