ABSTRACT
Parasites of the Babesia divergens Asia lineage, which are closely related to B. divergens in Europe and Babesia sp. strain MO1 in the United States, were recently reported in sika deer (Cervus nippon) in eastern Japan. To identify the tick vector(s) for this parasite, we conducted a field survey in Hokkaido, Japan, where the infection rate in sika deer is the highest in the country. A specific PCR system which detects and discriminates between lineages within B. divergens and between those lineages and Babesia venatorum showed that Ixodes persulcatus (11/822), but not sympatric Ixodes ovatus (0/595) or Haemaphysalis sp. (0/163) ticks, carried B. divergens Asia lineage. Genomic DNA was archived from salivary glands of partially engorged I. persulcatus females and three isolates of B. divergens Asia lineage were newly described. The 18S rRNA gene sequence of the isolates formed the Asia lineage cluster with those previously described in sika deer isolates. One salivary gland also contained parasites of Babesia microti U.S. lineage, which were subsequently isolated in a hamster in vivo. B. venatorum (strain Etb5) was also detected in one I. persulcatus tick. The 18S rRNA sequence of Etb5 was 99.7% identical to that of B. venatorum (AY046575) and was phylogenetically positioned in a taxon composed of B. venatorum isolates from Europe, China, and Russia. The geographical distribution of I. persulcatus is consistent with that of B. divergens in sika deer in Japan. These results suggest that I. persulcatus is a principal vector for B. divergens in Japan and Eurasia, where I. persulcatus is predominantly distributed.
IMPORTANCE The Babesia divergens Asia lineage of parasites closely related to B. divergens in Europe and Babesia sp. MO1 in the United States was recently reported in Cervus nippon in eastern Japan. In this study, specific PCR for the Asia lineage identified 11 positives in 822 host-seeking Ixodes persulcatus ticks, a principal vector for many tick-borne disease agents. Gene sequences of three isolates obtained from DNA in salivary glands of female ticks were identical to each other and to those in C. nippon. We also demonstrate the coinfection of B. divergens Asia lineage with Babesia microti U.S. lineage in a tick salivary gland and, furthermore, isolated the latter in a hamster. These results suggest that I. persulcatus is the principal vector for B. divergens as well as for B. microti, and both parasites may be occasionally cotransmitted by I. persulcatus. This report will be important for public health, since infection may occur through transfusion.
KEYWORDS: Babesia, Ixodes persulcatus, tick-borne pathogens
INTRODUCTION
Human babesiosis is caused by intraerythrocytic protozoa belonging to the genus Babesia, which is maintained between ixodid ticks and various mammals in nature. Human babesiosis was first described in 1957 in a splenectomized Yugoslavian farmer (1). The cattle piroplasm Babesia divergens was subsequently identified as the causative agent, based on similar morphological features under light microscopy and successful transmission of human isolates to bovines (2). To date, approximately 40 human cases have been reported from Europe and attributed to B. divergens infection, although not all cases were diagnosed based on molecular methods. Almost all patients were asplenic and/or were immunocompromised at the time of infection, in which case the disease tends to be a life-threatening event, resulting in a high mortality rate of 42% (3). However, recent studies suggest that a wider range of infection has occurred in humans than previously recognized. Martinot et al. reported that an influenza-like infection was caused by B. divergens in immunocompetent patients in France (4). Furthermore, the existence of antibodies against B. divergens was evident in blood donors in Austria (2.1%) (5), in tick-exposed patients in Germany (3.6%) (6) and Belgium (33.2%) (7), and in forestry workers in France (0.1%) (8). In areas where B. divergens is endemic, Ixodes ricinus, which is distributed in many European countries and from North Africa to Scandinavia, is regarded as its vector. Vector competence is evidenced by experimental transstadial and transovarial transmission to susceptible animals (bovine and gerbil) (9–11), and by molecular epidemiology that indicates overlapping geographical distribution of this tick species and human cases (12–15).
Recently the view of human babesiosis has been changed to an emerging tick-borne disease, since patients infected with novel Babesia spp., as well as with piroplasms genetically related but not identical to well-known zoonotic Babesia spp., including B. divergens, are increasingly reported worldwide. In the United States, three cases of human babesiosis caused by B. divergens-like parasites were documented from Missouri (Babesia sp. strain MO1), Washington, and Kentucky (Babesia sp. strain KY). All three patients were previously splenectomized, and one babesiosis case was fatal (16–18). The piroplasm 18S rRNA gene sequences from all three patients were identical to each other and close to that of European B. divergens (U16370), with a sequence identity between them of 99.8% (1,721/1,724 bp) (16). Parasites with identical 18S rRNA gene sequences to the B. divergens-like species were subsequently isolated from eastern cottontail rabbits (Sylvilagus floridanus) on Nantucket Island, Massachusetts (19), and antibodies against Babesia sp. MO1 were evident in eastern cottontail rabbits in Tennessee (20) as well, suggesting that this animal is a reservoir for this Babesia sp.
Very recently, we reported that sika deer (Cervus nippon) in Japan carried a Babesia parasite genetically closely related to but different from European (EU) B. divergens and Babesia sp. MO1 (in the United States [U.S.] lineage) (21). In phylogenetic analysis, these parasites assembled into a monophyletic clade (B. divergens/B. capreoli group) and were divided into distinct lineages that reflected geographical origin of the parasites (EU, U.S., and Asia lineages). These results raised concern of possible infection in humans and the necessity of elucidating the vector ticks for the parasites in this country. However, neither of the (suspected) vectors for B. divergens in Europe and the United States, I. ricinus and Ixodes dentatus (22), respectively, is distributed in Japan.
To examine a large number of ticks, we established a PCR system which could detect and discriminate between EU/U.S. B. divergens and B. divergens Asia lineages, and between the B. divergens parasites and Babesia venatorum (formerly Babesia sp. strain EU1). B. venatorum is genetically similar to B. divergens and recently emerged in patients in Europe (23) and China (24). Since both zoonotic parasites have been frequently detected in sympatric ticks (12, 14, 15, 25–27), we conducted a field survey on ticks collected in Hokkaido, Japan, where deer infected with B. divergens are most abundant in this country, and screened DNA from the ticks using the discriminatory PCR we developed.
RESULTS
Development of specific PCR.
By designing primers specific for the 18S rRNA genes of B. divergens Asia and EU/U.S. lineages and B. venatorum parasites, we developed a type-specific nested PCR. The specificity of this PCR system was examined by using plasmids carrying the 18S rRNA gene sequence of each parasite. The results are shown in Fig. 1C and demonstrate formation of a single positive signal that was specific for each parasite. Specificity was further tested by mixing two plasmid types together (1 × 108 copies of each) for every possible combination. In each case, only the target species was amplified (data not shown).
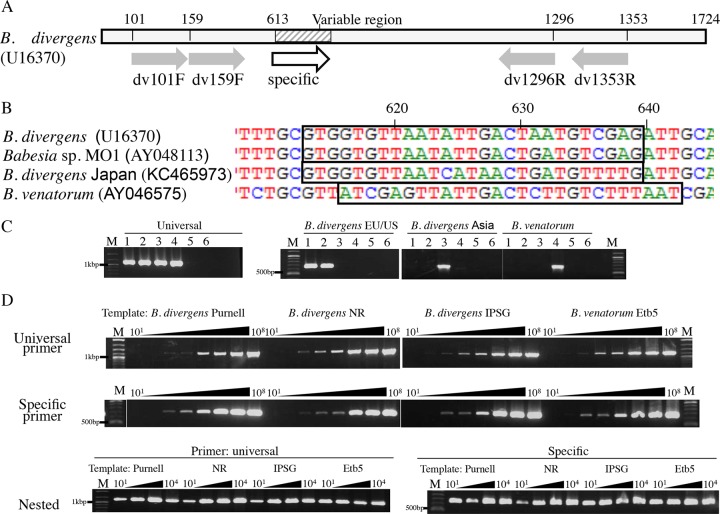
FIG 1.
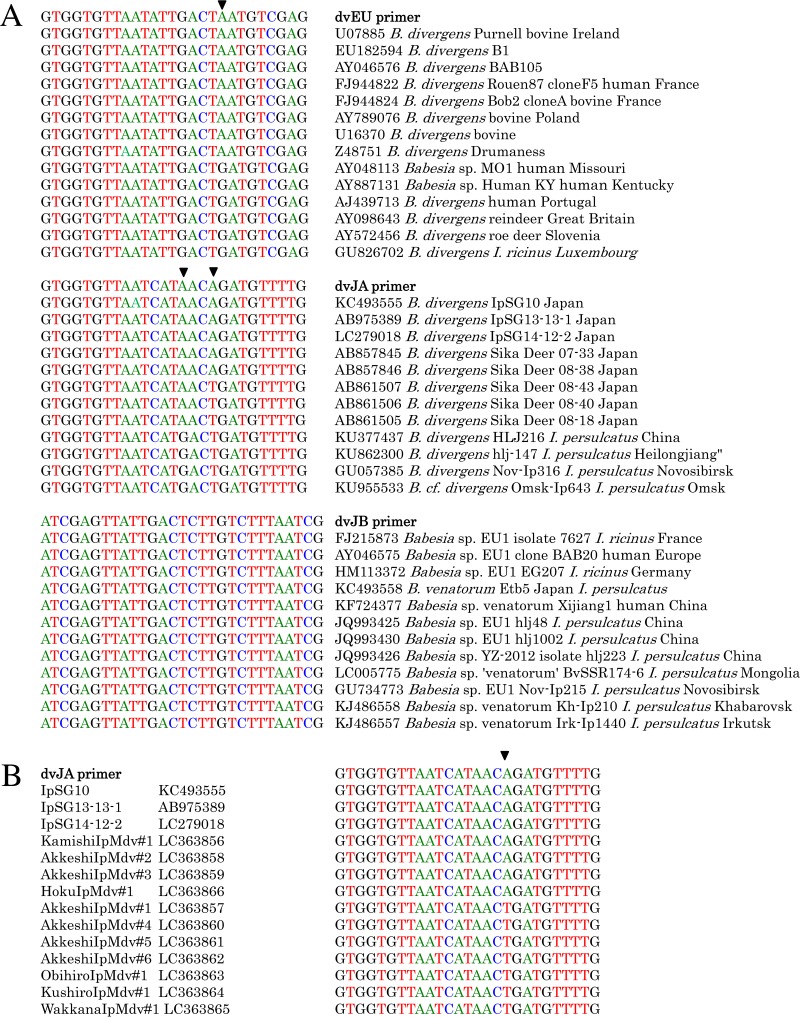
Type-specific PCR targeting the 18S rRNA genes of B. divergens and B. venatorum. (A) Schematic diagram of the PCR primer design. Numbers above the bar indicate nucleotide positions of designed primers in B. divergens (U16370). Gray and white arrows show positions of universal and specific primers, respectively. (B) Sequences of each lineage/species with boxes indicating the positions of the primers for dvEU (B. divergens and Babesia sp. MO1), dvJA (B. divergens, Japan) and dvJB (B. venatorum). (C) Specificity of the type-specific PCR. PCR amplification using universal primers dv159F and dv1296R or specific primers dvEU, dvJA, or dvJB/dv1296R. Plasmids carrying the 18S rRNA gene of B. divergens Purnell strain (EU lineage, lane 1), Babesia sp. NR strain (U.S. lineage, lane 2), B. divergens IpSG10 (Asia lineage, lane 3), B. venatorum Etb5 (lane 4), and negative controls (B. microti, lane 5, and Theileria sp., lane 6) were used as the template. M, marker. (D) Sensitivity of specific nested PCR. Conventional PCR using universal primers dv101F and dv1353R (upper panel) or specific primers dvEU, dvJA, or dvJB/dv1296R (middle panel) with the plasmid carrying the 18S rRNA gene of B. divergens strain Purnell (EU lineage), Babesia sp. strain NR (U.S. lineage), B. divergens IpSG10 (Asia lineage), and B. venatorum Etb5. Plasmids were diluted 10-fold from 1 × 101 to 1 × 108 copies and used (lanes 101 to 108). M, marker. Lower panel shows nested PCR amplification on the first 101 to 104 copy template PCR products from the universal primers shown above. Universal nested primers dv159F/dv1296R or specific nested primers dvEU, dvJA, or dvJB/dv1296R were used.
Sensitivity of the type-specific nested PCR was also examined by using 10-fold dilutions of plasmid (1 × 108 to 1 × 101 copies in a PCR mixture) as the template in the first-round PCR (Fig. 1D). Using either universal (top panel) or specific (middle panel) primers, products were observed at all template concentrations except at 1 × 101 and 1 × 102 copies of plasmid. In the nested PCR, in which first-round PCR products (universal primer) were used as the template, amplicons were visible at these lower concentrations (Fig. 1D, bottom panels).
The effectiveness of the dvJA primer was also evaluated in the presence of excess tick DNA by spiking tick DNA into PCR mixtures containing template B. divergens IpSG10 (Asia lineage) at a low copy number. Amplification of the target gene occurred in all samples whether or not excess tick DNA was present (not shown).
Detection of B. divergens and B. venatorum in Ixodes persulcatus.
The ticks collected at the 7 areas—Kamishihoro, Akkeshi, Obihiro, Kushiro, Kiyosato, Esashi and Wakkanai—in Hokkaido (Fig. 2, Table 1) were all adults. The ticks were comprised of 3 species, Ixodes persulcatus, I. ovatus, and Haemaphysalis spp. Almost all collected ticks, except for I. persulcatus females, were processed for PCR examination. Broad-spectrum PCR targeting the 18S rRNA genes of B. divergens and B. venatorum (Fig. 1) (21) revealed that 11 of 822 examined I. persulcatus males were positive, while all DNA samples from I. ovatus (n = 595) and Haemaphysalis spp. (n = 163) were negative (Table 1, “Universal”). Specific PCR, which discriminates B. divergens (EU/U.S. and Asia lineages) and B. venatorum, revealed that the 11 positive I. persulcatus males carried B. divergens Asia lineage DNA (Table 1). Infection rates calculated from I. persulcatus males in four areas with positive results ranged from 0.5% (1/180, Kamishihoro) to 7.6% (1/13, Wakkanai). All positive samples examined by PCR using the dvJA primer were confirmed by sequencing the 18S rRNA, β-tubulin, and CCT7 genes to be of the B. divergens Asia lineage (see Table 3).
FIG 2.
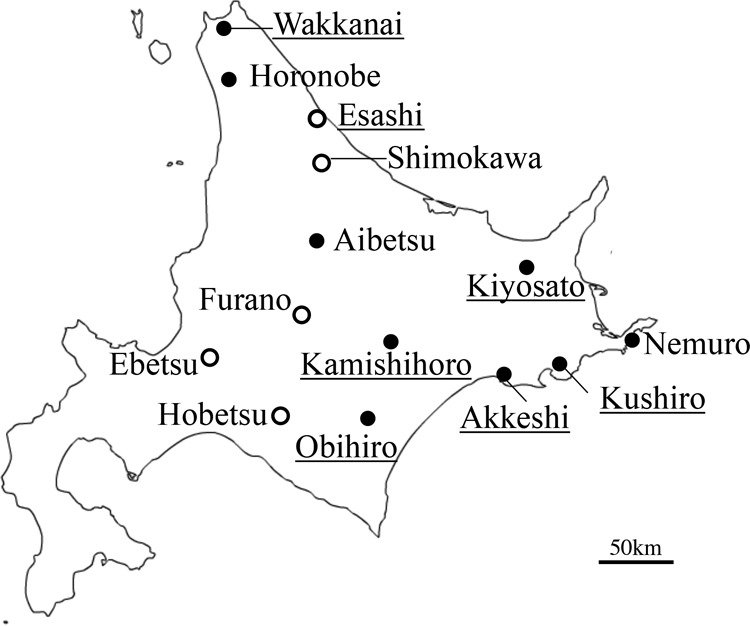
Tick survey areas in Hokkaido Island, Japan. Black and white circles show the areas where I. persulcatus ticks were PCR positive and negative, respectively, for B. divergens or B. venatorum. The areas where the ticks were collected during this study are underlined.
TABLE 1.
Detection of Babesia divergens and B. venatorum in field-collected Ixodes persulcatus ticks
| Area |
I. persulcatus |
I. ovatus adult |
Haemaphysalis sp. adult |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adult male (whole body) |
Adult female (salivary glands) |
|||||||||||||||||
| No. collected | No. examined | No. of positive PCR results for: |
No. collected | No. examined | No. of positive PCR results for: |
No. collected | No. examined | No. of positive PCR results, universal | No. collected | No. examined | No. of positive PCR results, universal | |||||||
| Universal | B. divergens Asia | B. divergens EU/U.S. | B. venatorum | Universal | B. divergens Asia | B. divergens EU/U.S. | B. venatorum | |||||||||||
| Kamishihoroa | 184 | 180 | 1 | 1 | 0 | 0 | 166 | 31 | 1b | 1b | 0 | 0 | 78 | 78 | 0 | 1 | 1 | 0 |
| Akkeshia | 335 | 334 | 7 | 7 | 0 | 0 | 321 | 34 | 1b | 1b | 0 | 0 | 309 | 309 | 0 | 142 | 142 | 0 |
| Obihiroa | 162 | 162 | 1 | 1 | 0 | 0 | 120 | 33 | 1b,c | 1b,c | 0 | 0 | 146 | 146 | 0 | 3 | 3 | 0 |
| Kushiro | 68 | 68 | 1 | 1 | 0 | 0 | 69 | NDd | ND | ND | ND | ND | 38 | 38 | 0 | 10 | 10 | 0 |
| Kiyosato | 49 | 49 | 0 | 0 | 0 | 0 | 47 | ND | ND | ND | ND | ND | 24 | 24 | 0 | 7 | 7 | 0 |
| Esashi | 18 | 16 | 0 | 0 | 0 | 0 | 14 | ND | ND | ND | ND | ND | 0 | 0 | 0 | 0 | 0 | 0 |
| Wakkanai | 13 | 13 | 1 | 1 | 0 | 0 | 16 | ND | ND | ND | ND | ND | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 829 | 822 | 11 | 11 | 0 | 0 | 753 | 98 | 3 | 3 | 0 | 0 | 595 | 595 | 0 | 163 | 163 | 0 |
Samples from Zamoto-Niikura et al. (2016) (29) were included.
Positive results were confirmed in salivary glands from partially engorged I. persulcatus.
B. microti U.S. lineage parasites from tick salivary glands were isolated in a hamster.
ND, not done.
TABLE 3.
Accession numbers determined in this study
| Species | Strain | Origin | Accession no. for: |
||
|---|---|---|---|---|---|
| 18S rRNA | β-Tubulin | CCT7 | |||
| B. divergens | IpSG10 | I. persulcatus female | KC493555 | KC493556 | AB975388 |
| IpSG13-13-1 | I. persulcatus female | AB975389 | AB975390 | AB975391 | |
| IpSG14-12-2 | I. persulcatus female | LC279018 | LC279019 | LC279020 | |
| KamishihoroIpMdv#1 | I. persulcatus male | LC363856 | LC363867 | LC363878 | |
| AkkeshiIpMdv#1 | I. persulcatus male | LC363857 | LC363868 | LC363879 | |
| AkkeshiIpMdv#2 | I. persulcatus male | LC363858 | LC363869 | LC363880 | |
| AkkeshiIpMdv#3 | I. persulcatus male | LC363859 | LC363870 | LC363881 | |
| AkkeshiIpMdv#4 | I. persulcatus male | LC363860 | LC363871 | LC363882 | |
| AkkeshiIpMdv#5 | I. persulcatus male | LC363861 | LC363872 | LC363883 | |
| AkkeshiIpMdv#6 | I. persulcatus male | LC363862 | LC363873 | LC363884 | |
| ObihiroIpMdv#1 | I. persulcatus male | LC363863 | LC363874 | LC363885 | |
| KushiroIpMdv#1 | I. persulcatus male | LC363864 | LC363875 | LC363886 | |
| WakkanaIpMdv#1 | I. persulcatus male | LC363865 | LC363876 | LC363887 | |
| HokuIpMdv#1 | I. persulcatus male | LC363866 | LC363877 | LC363888 | |
| NR Halpha1 | Eastern cottontail rabbit | NDa | MG229672 | MG229673 | |
| B. venatorum | Etb5 | I. persulcatus male | KC493558 | KC493557 | ND |
| B. gibsoni | Canine | ND | KC493559 | ND | |
| B. microti | IpSG14-12-2 | I. persulcatus female | ND | ND | LC333115 |
ND, no data.
To investigate additional ticks collected in other areas in Hokkaido (Fig. 2), archived DNA samples from I. persulcatus (previously tested for B. microti DNA; 28) were screened. By the broad-spectrum piroplasm PCR, 6 samples were found to be positive for Babesia DNA (Table 2). The specific nested PCR revealed that 5 samples and 1 sample were positive for B. divergens Asia lineage and B. venatorum (designated Etb5), respectively.
TABLE 2.
Detection of B. divergens and B. venatorum in DNA samples from ticks previously collected in additional areas in Hokkaidoa
| Collection area | Tick stage | n | Positive PCR result |
|||
|---|---|---|---|---|---|---|
| Universal |
B. divergens lineage |
B. venatorum | ||||
| Asia | EU/U.S. | |||||
| Nemuro | Adult | 139 | 1 | 1 | 0 | 0 |
| Nymph | 196 | 0 | 0 | 0 | 0 | |
| Horonobe | Adult | 42 | 2 | 2 | 0 | 0 |
| Kiyosato | Adult | 105 | 1 | 0 | 0 | 1 |
| Shimokawa | Adult | 15 | 0 | 0 | 0 | 0 |
| Aibetsu | Adult | 85 | 2 | 2 | 0 | 0 |
| Furano | Adult | 44 | 0 | 0 | 0 | 0 |
| Hobetsu | Adult | 36 | 0 | 0 | 0 | 0 |
| Ebetsu | Adult | 15 | 0 | 0 | 0 | 0 |
| Total | 677 | 6 | 5 | 0 | 1 | |
PCR was performed on the DNA samples previously prepared in Zamoto-Niikura et al. (2012) (28).
In all, I. persulcatus carrying B. divergens Asia lineage was detected as widely distributed across Hokkaido in 8 of the 14 sites examined (Fig. 2).
Genetic isolation of B. divergens Asia lineage from salivary glands of I. persulcatus.
To investigate whether I. persulcatus was competent for transstadial transmission of B. divergens, we attempted to genetically isolate mature sporozoites, which are produced in the salivary glands when the ticks feed. A total of 98 salivary glands from I. persulcatus females collected in Kamishihoro, Akkeshi, and Obihiro were examined after the ticks fed on gerbils (Table 1, “Adult female”). The specific PCR on DNAs extracted from the salivary glands revealed that 1 tick from each area carried B. divergens Asia lineage, i.e., IpSG13-13-1 (Kamishihoro), IpSG10 (Akkeshi), and IpSG14-12-2 (Obihiro).
B. divergens 18S rRNA, β-tubulin, and CCT7 gene sequences from I. persulcatus.
Partial 18S rRNA, β-tubulin, and CCT7 gene sequences of the B. divergens detected in I. persulcatus males (whole body) and females (salivary glands) were successfully amplified and sequenced. Comparison of the partial 18S rRNA genes (1,567 bp) of the isolates revealed that they were identical to each other, except for single nucleotide substitution at positions 256, 630, and 663 (position numbering based on GenBank reference sequence U16370). The 18S rRNA gene sequences were also identical to those of B. divergens in sika deer in Japan (GenBank accession numbers KC465973 to KC465977, AB857845, AB857846, and AB861504 to AB861507 [28]), except for the single-nucleotide polymorphism (SNP) noted above. In a BLAST search (July 2017), the 18S rRNA gene sequence of B. divergens isolate Nov-Ip316 in Russia (GenBank accession number GU057385, I. persulcatus origin) showed high sequence similarity (99.75%, 1,180/1,183 bp) to sequences from the B. divergens in I. persulcatus in Japan.
Partial β-tubulin sequences (1,347 bp, intron and exon) of all isolates in this study were identical to each other. Identical sequences were also found in sika deer in Japan (GenBank reference numbers KC465968 to KC465970 and AB861508 to AB861514).
CCT7 sequences (intron and exon) of B. divergens in I. persulcatus showed high identity, although SNPs (36 base substitutions in 1,647 bp) were observed. Among the 36 substitutions, 29 and 7 SNPs were seen in the exon and intron, respectively. All substitutions in the exon were in the third nucleotide position of the triplet genetic code, except for one substitution which occurred in the first position of the codon (position 669 in GenBank reference sequence AB367925). As a result, the CCT7 amino acid sequences of the isolates were identical except for one amino acid residue at position 190 (serine or glycine).
18S rRNA and β-tubulin sequences of B. venatorum Etb5.
By nested PCR, approximately 1,600 bp of 18S rRNA and 1,200 bp of β-tubulin gene sequences of B. venatorum Etb5 were successfully amplified and directly sequenced. A BLAST search of the partial 18S rRNA gene sequence (1,588 bp) revealed a similarity of 99.7% (1,584/1,588 bp) to those of B. venatorum (formerly Babesia sp. EU1) from a human (GenBank accession number AY046575), deer (GQ888709), and I. ricinus ticks (HM113372 and AY553915) in Europe and to B. venatorum from a human (KF724377) in China and I. persulcatus (LC005775) in Mongolia. A BLAST search of the Etb5 β-tubulin sequence (1,237 bp) identified sequences of B. odocoilei (GenBank accession number KC465972; 91%, 1,006/1,105 bp) and B. divergens (KC465967; 90%, 980/1,086 bp) as having the highest similarity.
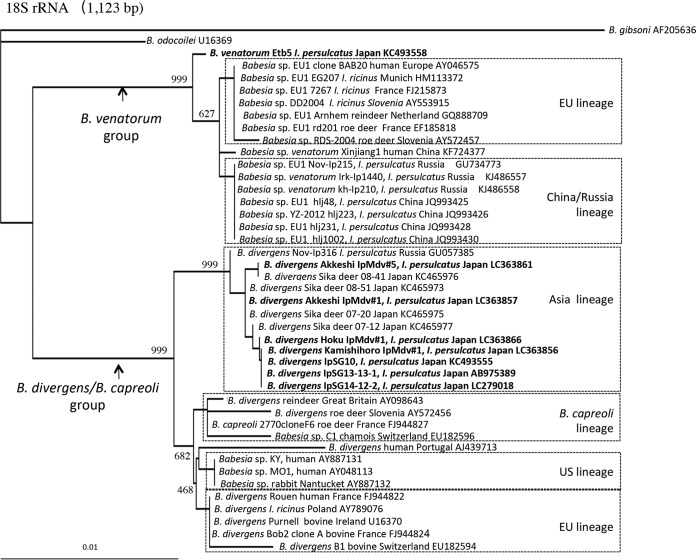
Phylogenetic analysis based on 18S rRNA gene sequences.
A neighbor-joining phylogenetic tree based on the 18S rRNA gene sequences (1,123 bp) obtained herein and those of related parasites available in GenBank was constructed (Fig. 3). The phylogenetic tree contained two large clades. One included B. venatorum Etb5, detected in this study, and B. venatorum (formerly Babesia sp. EU1) from Eurasian countries (B. venatorum group), and another clade (B. divergens/B. capreoli group) included B. divergens, B. capreoli, and Babesia sp. MO1. The newly identified B. divergens parasites from I. persulcatus adults in Japan fell into the B. divergens Asia lineage cluster and were closely related to isolates from sika deer in Japan (Fig. 3).
FIG 3.
Neighbor-joining phylogenetic tree based on 18S rRNA gene sequences. Names in bold indicate parasites isolated in this study. The number on each branch shows the occurrence in 1,000 bootstrap replicates.
The B. venatorum clade consisted of 2 or more lineages correlating with geographical origin, namely, EU and China/Russia lineages (Fig. 3). The EU lineage included B. venatorum (formerly Babesia sp. EU1) from the first index patient (GenBank accession number AY046575), I. ricinus ticks, and deer in Europe. The China/Russia lineage included B. venatorum from I. persulcatus in Russia and China. The newly identified B. venatorum Etb5 formed a distinct lineage from the EU and China/Russia lineages, with a high bootstrap value.
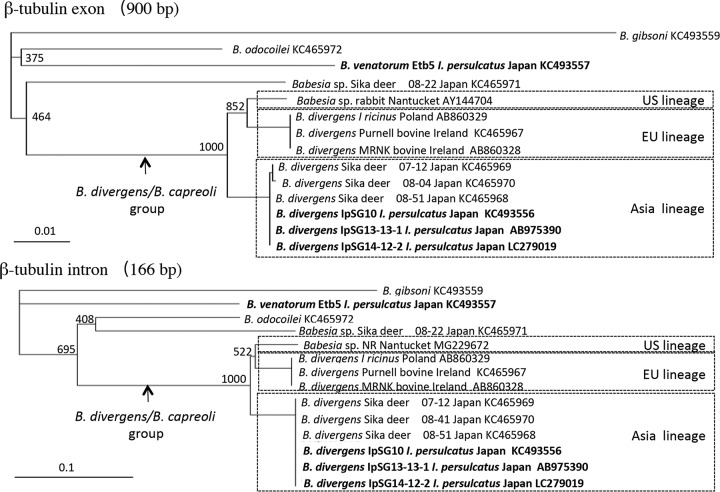
Phylogenetic analysis based on β-tubulin gene sequences.
To further investigate the phylogenetic placement of the Babesia parasites found in I. persulcatus in this study, a neighbor-joining phylogenetic tree was constructed based on the β-tubulin gene sequences (Fig. 4). The phylogenetic trees of the intron and exon sequences both showed similar topologies, with high bootstrap values compared to those in the tree based on the 18S rRNA gene (Fig. 4). A distinct branch comprising B. divergens and Babesia sp. MO1 (B. divergens/B. capreoli group) β-tubulin separated into 3 lineages that reflect the geographic distribution of the parasites (United States, Europe, and Japan). Comparing the lengths of the distance bar and bootstrap values in the β-tubulin tree to those for the 18S rRNA gene, these lineages appear distinct.
FIG 4.
Neighbor-joining phylogenetic tree based on β-tubulin gene sequences. Names in bold indicate parasites isolated in this study. The number on each branch shows the occurrence in 1,000 bootstrap replicates.
Isolation of B. microti U.S. lineage IpSG14-12-2.
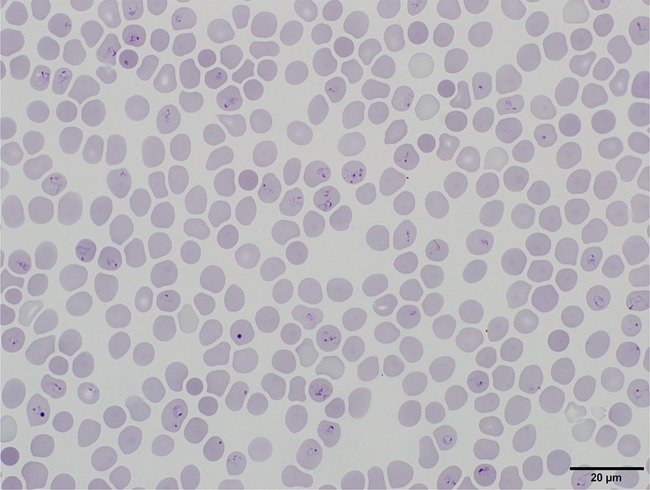
Salivary glands containing B. divergens IpSG14-12-2 were positive for B. microti U.S. lineage when examined by PCR (Fig. 5), and therefore a portion of the homogenate was inoculated into a naive hamster. Ten days after inoculation, parasites were observed in hamster erythrocytes in a stained blood film under microscopy (Fig. 6). The parasite was subsequently isolated (B. microti strain IpSG14-12-2) and confirmed to be genetically of B. microti U.S. lineage by sequencing the CCT7 gene (LC333115).
FIG 5.
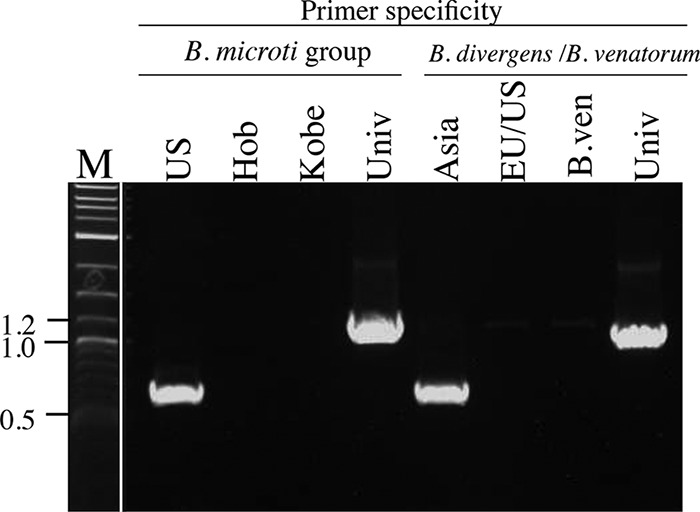
IpSG14-12-2, isolated from salivary glands of an I. persulcatus tick, contained both B. microti and B. divergens DNA, detected by type-specific PCR targeting 18S rRNA genes of B. microti, B. divergens, and B. venatorum. Specific primers for U.S., Hobetsu, and Kobe lineages within the B. microti group and universal primers for all lineages were used (lanes US, Hob, Kobe and Univ, respectively). Specific primers for Asia and EU/U.S. lineages of B. divergens and B. venatorum within the B. divergens/B. venatorum group and universal primers were used (lanes: Asia, EU/US, B.ven and Univ, respectively). M, marker.
FIG 6.
Intraerythrocytic B. microti U.S. lineage parasites (IpSG14-12-2) isolated in a hamster from infected I. persulcatus salivary glands in Japan. Giemsa-stained blood film. Bar, 20 μm.
Genetic stability of the specific primer region of the 18S rRNA gene.
Since specific primers were designed from a variable region of the 18S rRNA gene, we examined genetic stability by comparing this region in sequences available in GenBank. In the region where dvEU and dvJA primers were derived, 1 and 2 SNPs, respectively, were found (Fig. 7A). Guanine replaced an adenine at position 19 of dvEU and at position 15 in dvJA. Thymidine substituted for adenine at position 18 in dvJA. The latter SNP was also found in some of the B. divergens Asia lineage sequences determined in this study from I. persulcatus (Fig. 7B).
FIG 7.
SNPs in the 18S rRNA gene region where specific primers were designed. (A) Corresponding regions of B. divergens and B. venatorum available in GenBank. (B) Corresponding region of the B. divergens Asia lineage detected in I. persulcatus, Japan.
DISCUSSION
We describe herein I. persulcatus in Japan carrying parasites of the B. divergens Asia lineage, which was recently identified from sika deer in this country (21). By using a specific PCR system established in this study (Fig. 1), I. persulcatus ticks carrying the parasite were identified as widely distributed over the north, central, and eastern parts of Hokkaido, while samples from I. ovatus, a sympatric and abundant tick, were all negative for this parasite (Fig. 2, Tables 1 and 2). The 18S rRNA and β-tubulin gene sequences of the B. divergens Asia lineage isolated from salivary glands of I. persulcatus were identical to those found in sika deer blood (Fig. 3 and 4), suggesting that the parasite in Japan, and perhaps also throughout the temperate zones of the eastern Eurasian continent, is maintained in an enzootic cycle involving I. persulcatus ticks and sika deer.
We modified a previously established PCR system (Fig. 1) to specifically detect and easily classify the 18S rRNA gene sequences of B. divergens Asia and EU/U.S. lineages and B. venatorum. In Hokkaido these Babesia species, as well as B. microti U.S. and Hobetsu lineages, are carried in ixodid ticks (28, 29). Therefore, a specific PCR system was crucial in this study for detecting B. divergens Asia lineage in ticks without the need for sequencing. Furthermore, this system will be greatly advantageous for large-scale epidemiological surveys for B. divergens, especially in investigating sika deer, because Theileria infection is also abundant in this host (30, 31). In most cases B. divergens infection in sika deer is found as a coinfection with Theileria, with the Theileria parasitemia generally much higher. The massive Theileria infection may consequently mask the B. divergens infection so that PCR using broad primers designed for piroplasm DNA often fails to detect the more limited B. divergens sequences (unpublished data).
I. persulcatus carrying B. divergens Asia lineage was proven to be spread over north, central, and eastern parts of Hokkaido (8/14 areas; Fig. 2 and Tables 1 and 2). This tick species is also distributed broadly over the eastern half of Japan, where sika deer carrying the lineage are frequently found (21), suggesting that I. persulcatus is the main vector for this parasite in this country. Rar et al. (25) reported that I. persulcatus collected in the Novosibirsk region, Russia, also carried B. divergens (Nov-Ip316). In our analysis, the 18S rRNA gene sequence of the Russian isolate Nov-Ip316 (GenBank accession number GU057385) was nearly identical to that of B. divergens from I. persulcatus in Japan (except for 3 out of 1,183 bases), and it consequently fell into the Asia lineage clade in the phylogenetic tree (Fig. 3). Thus, it is very likely that the Asia lineage of B. divergens is transmitted primarily by I. persulcatus throughout the temperate zones of the eastern Eurasian continent where I. persulcatus is commonly distributed.
Evidence showing an association of B. divergens infection with ticks that bite people raises a concern about the emergence of human babesiosis in the region. Notably, Qi et al. reported B. divergens infection in anemic patients in China (Shandong province) examined by PCR and sequencing (32). Although we were unable to include the Chinese B. divergens isolate in our genetic analysis due to its short reported sequence, nonetheless this information suggests that B. divergens in Asia may be infectious to humans and cause mild illness.
The phylogenetic tree based on the 18S rRNA gene sequences revealed that B. divergens is a complex composed of four lineages: the Asia, EU, and U.S. lineages of B. divergens, and one minor assemblage (Fig. 3 and 4), which corresponds to a previous study (21). The minor assemblage (B. capreoli lineage) includes parasites of different names, such as B. capreoli, Babesia sp., and B. divergens. The phylogenetic tree based on the β-tubulin (exon and intron) gene sequences also supports the separation of B. divergens into three geographic lineages, U.S., EU, and Asia.
The various lineages are related to different vector tick species. The EU lineage of B. divergens, a main causative agent of human babesiosis in Europe, is transmitted primarily by I. ricinus ticks. In North America, I. dentatus has been reported to carry the U.S. lineage of B. divergens (22). For the Asia lineage of B. divergens, I. persulcatus is found to be the principal vector (Tables 1 and 2). All of these tick species, including I. persulcatus, are genetically very closely related (I. ricinus species complex) and are found across the temperate zones of the Northern Hemisphere with distinct geographic distributions (33). We speculate that an ancestor of the I. ricinus complex was infected with B. divergens and expanded its habitat. As a result of recent coevolution between the parasites and vector ticks, we observe that each of the three lineages of B. divergens is primarily vectored by regional ticks belonging to the I. ricinus species complex. A similar coevolutionary perspective has been described to explain the close relationship between the three sublineages in the B. microti phylogroup (34, 35). The Europe-Central Asia, East Asia, and North American B. microti sublineages are primarily transmitted by I. ricinus, I. persulcatus (28), and I. scapularis (I. dammini) (36), respectively. To elucidate their precise coevolutionary history, transmission studies and analysis of a greater number of samples from the various geographical origins and various tick species are needed.
The speculation of the coevolutionary history described above is also supported by the fact that B. divergens Asia lineage is specifically transmitted by I. persulcatus. In this survey, the majority of the ticks collected in Hokkaido were I. persulcatus and I. ovatus, which accounted for about 90% of all ticks collected (Table 1). This collection was advantageous for the purpose of the survey, since these species are the common ticks that feed on sika deer (37). Furthermore, the geographical distributions of both ticks largely overlap in Hokkaido (37–40). Therefore, it is considered likely that these two tick species have an equal opportunity to ingest deer blood infected with B. divergens. However, PCR examination revealed that only I. persulcatus carried B. divergens Asia lineage. None of the I. ovatus ticks tested positive, even in the Akkeshi and Obihiro areas where the number of I. ovatus ticks examined was comparable to that of I. persulcatus (Table 1). These results strongly suggest a species-specific interaction between the tick and parasite in nature. I. ovatus (subgenus Partipalpiger), an ancient type of ixodid tick, is genetically different from the I. ricinus complex (41, 42). We speculate that the ancestral I. ricinus complex has been infected recently with B. divergens, long after I. ricinus evolved and separated from the I. ovatus clade.
In this study, we demonstrated that a tick could be coinfected with B. divergens and B. microti. When B. divergens-positive salivary gland homogenate (IpSG14-12-2) from an I. persulcatus female that also tested positive for B. microti was inoculated into a naive hamster, protozoa emerged in the erythrocytes (Fig. 6). Based on the sequence of the CCT7 gene, the parasite was genetically identical to the B. microti U.S. lineage, which is maintained between rodents and I. persulcatus in Japan (29). Although we did not exclude the possibility that the I. persulcatus female ingested both B. divergens and B. microti at same time as a nymph, the reservoirs for the two piroplasms—sika deer and rodents, respectively—are quite different. Rather, we speculate that a persistently B. divergens-infected I. persulcatus tick fed on a rodent infected with B. microti U.S. lineage as a nymph, since B. divergens is reported to survive through all three stages of a tick generation. Donnelly and Peirce (9) demonstrated that all stages of the F1 generation resulting from an infected I. ricinus female tick could transmit B. divergens to cattle. Furthermore, in some cases the infection persisted until the F2 larval stage. Bonnet et al. (10) also showed that B. divergens could persist in I. ricinus beyond more than one molt by detecting B. divergens DNA in the nymphal stage after feeding on nonparasitized blood as infected larvae. On the other hand, B. microti is unable to persist in I. ricinus ticks beyond one instar (43).
Another possible mechanism for the mixed infection is that there may be an unknown reservoir(s) which is susceptible to both B. divergens and B. microti. In the United States, parasites identical to B. divergens sp. MO1 (U.S. lineage) were isolated from an unexpected host, the cottontail rabbit (44). In Hokkaido, Japan, lagomorphs, including the pika (Ochotona hyperborea) and mountain hare (Lepus timidus), are also distributed (45), but they have not been investigated for piroplasm infections. It may be worthwhile to survey such wild animals to elucidate the life cycle of piroplasms, despite the rare opportunity to do so.
B. venatorum, formerly Babesia sp. EU1, was first described in 2003 as the agent infecting asplenic patients in Italy and Austria (23). An additional human case was reported in Germany (46). Through epidemiological surveys in Europe (47–49) and successful transmission experiments (50, 51), I. ricinus was recognized as a competent vector for B. venatorum. In this study, specific PCR and sequencing identified the presence of B. venatorum in I. persulcatus in Japan. Although only one B. venatorum-infected I. persulcatus tick was detected in this study, very closely related sequences were also reported from I. persulcatus in the Novosirsk region of Russia (GenBank accession number GU734773; 25) and in China (JQ993425, JQ993428, and JQ993430) (Fig. 3), suggesting geographically wide distribution of the parasite over eastern Eurasia where I. persulcatus exists.
The genetic regions where the specific 18S rRNA gene primers were designed are variable, and a few SNPs were noted when the corresponding regions of the 18S rRNA genes of various B. divergens isolates were aligned with primers dvJA and dvEU. Nevertheless, amplicons were generated from the Babesia sp. NR813 strain, whose sequence is identical to those of Babesia sp. MO1 and Babesia sp. KY, using primer dvEU in the PCR system (Fig. 1C and D). Similarly, 18S rRNA gene sequences of B. divergens Asia lineage were newly amplified in this study by nested specific PCR using the dvJA primer, even though some of these possessed a single nucleotide substitution (Fig. 7A and B). These results suggested that single substitution in those positions had little effect, possibly because the SNPs were internal and not at the critical 3′ end. Future studies might include modification of the primers or conditions (e.g., different annealing temperature) to safeguard against possible effects attributable to the primer sequence.
MATERIALS AND METHODS
Field collections.
Unfed host-seeking ticks were collected by flagging vegetation alongside trails in forests on Hokkaido Island, where sika deer previously were found to be most prevalently infected with B. divergens (33%) in Japan (21). The survey areas in this study (Kamishihoro, Akkeshi, Obihiro, Kushiro, Kiyosato, Esashi, and Wakkanai) (Table 1) are shown in Fig. 2. Species identification was performed by morphological examination under microscopy of the collected ticks, as described by Takada (38) and Ehara (39). In addition, genetic identification (mitochondrial 16S rRNA and cytochrome oxidase gene sequencing) was performed on ticks which were morphologically suspect or positive for Babesia spp. We also examined DNA extracted from ticks (either individual or pooled samples from 2 to 5 ticks) that were collected from other areas in Hokkaido in a previous study (28) (Fig. 2, Table 2).
Extraction of DNA from ticks.
Ticks were individually crushed with a pestle homogenizer (Scientific Specialties, Inc.) and suspended in 300 μl of TNE buffer (10 mM Tris-HCl, 150 mM NaCl, and 100 mM EDTA; pH 8.0) containing 0.1% sodium dodecyl sulfate (SDS). The suspensions were digested with 100 μg/ml proteinase K at 55°C overnight. DNAs were purified by phenol extraction followed by ethanol precipitation. To facilitate visualization of the DNA pellets, Glyco blue (Ambion) was used as the carrier. Pellets were resuspended in 50 μl of TE buffer (10 mM Tris-HCl and 1 mM EDTA; pH 7.5). The final double-stranded DNA (dsDNA) concentration of the samples was approximately 2 × 103 ng/μl (Qubit; Thermo Fisher Scientific).
Type-specific PCR.
To detect and discriminate between B. divergens Asia and EU/U.S. lineages (21) and between B. divergens and B. venatorum, a PCR for specific amplification was developed based on 18S rRNA gene sequences (Fig. 1) and modification of previously described primers (primers dvEU and dvJB are 8 bases longer, respectively, than the primers BDV and BOD described by Duh et al. [52]). The forward primers dvEU (5′-GTGGTGTTAATATTGACTAATGTCGAG-3′; specific for the EU and U.S. lineages within B. divergens), dvJA (5′-GTGGTGTTAATCATAACAGATGTTTTG-3′; specific for the Asia lineage in B. divergens), and dvJB (5′-ATCGAGTTATTGACTCTTGTCTTTAATCG-3′; specific for B. venatorum) were designed and used with reverse primer dv1296R (5′-CGGACGAACCTTTTTACGGACACTAG-3′) (21) (Fig. 1).
Questing ticks carried parasites at levels too low for detection in the first-round PCR, so for the epidemiological study nested specific PCR was performed on the first-round products using primers dv101F (5′-ACAACAGTTATAGTTTCTTTGGTATTCG-3′) and dv1353R (5′-GCCTTAAACTTCCTTGCGGCTTAGAGC-3′) (21), which broadly anneal to the 18S rRNA gene sequence of B. divergens and B. venatorum DNA (Fig. 1).
Parasites in the activated salivary glands could be detected by conventional PCR.
18S rRNA, β-tubulin, and chaperonin containing TCP1 subunit eta (CCT7) gene amplification and sequencing.
Nested PCR using the primers dv101F/dv1353R and then dv159F/dv1296R was performed to universally amplify the 18S rRNA gene of B. divergens and closely related zoonotic parasites (B. venatorum) according to Zamoto-Niikura et al. (21) (Fig. 1). Sequences of β-tubulin and chaperonin containing TCP1 subunit eta (CCT7) genes of B. divergens were amplified according to Zamoto-Niikura et al. (21).
Each PCR mixture contained 200 μM each of deoxynucleoside triphosphate (dNTP), 0.4 μM of each primer, 1 μl of DNA, and 0.5 U of Ex Taq DNA polymerase (TaKaRa Bio) in 20 μl total volume. Positive controls were purified amplicons of the target sequence, and negative controls contained no DNA. Thermal cycling was carried out in a Mastercycler ep (Eppendorf) with 30 cycles of denaturation at 94°C for 10 s, annealing at 55°C for 30 s, and extension at 72°C for 90 s, and final extension at 72°C for 5 min. Nucleotide sequences of the 18S rRNA, β-tubulin and CCT7 gene amplicons were determined directly on the PCR products with the primers used for amplification and additional internal primers for CCT7, i.e., BdivCCTSQ1F (5′-TTTACAGGTCCKAGGGGCATGGACAAGC-3′), BdivCCTSQ2F (5′-GCTGAGACGTCACTAAATTCRAAGCTACT-3′), BdivCCTSQ3F (5′-AAGGCCACYGGAGCRTCCATACAGACCAC-3′), and BdivCCTSQ1R (5′-ACAAGCMGYGCGGAAGTACTTAATGATGACTTGTG-3′).
Specificity and sensitivity of the type-specific PCR.
Partial 18S rRNA gene sequences from B. divergens strain Purnell (53), Babesia sp. (B. divergens) strain NR813 (44), B. divergens IpSG10, B. microti U.S. lineage isolated from I. persulcatus in Japan (strain IpSG13-1-2) (29), B. venatorum strain Etb5, and Theileria sp. (GenBank accession number AB012199) from I. persulcatus in Japan were amplified by PCR (29) and cloned individually into pCR2.1 (Thermo Fisher Scientific). After sequence confirmation, the concentration of each plasmid was diluted to 1 × 108 copies/μl. The copy number of the plasmid was calculated based on the concentration and length of plasmid using an online copy number calculator (http://cels.uri.edu/gsc/cndna.html).
The specificity of the type-specific primers was examined on all of the plasmids generated above containing 18S rRNA gene inserts for the various piroplasms, using 108 copies of the plasmid as the template in a PCR mixture (Fig. 1C). The PCR procedure was as described above. Specificity was further evaluated by PCR using a mixture of 108 copies of B. divergens strain Purnell plasmid with 108 copies of B. microti, B. venatorum strain Etb5, or Theileria sp. plasmid as the template with dvEU and dv1296R primers. PCR was also similarly conducted with the specific primers for B. divergens IpSG or B. venatorum, using mixtures of two plasmids as the template. Finally, specificity was evaluated by mixing plasmid-carrying B. divergens IpSG10 (Asia lineage) at 1 × 101, 1 × 102, and 1 × 103 copies with 6 × 103 ng of tick DNA (3 times as concentrated as normal) for the template in nested PCR and using the dvJA primer in the second PCR.
Serial 10-fold dilutions were prepared in water from the 108 copy stocks, and 1 μl of each dilution was used in a PCR mixture to examine the sensitivity of the type-specific PCR for B. divergens strain Purnell, B. divergens NR813, B. divergens IpSG, and B. venatorum strain Etb5 (Fig. 1D). The PCR procedure was as described above.
Isolation of B. divergens genomic DNA from salivary glands of I. persulcatus females.
I. persulcatus female ticks collected in areas of Akkeshi, Kamishihoro, and Obihiro where Babesia is endemic were used for genetic isolation of B. divergens. Ticks were fed on gerbils (a noncompetent host) for 4 days to activate quiescent salivary gland sporozoites to multiply. Thereafter, the partially engorged ticks were removed manually. Salivary glands were dissected from ticks individually under a stereomicroscope and homogenized in cold phosphate-buffered saline (PBS) with a tissue grinder (glass wall tissue grinder; Radnoti) (54). DNA extraction from the homogenates and subsequent PCRs were performed according to the methods described above.
Detection and isolation of B. microti U.S. lineage from tick salivary glands.
Since I. persulcatus is demonstrated to be a vector for B. microti U.S. lineage in Japan (29), DNA extracted from salivary glands was examined for the presence of this parasite by specific PCR based on the β-tubulin gene sequence as previously described (55). Salivary gland homogenates testing positive for B. microti were inoculated into hamsters as previously described (29).
Phylogenetic analysis.
Partial 18S rRNA (1,123 bp) and β-tubulin (exon, 900 bp; intron, 166 bp) gene sequences of B. divergens from salivary glands of I. persulcatus females (IpSG10, IpSG13-13-1, and IpSG14-12-2) and B. venatorum (Etb5) from an I. persulcatus male were aligned with closely related sequences available from GenBank. 18S rRNA gene sequences of B. divergens from I. persulcatus males were also included (the sequences identical to the ones used were not included). Phylogenetic trees of the β-tubulin gene were constructed separately, based on the exon and intron sequences of the gene. Accession numbers are shown in the phylogenetic trees (Fig. 3 and 4). Babesia gibsoni was used as the outgroup. The multiple sequence alignments and construction of the phylogenetic trees using the neighbor-joining method were done by ClustalW (56). Consensus phylogenetic trees were built from 1,000 bootstrap repetitions.
Genetic stability of the 18S rRNA variable region gene sequences used in primer design.
Specific species primers were designed from the 18S rRNA gene sequence spanning a variable region (Fig. 1A) for B. venatorum and the various B. divergens strains. Corresponding sequences of related isolates from geographically distant countries (or regions) of origin were downloaded from GenBank, aligned and compared (Fig. 7).
Laboratory animals.
A specific-pathogen-free (SPF) gerbil 30 weeks of age (MON/JmsGbsSlc, retired) and Syrian hamster (slc:syrian) 3 weeks of age were purchased from Japan SLC, Inc.
Reference strains.
For sequence comparison, genomic DNA from B. divergens strain MRNK (21), B. divergens from I. ricinus collected in Poland (21), and Babesia sp. strain NR (44) was used.
Accession number(s).
Sequences determined in this study were deposited in the International Nucleotide Sequence Database (INSD) (GenBank or DDBJ) under the accession numbers listed in Table 3.
ACKNOWLEDGMENTS
We thank Haruyuki Hirata (Rakuno-Gakuen University) for his assistance. The research was supported by AMED under grant number JP17fk0108217.
REFERENCES
- 1.Skrabalo Z, Deanovic Z. 1957. Piroplasmosis in man; report of a case. Doc Med Geogr Trop 9:11–16. [PubMed] [Google Scholar]
- 2.Lewis D, Purnell RE, Shaw SR. 1980. The isolation and characterization of human and bovine strains of Babesia divergens from Drumnadrochit, Scotland. Parasitology 81:145–155. doi: 10.1017/S0031182000055116. [DOI] [PubMed] [Google Scholar]
- 3.Gorenflot A, Moubri K, Precigout E, Carcy B, Schetters TP. 1998. Human babesiosis. Ann Trop Med Parasitol 92:489–501. [DOI] [PubMed] [Google Scholar]
- 4.Martinot M, Zadeh M, Hansmann Y, Grawey I, Christmann D, Aguillon S, Jouglin M, Chauvin A, De Briel D. 2011. Babesiosis in immunocompetent patients, Europe. Emerg Infect Dis 17:114–116. doi: 10.3201/eid1701.100737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnleitner S, Fritz J, Bednarska M, Baumgartner R, Simeoni J, Zelger R, Schennach H, Lass Flörl C, Edelhofer R, Pfister K, Milhakov A, Walder G. 2014. Risk assessment of transfusion-associated babesiosis in Tyrol: appraisal by seroepidemiology and polymerase chain reaction. Transfusion 54:1725–1732. doi: 10.1111/trf.12606. [DOI] [PubMed] [Google Scholar]
- 6.Hunfeld KP, Lambert A, Kampen H, Albert S, Epe C, Brade V, Tenter AM. 2002. Seroprevalence of Babesia infections in humans exposed to ticks in midwestern Germany. J Clin Microbiol 40:2431–2436. doi: 10.1128/JCM.40.7.2431-2436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lempereur L, Shiels B, Heyman P, Moreau E, Saegerman C, Losson B, Malandrin L. 2015. A retrospective serological survey on human babesiosis in Belgium. Clin Microbiol Infect 21:96.e1–96.e7. doi: 10.1016/j.cmi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Rigaud E, Jaulhac B, Garcia Bonnet N, Hunfeld KP, Féménia F, Huet D, Goulvestre C, Vaillant V, Deffontaines G, Abadia Benoist G. 2016. Seroprevalence of seven pathogens transmitted by the Ixodes ricinus tick in forestry workers in France. Clin Microbiol Infect 22:735.e1–735.e9. doi: 10.1016/j.cmi.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly J, Peirce MA. 1975. Experiments on the transmission of Babesia divergens to cattle by the tick Ixodes ricinus. Int J Parasitol 5:363–367. doi: 10.1016/0020-7519(75)90085-5. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet S, Jouglin M, Malandrin L, Becker C, Agoulon A, L'Hostis M, Chauvin A. 2007. Transstadial and transovarial persistence of Babesia divergens DNA in Ixodes ricinus ticks fed on infected blood in a new skin-feeding technique. Parasitology 134:197–207. doi: 10.1017/S0031182006001545. [DOI] [PubMed] [Google Scholar]
- 11.Lewis D, Young ER. 1980. The transmission of a human strain of Babesia divergens by Ixodes ricinus ticks. J Parasitol 66:359–360. doi: 10.2307/3280841. [DOI] [PubMed] [Google Scholar]
- 12.Lempereur L, Wirtgen M, Nahayo A, Caron Y, Shiels B, Saegerman C, Losson B, Linden A. 2012. Wild cervids are host for tick vectors of Babesia species with zoonotic capability in Belgium. Vector Borne Zoonotic Dis 12:275–280. doi: 10.1089/vbz.2011.0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gigandet L, Stauffer E, Douet V, Rais O, Moret J, Gern L. 2011. Prevalence of three zoonotic Babesia species in Ixodes ricinus (Linne, 1758) nymphs in a suburban forest in Switzerland. Vector Borne Zoonotic Dis 11:363–366. doi: 10.1089/vbz.2010.0195. [DOI] [PubMed] [Google Scholar]
- 14.Øines Ø, Radzijevskaja J, Paulauskas A, Rosef O. 2012. Prevalence and diversity of Babesia spp. in questing Ixodes ricinus ticks from Norway. Parasit Vectors 5:156–156. doi: 10.1186/1756-3305-5-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaschitz M, Narodoslavsky Gföller M, Kanzler M, Stanek G, Walochnik J. 2008. Babesia species occurring in Austrian Ixodes ricinus ticks. Appl Environ Microbiol 74:4841–4846. doi: 10.1128/AEM.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beattie JF, Michelson ML, Holman PJ. 2002. Acute babesiosis caused by Babesia divergens in a resident of Kentucky. N Engl J Med 347:697–698. doi: 10.1056/NEJM200208293470921. [DOI] [PubMed] [Google Scholar]
- 17.Herwaldt B, Persing DH, Precigout EA, Goff WL, Mathiesen DA, Taylor PW, Eberhard ML, Gorenflot AF. 1996. A fatal case of babesiosis in Missouri: identification of another piroplasm that infects humans. Ann Intern Med 124:643–650. doi: 10.7326/0003-4819-124-7-199604010-00004. [DOI] [PubMed] [Google Scholar]
- 18.Herwaldt BL, de Bruyn G, Pieniazek NJ, Homer M, Lofy KH, Slemenda SB, Fritsche TR, Persing DH, Limaye AP. 2004. Babesia divergens-like infection, Washington State. Emerg Infect Dis 10:622–629. doi: 10.3201/eid1004.030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holman PJ, Spencer AM, Telford SR III, Goethert HK, Allen AJ, Knowles DP, Goff WL. 2005. Comparative infectivity of Babesia divergens and a zoonotic Babesia divergens-like parasite in cattle. Am J Trop Med Hyg 73:865–870. [PubMed] [Google Scholar]
- 20.Fritzen C, Mosites E, Applegate R, Telford S, Huang J, Yabsley M, Carpenter LR, Dunn J, Moncayo A. 2014. Environmental investigation following the first human case of babesiosis in Tennessee. J Parasitol 100:106–109. doi: 10.1645/12-158.1. [DOI] [PubMed] [Google Scholar]
- 21.Zamoto-Niikura A, Tsuji M, Imaoka K, Kimura M, Morikawa S, Holman P, Hirata H, Ishihara C. 2014. Sika deer carrying Babesia parasites closely related to B. divergens, Japan. Emerg Infect Dis 20:1398–1400. doi: 10.3201/eid2008.130061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goethert HK, Telford SR III. 2003. Enzootic transmission of Babesia divergens among cottontail rabbits on Nantucket Island, Massachusetts. Am J Trop Med Hyg 69:455–460. [PubMed] [Google Scholar]
- 23.Herwaldt BL, Caccio S, Gherlinzoni F, Aspock H, Slemenda SB, Piccaluga P, Martinelli G, Edelhofer R, Hollenstein U, Poletti G, Pampiglione S, Loschenberger K, Tura S, Pieniazek NJ. 2003. Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg Infect Dis 9:942–948. doi: 10.3201/eid0908.020748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J-F, Zheng Y-C, Jiang R-R, Li H, Huo Q-B, Jiang B-G, Sun Y, Jia N, Wang Y-W, Ma L, Liu H-B, Chu Y-L, Ni X-B, Liu K, Song Y-D, Yao N-N, Wang H, Sun T, Cao W-C. 2015. Epidemiological, clinical, and laboratory characteristics of 48 cases of “Babesia venatorum” infection in China: a descriptive study. Lancet Infect Dis 15:196–203. doi: 10.1016/S1473-3099(14)71046-1. [DOI] [PubMed] [Google Scholar]
- 25.Rar VA, Epikhina TI, Livanova NN, Panov VV. 2011. Genetic diversity of Babesia in Ixodes persulcatus and small mammals from North Ural and West Siberia, Russia. Parasitology 138:175–182. doi: 10.1017/S0031182010001162. [DOI] [PubMed] [Google Scholar]
- 26.Katargina O, Geller J, Vasilenko V, Kuznetsova T, Jarvekulg L, Vene S, Lundkvist A, Golovljova I. 2011. Detection and characterization of Babesia species in Ixodes ticks in Estonia. Vector Borne Zoonotic Dis 11:923–928. doi: 10.1089/vbz.2010.0199. [DOI] [PubMed] [Google Scholar]
- 27.Hilpertshauser H, Deplazes P, Schnyder M, Gern L, Mathis A. 2006. Babesia spp. identified by PCR in ticks collected from domestic and wild ruminants in southern Switzerland. Appl Environ Microbiol 72:6503–6507. doi: 10.1128/AEM.00823-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamoto-Niikura A, Tsuji M, Qiang W, Nakao M, Hirata H, Ishihara C. 2012. Detection of two zoonotic Babesia microti lineages, the Hobetsu and U.S. lineages, in two sympatric tick species, Ixodes ovatus and Ixodes persulcatus, respectively, in Japan. Appl Environ Microbiol 78:3424–3430. doi: 10.1128/AEM.00142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zamoto-Niikura A, Morikawa S, Hanaki KI, Holman PJ, Ishihara C. 2016. Ixodes persulcatus ticks as vectors for the Babesia microti U.S. lineage in Japan. Appl Environ Microbiol 82:6624–6632. doi: 10.1128/AEM.02373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata S. 2010. Undergraduate thesis. Rakuno Gakuen University, Hokkaido, Japan. [Google Scholar]
- 31.Inokuma H, Tsuji M, Kim S-J, Fujimoto T, Nagata M, Hosoi E, Arai S, Ishihara C, Okuda M. 2004. Phylogenetic analysis of Theileria sp. from sika deer, Cervus nippon, in Japan. Vet Parasitol 120:339–345. doi: 10.1016/j.vetpar.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Qi C, Zhou D, Liu J, Cheng Z, Zhang L, Wang L, Wang Z, Yang D, Wang S, Chai T. 2011. Detection of Babesia divergens using molecular methods in anemic patients in Shandong Province, China. Parasitol Res 109:241–245. doi: 10.1007/s00436-011-2382-8. [DOI] [PubMed] [Google Scholar]
- 33.Chao LL, Wu WJ, Shih CM. 2011. Species identification of Ixodes granulatus (Acari: Ixodidae) based on internal transcribed spacer 2 (ITS2) sequences. Exp Appl Acarol 54:51–63. doi: 10.1007/s10493-010-9419-z. [DOI] [PubMed] [Google Scholar]
- 34.Fujisawa K, Nakajima R, Jinnai M, Hirata H, Zamoto-Niikura A, Kawabuchi-Kurata T, Arai S, Ishihara C. 2011. Intron sequences from the CCT7 gene exhibit diverse evolutionary histories among the four lineages within the Babesia microti-group, a genetically related species complex that includes human pathogens. Jpn J Infect Dis 64:403–410. [PubMed] [Google Scholar]
- 35.Nakajima R, Tsuji M, Oda K, Zamoto-Niikura A, Wei Q, Kawabuchi-Kurata T, Nishida A, Ishihara C. 2009. Babesia microti-group parasites compared phylogenetically by complete sequencing of the CCTeta gene in 36 isolates. J Vet Med Sci 71:55–68. doi: 10.1292/jvms.71.55. [DOI] [PubMed] [Google Scholar]
- 36.Spielman A, Wilson ML, Levine JF, Piesman J. 1985. Ecology of Ixodes dammini-borne human babesiosis and Lyme disease. Annu Rev Entomol 30:439–460. doi: 10.1146/annurev.en.30.010185.002255. [DOI] [PubMed] [Google Scholar]
- 37.Isogai E, Isogai H, Masuzawa T, Postic D, Baranton G, Kamewaka Y, Kimura K, Nishikawa T, Fuji N, Ishii N, Ohno S, Yamaguti N. 1996. Borrelia burgdorferi sensu lato in an endemic environment: wild sika deer (Cervus nippon yesoensis) with infected ticks and antibodies. Microbiol Immunol 40:13–19. doi: 10.1111/j.1348-0421.1996.tb03311.x. [DOI] [PubMed] [Google Scholar]
- 38.Takada N. 1990. A pictorial review of medical acarology in Japan. Kinpodo, Kyoto, Japan: (In Japanese.) [Google Scholar]
- 39.Ehara S. 1980. Family Ixodidae. In Ehara S. (ed), Illustrations of the mites and ticks in Japan. Zenkoku Noson Kyoiku Kyokai, Tokyo, Japan: (In Japanese.) [Google Scholar]
- 40.Yamaguti N, Tipton VJ, Keegan HL, Toshioka S. 1971. Ticks of Japan, Korea, and the Ryukyu Islands. Brigham Young Univ Sci Bull Biol Ser 15:1–226. [Google Scholar]
- 41.Fukunaga M, Yabuki M, Hamase A, Oliver JH Jr, Nakao M. 2000. Molecular phylogenetic analysis of ixodid ticks based on the ribosomal DNA spacer, internal transcribed spacer 2, sequences. J Parasitol 86:38–43. doi: 10.1645/0022-3395(2000)086[0038:MPAOIT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 42.Dunlop J, Apanaskevich D, Lehmann J, Hoffmann R, Fusseis F, Ehlke M, Zachow S, Xiao X. 2016. Microtomography of the Baltic amber tick Ixodes succineus reveals affinities with the modern Asian disease vector Ixodes ovatus. BMC Evol Biol 16:203. doi: 10.1186/s12862-016-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray J, von Stedingk L, Gürtelschmid M, Granström M. 2002. Transmission studies of Babesia microti in Ixodes ricinus ticks and gerbils. J Clin Microbiol 40:1259–1263. doi: 10.1128/JCM.40.4.1259-1263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holman PJ, Spencer AM, Droleskey RE, Goethert HK, Telford SR III. 2005. In vitro cultivation of a zoonotic Babesia sp. isolated from eastern cottontail rabbits (Sylvilagus floridanus) on Nantucket Island, Massachusetts. J Clin Microbiol 43:3995–4001. doi: 10.1128/JCM.43.8.3995-4001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe H. 1994. A pictorial guide to the mammals of Japan. Japan Wildlife Research Center, Tokyo, Japan: (In Japanese.) [Google Scholar]
- 46.Haselbarth K, Tenter AM, Brade V, Krieger G, Hunfeld KP. 2007. First case of human babesiosis in Germany—clinical presentation and molecular characterisation of the pathogen. Int J Med Microbiol 297:197–204. doi: 10.1016/j.ijmm.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Duh D, Petrovec M, Avsic-Zupanc T. 2005. Molecular characterization of human pathogen Babesia EU1 in Ixodes ricinus ticks from Slovenia. J Parasitol 91:463–465. doi: 10.1645/GE-394R. [DOI] [PubMed] [Google Scholar]
- 48.Welc-Faleciak R, Bajer A, Paziewska-Harris A, Baumann-Popczyk A, Sinski E. 2012. Diversity of Babesia in Ixodes ricinus ticks in Poland. Adv Med Sci 57:364–369. doi: 10.2478/v10039-012-0023-9. [DOI] [PubMed] [Google Scholar]
- 49.Becker CA, Bouju-Albert A, Jouglin M, Chauvin A, Malandrin L. 2009. Natural transmission of zoonotic Babesia spp. by Ixodes ricinus ticks. Emerg Infect Dis 15:320–322. doi: 10.3201/eid1502.081247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonnet S, Brisseau N, Hermouet A, Jouglin M, Chauvin A. 2009. Experimental in vitro transmission of Babesia sp. (EU1) by Ixodes ricinus. Vet Res 40:21. doi: 10.1051/vetres/2009004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnet S, Jouglin M, L'Hostis M, Chauvin A. 2007. Babesia sp. EU1 from roe deer and transmission within Ixodes ricinus. Emerg Infect Dis 13:1208–1210. doi: 10.3201/eid1308.061560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duh D, Petrovec M, Bidovec A, Avsic-Zupanc T. 2005. Cervids as Babesiae hosts, Slovenia. Emerg Infect Dis 11:1121–1123. doi: 10.3201/eid1107.040724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holman PJ, Madeley J, Craig TM, Allsopp BA, Allsopp MT, Petrini KR, Waghela SD, Wagner GG. 2000. Antigenic, phenotypic and molecular characterization confirms Babesia odocoilei isolated from three cervids. J Wildl Dis 36:518–530. doi: 10.7589/0090-3558-36.3.518. [DOI] [PubMed] [Google Scholar]
- 54.Zamoto-Niikura A, Tsuji M, Qiang W, Hirata H, Nakajima R, Morikawa S, Holman PJ, Ishihara C. 2015. Field survey and molecular analysis of Babesia microti group. In Epidemiology II: theory, research and practice, 1st ed iConcept Press Ltd, Kowloon, Hong Kong. [Google Scholar]
- 55.Zamoto A, Tsuji M, Kawabuchi T, Wei Q, Asakawa M, Ishihara C. 2004. U.S.-type Babesia microti isolated from small wild mammals in Eastern Hokkaido, Japan. J Vet Med Sci 66:919–926. doi: 10.1292/jvms.66.919. [DOI] [PubMed] [Google Scholar]
- 56.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]