ABSTRACT
Chronic, biofilm-based bacterial infections are exceptionally difficult to eradicate due to the high degree of antibiotic recalcitrance exhibited by cells in biofilm communities. In the opportunistic pathogen Pseudomonas aeruginosa, biofilm recalcitrance is multifactorial and arises in part from the preferential expression of resistance genes in biofilms compared to exponential-phase planktonic cells. One such mechanism involves ndvB, which we have previously shown to be expressed specifically in biofilms. In this study, we investigated the regulatory basis of this lifestyle-specific expression by developing an unstable green fluorescent protein (GFP) transcriptional reporter to observe the expression pattern of ndvB. We found that in addition to its expression in biofilms, ndvB was upregulated in planktonic cells as they enter stationary phase. The transcription of ndvB in both growth phases was shown to be dependent on the stationary-phase sigma factor RpoS, and mutation of a putative RpoS binding site in the ndvB promoter abolished the activity of the promoter in stationary-phase cells. Overall, we have expanded our understanding of the temporal expression of ndvB in P. aeruginosa and have uncovered a regulatory basis for its growth phase-dependent expression.
IMPORTANCE Bacterial biofilms are more resistant to antibiotics than free-living planktonic cells, and understanding the mechanistic basis of this resistance can inform treatments of biofilm-based infections. In addition to chemical and structural barriers that can inhibit antibiotic entry, the upregulation of specific genes in biofilms contributes to the resistance. We investigated this biofilm-specific gene induction by examining expression patterns of ndvB, a gene involved in biofilm resistance of the opportunistic pathogen Pseudomonas aeruginosa. We characterized ndvB expression in planktonic and biofilm growth conditions with an unstable green fluorescent protein (GFP) reporter and found that the expression of ndvB in biofilms is dependent on the stationary-phase sigma factor RpoS. Overall, our results support the physiological similarity between biofilms and stationary-phase cells and suggest that the induction of some stationary-phase genes in biofilms may contribute to their increased antibiotic resistance.
KEYWORDS: Pseudomonas aeruginosa, antibiotic resistance, biofilms, ndvB, RpoS, stationary phase
INTRODUCTION
Antibiotic resistance in bacterial pathogens is a continuing public health concern that severely impacts the ability to treat infections (1–3). Increased resistance in bacterial populations is caused by the acquisition of resistance genes through horizontal gene transfer and by selection of resistance mutations (4, 5). In addition, numerous structural and functional traits contribute to the underlying intrinsic resistance of microbes, which can reduce the spectrum and efficiency of antibiotics used in treatments (3, 6). Identifying the genetic and physiological mechanisms of intrinsic resistance in microbes can potentially inform the therapeutic use of already proven antibiotics.
Antibiotic resistance is especially problematic for cystic fibrosis patients harboring chronic biofilm-based Pseudomonas aeruginosa lung infections. P. aeruginosa has high intrinsic resistance to multiple antibiotics that, combined with its ability to readily acquire elevated resistance by de novo mutation, can make infections impossible to eliminate (7). The intrinsic resistance is mediated by drug efflux pumps, stress responses, low outer membrane permeability, and other poorly understood genetic functions (8–10). Resistance is additionally increased in biofilms, the predominant lifestyle of P. aeruginosa in cystic fibrosis airway infections (11). Bacterial biofilms are a physiological state characterized by bacterial adhesion to a substrate and the production of an extracellular matrix composed of polysaccharides, protein, and DNA (12, 13). Multiple physical and physiological factors contribute to the increased antibiotic resistance of P. aeruginosa biofilms compared to free-living planktonic cultures, including reduced penetration of the antibiotic through the extracellular matrix and the presence of persister cells, which exist in a metabolically inactive state that protects against antibiotic action (14–17).
Biofilm resistance is also mediated by several genetic mechanisms that are specifically expressed in biofilms but not in exponential-phase planktonic cells (18–23). One of these mechanisms requires the gene ndvB, which encodes a glucosyltransferase enzyme that catalyzes the synthesis of periplasmic β-(1→3)-cyclic glucans (18). The glucans are thought to promote aminoglycoside resistance by sequestering antibiotics in the periplasm away from their cellular target (18, 24). In addition, ndvB is required, via an uncharacterized mechanism, for the biofilm-specific expression of genes that are involved in ethanol oxidation, and these ndvB-regulated genes may also play a role in biofilm resistance (25). The expression of ndvB is elevated in biofilms in comparison to that in exponentially growing planktonic cultures (18); however, the regulatory mechanism(s) responsible for this upregulation is unknown. Investigation of the genetic and physiological requirements of ndvB transcription may uncover the basis of its biofilm-specific expression and potentially identify conditions that diminish ndvB expression and reduce antibiotic resistance of biofilms.
In the present study, we examined transcriptional activity of ndvB and investigated the environmental and genetic factors governing its regulation. We used an unstable green fluorescent protein (GFP) variant as a reporter of ndvB promoter activity to examine the gene's temporal expression in live cells (26, 27). Importantly, GFP is functional in bacterial biofilms and has been extensively used to visualize P. aeruginosa cells in biofilms (28–31). The resulting observations motivated genetic tests that demonstrated the requirement of the stationary-phase sigma factor RpoS for ndvB induction in biofilms.
RESULTS
ndvB transcriptional fusions to unstable gfp variants.
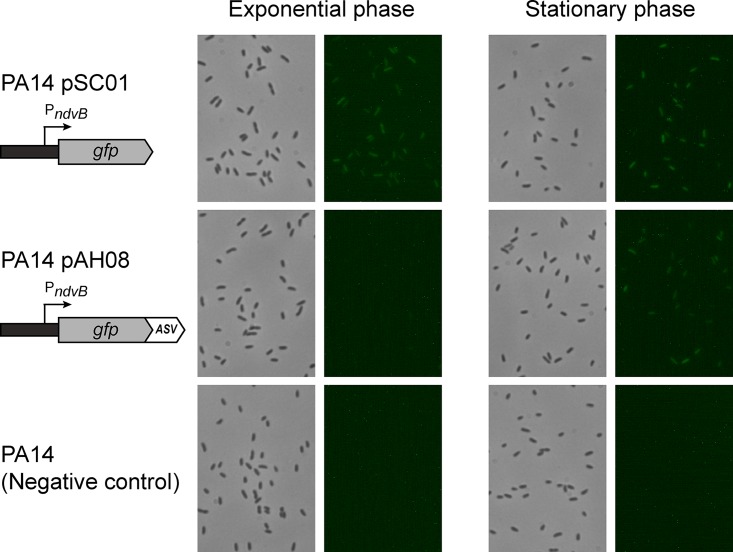
We constructed a gfp transcriptional reporter to visualize the expression of ndvB in P. aeruginosa cells. The 500-bp sequence located immediately upstream of the ndvB translational start site was inserted upstream of the gfp gene in the shuttle vector pMQ80 using a yeast recombination-based cloning method (see Materials and Methods) (32, 33). The transcriptional fusion was constructed such that the gfp reporter retained its own Shine-Dalgarno sequence that was present in pMQ80. The resulting vector (pSC01) was transformed into P. aeruginosa (PA14), and GFP expression in cell cultures was examined by fluorescence microscopy. Reverse transcriptase PCR experiments have shown that ndvB transcription is preferentially expressed in PA14 biofilms compared to exponential-phase planktonic cultures, which exhibit negligible ndvB expression (18). However, we observed constitutive fluorescence in exponential-phase planktonic cells carrying the pSC01 reporter (Fig. 1). Thus, while the PndvB-gfp reporter was clearly functional (i.e., GFP was actively expressed), its fluorescence poorly corresponded to established ndvB expression patterns.
FIG 1.
Expression of PndvB-gfp reporters in planktonic cultures. Cultures of the stable ndvB-gfp reporter (PA14/pSC01), the ASV-tagged reporter (PA14/pAH08), and a negative control (PA14) were diluted (OD600 = 0.05) in M63-arginine medium and incubated at 37°C with shaking. Samples were removed after 6 h (exponential phase) and 12 h (stationary phase) for phase-contrast (left panels) and fluorescence (right panels) microscopy at a magnification of ×630. The brightness was increased in each of the fluorescent panels for image clarity. The images are representative of at least three replicate cultures.
We suspected that the constitutive fluorescence of the pSC01 reporter was due to the long half-life of GFP, which has been estimated to be greater than 24 h in Escherichia coli and Pseudomonas fluorescens (34). High GFP stability in the ndvB reporter could cause steady-state fluorescence even in the absence of ndvB transcription. To address this possibility, we utilized unstable GFP variants which contain 13-amino-acid tags fused to the C terminus of GFP. These tags target proteins for degradation by tail-specific proteases (34, 35), and the rate of proteolysis is dependent on the sequences of the final three amino acids. Thus, by altering these sequences, GFP variants with a range of stabilities can be generated (34).
We fused four variants of the protease tag to the C terminus of GFP in the ndvB reporter vector (pSC01) by yeast recombination cloning (pAH05 to pAH08 [Table 1]). We transformed the four unstable GFP reporter plasmids (which differed only in the final three amino acids of the protease tag) into PA14 and examined fluorescence during planktonic growth. For each of the reporters, fluorescence was absent in exponential cultures, suggesting that the tags effectively reduced the half-life of GFP (Fig. 1 and data not shown). However, three of the variants (pAH05 to pAH07) failed to fluoresce under any growth conditions tested, including biofilms (a known inducing condition for ndvB expression), indicating that they were too unstable to visualize ndvB transcription. The fluorescence of the ASV-tagged GFP variant (pAH08) was not completely abolished, as evidenced by faint, but reproducible, fluorescence observed in stationary-phase cultures (Fig. 1). Although ndvB expression was not expected in this growth phase, the ASV-tagged variant was a more promising ndvB reporter candidate and was characterized further.
TABLE 1.
Strains and plasmids used in this study
| Name | Genotype or description | Reference |
|---|---|---|
| Strains | ||
| PA14 | P. aeruginosa burn wound isolate | 67 |
| TFM15 | PA14 ΔndvB; unmarked deletion of ndvB (codons 273–727 deleted) | 18 |
| TFM425 | PA14 ΔrpoS; unmarked deletion of rpoS (codons 15–272 deleted) | This study |
| TFM423 | PA14 attTn7::mini-Tn7T-Gm-lacZ; promoterless lacZ; Gmr | This study |
| TFM427 | PA14 attTn7::mini-Tn7T-Gm-PndvB-lacZ; PndvB-lacZ fusion; Gmr | This study |
| TFM456 | PA14 attTn7::mini-Tn7T-Gm-PndvB-mut-lacZ; PndvB-lacZ fusion with mutated RpoS binding site; Gmr | This study |
| DH5α | E. coli strain; supE44 ΔlacU169(ϕ80 lacZΔM15) recA1 hsdR17 thi-1 relA1 | 81 |
| S17-1 | E. coli strain; recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | 82 |
| YPH500 | Saccharomyces cerevisiae uracil auxotroph; MATα ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 | 83 |
| Plasmids | ||
| pMQ80 | araC-PBAD-gfpmut3 reporter on E. coli-S. cerevisiae-Pseudomonas shuttle vector; Gmr | 33 |
| pSC01 | PndvB-gfp stable gfp reporter; pMQ80 with ndvB promoter upstream of gfpmut3; Gmr | This study |
| pAH04 | araC-PBAD-gfp (ASV) reporter; pMQ80 with gfp protease tag from pJBA113; Gmr | This study |
| pAH05 | PndvB-gfp (LAA) reporter; pSC01 with gfp protease tag from pJBA110; Gmr | This study |
| pAH06 | PndvB-gfp (LVA) reporter; pSC01 with gfp protease tag from pJBA111; Gmr | This study |
| pAH07 | PndvB-gfp (AAV) reporter; pSC01 with gfp protease tag from pJBA112; Gmr | This study |
| pAH08 | PndvB-gfp (ASV) reporter; pSC01 with gfp protease tag from pJBA113; Gmr | This study |
| pJBA110 | pUC18Not-PA1/04/03-RBSII-gfp (LAA)-T0-T1; source of LAA protease tag; Apr | 34 |
| pJBA111 | pUC18Not-PA1/04/03-RBSII-gfp (LVA)-T0-T1; source of LVA protease tag; Apr | 34 |
| pJBA112 | pUC18Not-PA1/04/03-RBSII-gfp (AAV)-T0-T1; source of AAV protease tag; Apr | 34 |
| pJBA113 | pUC18Not-PA1/04/03-RBSII-gfp (ASV)-T0-T1; source of ASV protease tag; Apr | 34 |
| pEX18Gm | ||
| pEX18Gm::ΔrpoS | pEX18Gm derivative for unmarked deletion of rpoS; Gmr | This study |
| pUC18-mini-Tn7T-Gm-lacZ | Mini-Tn7T transcriptional lacZ fusion vector; Apr Gmr | 68 |
| pUC18-mini-Tn7T-Gm-PndvB-lacZ | pUC18-mini-Tn7T-Gm-lacZ carrying the sequence from −112 to −1 relative to the translational start site of ndvB; Apr Gmr | This study |
| pUC18-mini-Tn7T-Gm-PndvB-mut-lacZ | Same as pUC18-mini-Tn7T-Gm-PndvB-lacZ except that the sequence CTAGACT was mutated to AGAGACT; Apr Gmr | This study |
| pTNS2 | Transposase helper plasmid for mini-Tn7 integration; Apr | 68 |
| pRK2013 | Helper plasmid for triparental matings; Kmr | 72 |
| pUC18 | High-copy-number E. coli cloning/sequencing plasmid; Apr | 84 |
| pUCP19 | Broad-host-range cloning and expression vector; Cbr | 85 |
| pUCP19::rpoS+ | pUCP19 with rpoS under the control of its native promoter and terminator; Cbr | This study |
Stability of ASV-tagged GFP in PA14.
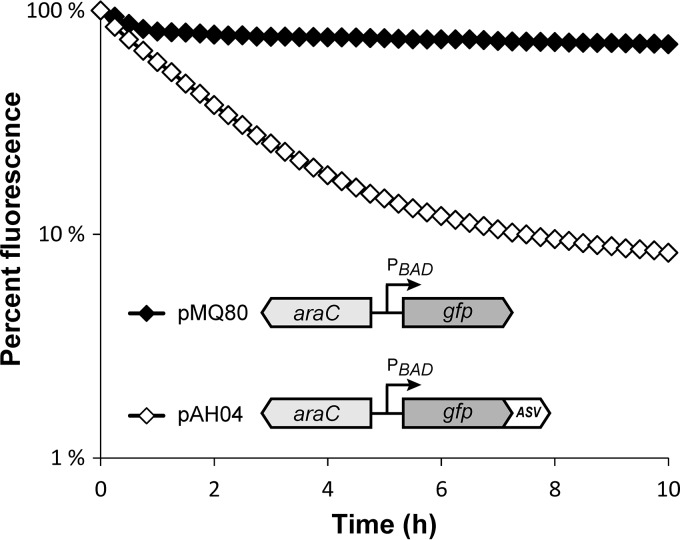
To directly measure the impact of the ASV tag on GFP stability, we examined the kinetics of GFP decay in PA14 using arabinose-inducible GFP constructs (pMQ80 and pAH04 [Table 1]). Exponential-phase planktonic cultures were grown in arabinose-containing rich medium to induce GFP expression and then shifted to arabinose-free minimal medium to block further expression. GFP stabilities were determined by quantifying the reduction in fluorescence over time with a plate reader. As shown in Fig. 2, cultures expressing the ASV-tagged GFP construct (pAH04) exhibited a pronounced decline in fluorescence in comparison to cultures with the untagged construct (pMQ80). We calculated the GFP half-lives from decay constants (λ) estimated during intervals in which the decline in fluorescence was exponential. The half-life of ASV-tagged GFP was approximately 1.9 h for the first 5 h after the downshift (λ = −0.386; R2 = 0.99) and increased to approximately 6.5 h for the next 5 h (time [T] = 5 h to 10 h; λ = −0.107; R2 = 0.96). In contrast, following a small reduction immediately after the downshift, untagged GFP was extremely stable over the course of the experiment, with an estimated half-life of 53 h (T = 1 h to 10 h; λ = −0.013; R2 = 0.97). These results confirmed that the introduction of the ASV tag substantially reduces the fluorescent half-life of GFP in PA14 planktonic cultures. Notably, the decay slowed at the later time points, suggesting that the nutrient availability and/or physiological state of the cultures can influence the proteolysis rate.
FIG 2.
Stability of unmodified and ASV-tagged GFP in P. aeruginosa. Exponential cultures of PA14 harboring arabinose-inducible unmodified GFP (pMQ80) and ASV-tagged GFP (pAH04) were treated with arabinose for 4 h to induce GFP expression. The cultures were washed to inhibit further induction, and GFP fluorescence was measured at intervals for 10 h. The fluorescence is given as percentage of initial fluorescence of eight replicate cultures (mean ± SEM; the error bars are smaller than the symbols).
Expression of the ASV-tagged ndvB-gfp reporter in biofilms.
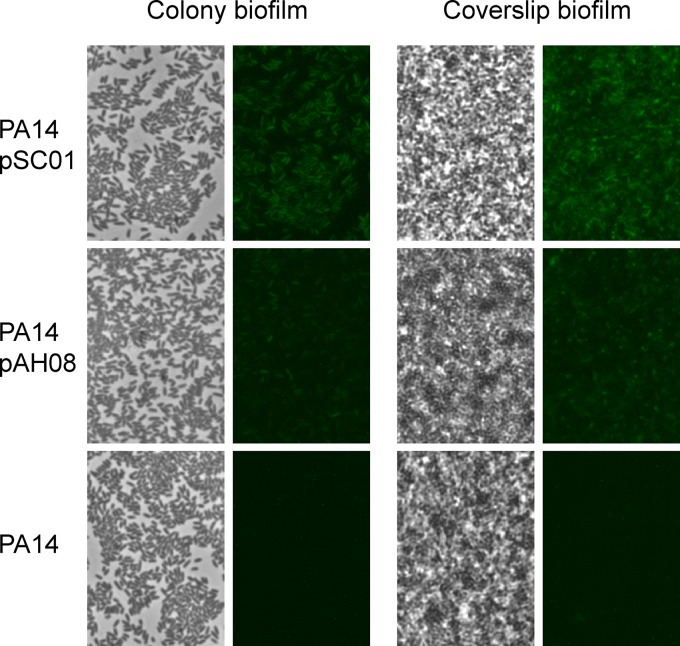
The reduced GFP half-life caused by the ASV protease tag enabled the visualization of ndvB transcriptional downregulation in planktonic exponential-phase cultures (Fig. 1). We next characterized the fluorescence of the ndvB reporters in biofilms, in which upregulation of ndvB transcription has been well documented (18, 25). Biofilms were grown on the air-liquid interface of glass coverslips submerged in growth medium or as colony biofilms on agar plates (36) (see Materials and Methods). Biofilms of the reporter strains were visually indistinguishable from wild-type biofilms, indicating that the vectors did not influence biofilm formation (Fig. 3; phase panels). Biofilm cells carrying the stable GFP reporter (pSC01) showed robust fluorescence (Fig. 3). Importantly, fluorescence was also evident for the unstable reporter (pAH08) but to a lesser degree, which was consistent with the reduced stability of ASV-tagged GFP (Fig. 3). Thus, based on the increased fluorescence of biofilms compared to exponential-phase planktonic cultures (Fig. 1), we concluded that the fluorescence of the ASV-tagged reporter conformed to previously established ndvB expression patterns.
FIG 3.
Expression of PndvB-gfp reporters in biofilms. Biofilms were grown at the air-liquid interface of coverslips partially submerged in M63-arginine medium (coverslip biofilms) and on semipermeable membranes on M63-arginine agar plates (colony biofilms). The coverslip biofilms were rinsed and the coverslips placed on a slide for phase-contrast (left panels) and fluorescence (right panels) microscopy at a magnification of ×630. The colony biofilms were stamped onto a slide for imaging. The images are representative of those from at least three replicate biofilm cultures.
ndvB transcription is induced in stationary-phase planktonic cultures.
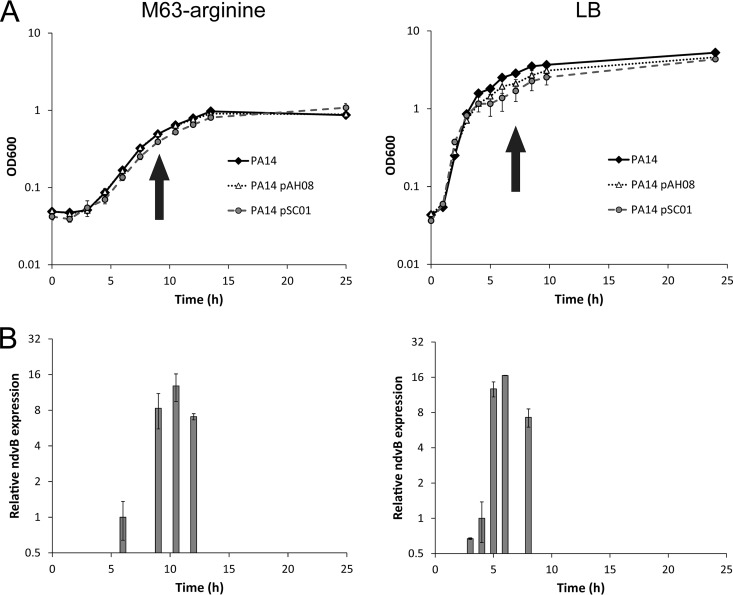
We next used the ndvB transcriptional reporter to explore the temporal regulation of ndvB expression. In particular, we were interested in identifying specific conditions that induce ndvB transcription, which might hint at the mechanism(s) underlying its regulation. As described for the planktonic expression experiments, we observed fluorescence in overnight cultures of PA14 carrying the ASV-tagged PndvB-gfp reporter (pAH08), suggesting that ndvB might be expressed in stationary-phase planktonic cultures in addition to biofilms (Fig. 1). We examined planktonic expression of the PndvB-gfp reporters in more detail by observing the fluorescence of planktonic cells throughout a growth curve (Fig. 4A). Subcultures of PA14 carrying the unstable GFP reporter (pAH08) showed little or no fluorescence during the lag and exponential phases of growth in both LB and minimal M63 media. However, at early stationary phase, fluorescence was detected in both media, indicating that ndvB promoter activity is induced at the onset of stationary phase (Fig. 4). This pattern was also observed with the stable GFP reporter (pSC01) as a transient reduction of fluorescence intensity in exponential-phase cells, which we interpret as dilution of existing GFP by cell division, followed by induction and increased fluorescence intensity at stationary phase (data not shown). The GFP induction was observed at the same phase of the growth curve in both LB and M63 growth media.
FIG 4.
ndvB is upregulated in stationary-phase planktonic cultures. (A) Growth curves of planktonic cultures of PA14 carrying the stable (pSC01) and ASV-tagged (pAH08) ndvB-gfp reporters grown in M63-arginine (left) or LB (right) medium. At each time point, samples were removed for optical density measurements and microscopy. Arrows indicate the earliest observation of fluorescence for the unstable reporter (pAH08), which coincides with the entry to stationary phase. (B) Expression of ndvB was measured by qPCR in PA14 planktonic cultures corresponding to the time points of the growth curves in panel A. Expression was quantified at four time points for the M63 cultures (6 h, 9 h, 10.5 h, and 12 h) and five time points for the LB cultures (3 h, 4 h, 5 h, 6 h, and 8 h). ndvB expression is given relative to the 6-h time point for M63 or the 4-h time point for LB. The mean and SEM are presented for the data from three replicate cultures assayed in triplicate qPCRs.
Although the fluorescence profile of the pAH08 reporter agreed with ndvB transcriptional patterns in exponential-phase and biofilm cultures, it was possible that the stationary-phase fluorescence might be an artifact of the reporter system. For example, GFP proteolysis might be compromised during stationary phase and allow accumulation of GFP in the absence of significant transcriptional upregulation. Therefore, we used qPCR to validate the microscopic observation by comparing relative ndvB mRNA levels at time points corresponding to different planktonic growth phases. The results corroborated those observed with the gfp reporter. The early stationary-phase cells contained greater ndvB transcript levels than exponential-phase cells in both LB and M63 cultures, and this induction corresponded to the phase of growth at which the pAH08 gfp reporter fluoresced (Fig. 4). Thus, ndvB expression was confirmed to be upregulated in stationary-phase planktonic cells as well as in biofilms.
ndvB expression is RpoS dependent.
Given that ndvB expression coincided with entry into stationary phase, we considered the possibility that the stationary-phase sigma factor RpoS might regulate ndvB transcription. RpoS levels rise at the onset of stationary phase (37), and RpoS is known to regulate the expression of a number of P. aeruginosa genes in stationary phase. The RpoS sigma factor regulon, or “sigmulome,” has been documented by two independent groups using microarray (38) and transcriptome sequencing (RNA-seq) (39) comparisons of wild-type and rpoS mutant strains. Data mining of the aforementioned transcriptomic studies revealed that while ndvB was listed as a member of the RpoS sigmulome in the study by Schulz et al. (39), ndvB expression was not affected by deletion of rpoS in the work done by Schuster et al. (38).
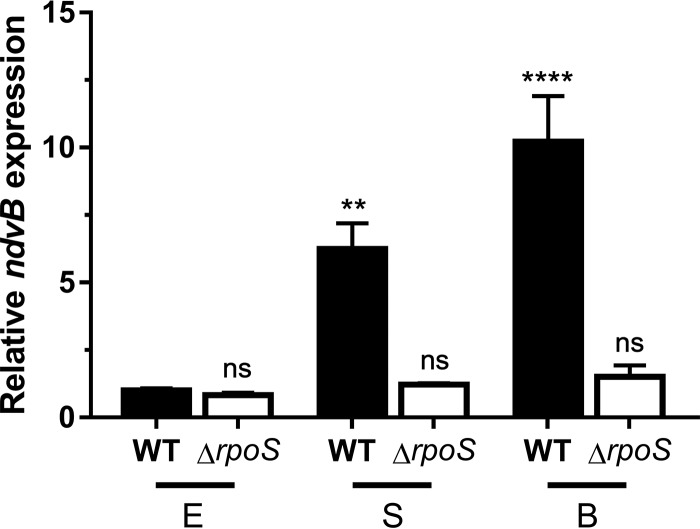
To clarify the role of RpoS in ndvB expression, we constructed an unmarked ΔrpoS deletion mutant and assayed ndvB expression of this mutant at different growth stages by quantitative PCR (qPCR). Consistent with our findings in Fig. 4, ndvB was significantly upregulated in wild-type stationary-phase and biofilm cells compared to wild-type exponential-phase cells (Fig. 5). Interestingly, however, induction of ndvB expression was not observed in ΔrpoS stationary-phase cells or in ΔrpoS biofilms (Fig. 5).
FIG 5.
RpoS is required for the upregulation of ndvB expression observed in stationary-phase and biofilm cells. The expression of ndvB (relative to that in exponential-phase wild-type cultures) was assessed by qPCR in wild-type (WT) PA14 and ΔrpoS strains grown planktonically to exponential (E) or stationary (S) phase or as biofilms (B). Data are expressed as means + SEMs for three biological replicates assayed in three technical replicates. Statistical significance was determined using one-way analysis of variance (ANOVA) with Dunnett's test (the exponential-phase wild type was used as the comparison control).
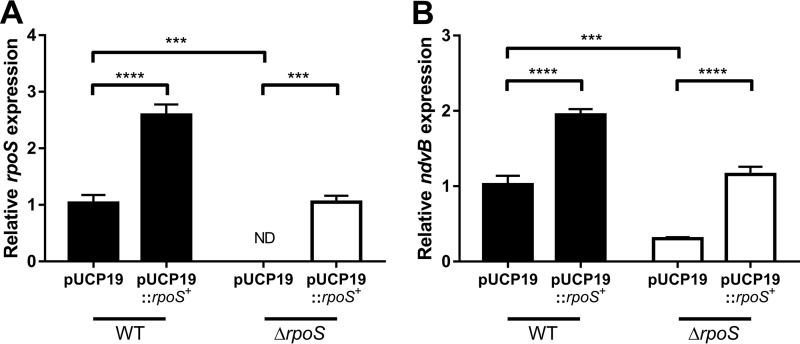
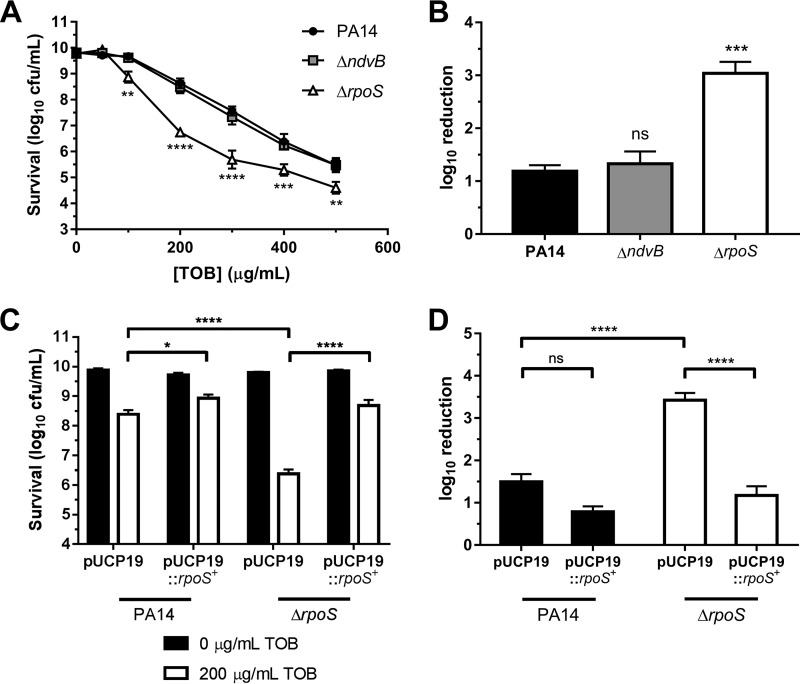
Introduction of an rpoS complementation plasmid (pUCP19::rpoS+) into the ΔrpoS mutant restored stationary-phase rpoS expression to wild-type levels (Fig. 6A). Importantly, stationary-phase expression of ndvB was also restored to wild-type levels in the ΔrpoS mutant carrying pUCP19::rpoS+ as determined by qPCR (Fig. 6B). Furthermore, overexpression of rpoS in wild-type cells led to overexpression of ndvB (Fig. 6B). Overall, these results indicate that the rpoS gene is essential for the upregulation of ndvB in both stationary-phase and biofilm cells.
FIG 6.
Complementation of the ΔrpoS strain restores rpoS and ndvB expression in stationary phase. Stationary-phase expression of rpoS (A) or ndvB (B) (relative to PA14 pUCP19) was assessed by qPCR in PA14 wild-type and ΔrpoS strains carrying pUCP19 or pUCP19::rpoS+. Data are presented as mean relative gene expression + SEMs for four biological replicates assayed in triplicate. Statistical significance was determined using one-way ANOVA with Tukey's multiple-comparison test. ND, not detected (the relative expression value was set to zero for statistical analyses).
RpoS regulates ndvB directly.
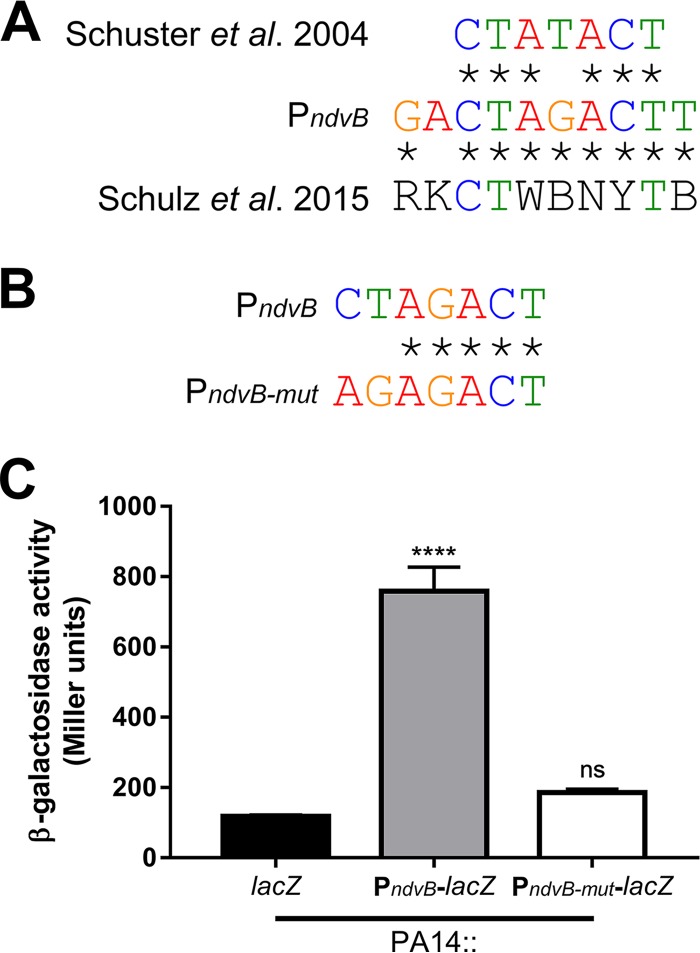
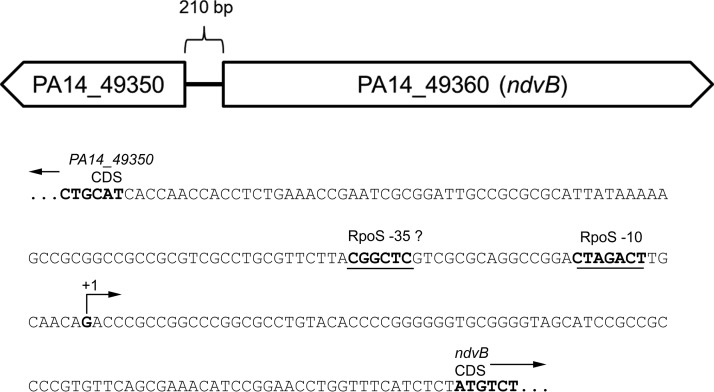
To determine whether the RpoS-dependent regulation of ndvB is direct, we searched for a sequence upstream of ndvB that matched the consensus −10 RpoS binding site. The consensus −10 RpoS binding site was previously defined as CTATACT by Schuster et al. (38), and this consensus motif was recently corroborated by Schulz et al. (39). To our knowledge, a consensus −35 RpoS binding motif has not been described for P. aeruginosa. Interestingly, we found that the ndvB promoter region contains the sequence CTAGACT (−109 to −103 relative to the translational start site; the underlined fourth position deviates from the consensus motif described by Schuster et al.), which closely matches the −10 consensus motifs (Fig. 7A). To investigate the importance of this motif for ndvB expression, a transcriptional reporter construct was generated in which part of the regulatory region upstream of the ndvB gene (from −112 to −1 relative to the ndvB translational start site) was cloned upstream of the lacZ reporter gene. Another reporter construct identical to the first but with engineered mutations in the putative −10 RpoS binding site (agAGACT; lowercase letters indicate the mutated positions) was also produced (Fig. 7B). The first and second positions were chosen for mutation because these positions are highly conserved in the consensus motifs and are, therefore, likely essential for RpoS binding to the ndvB promoter. These reporter constructs were chromosomally integrated into wild-type PA14, and β-galactosidase activity was measured in stationary-phase cultures. As shown in Fig. 7C, the activity of the mutated reporter (PndvB-mut-lacZ) was significantly decreased compared to that of the wild-type reporter (PndvB-lacZ). The activity of the PndvB-mut-lacZ reporter did not differ significantly from that of the promoterless lacZ control (Fig. 7C).
FIG 7.
The ndvB promoter contains a sequence that matches the RpoS consensus binding site. (A) Comparison of the proposed −10 RpoS binding site in the ndvB promoter to the consensus −10 RpoS elements independently determined by Schuster et al. (38) and by Schulz et al. (39). Sequence identity is shown using asterisks. (B) Comparison of the proposed −10 RpoS binding site in the ndvB promoter with the mutated site in the PndvB-mut-lacZ reporter. (C) β-Galactosidase activity of promoterless lacZ negative control (lacZ), PndvB-lacZ, and PndvB-mut-lacZ reporter constructs that were chromosomally integrated into wild-type PA14 at the attTn7 site. Data are expressed as mean + SEMs for three biological replicates. Statistical significance was determined using one-way ANOVA with Dunnett's test (promoterless lacZ was used as the comparison control).
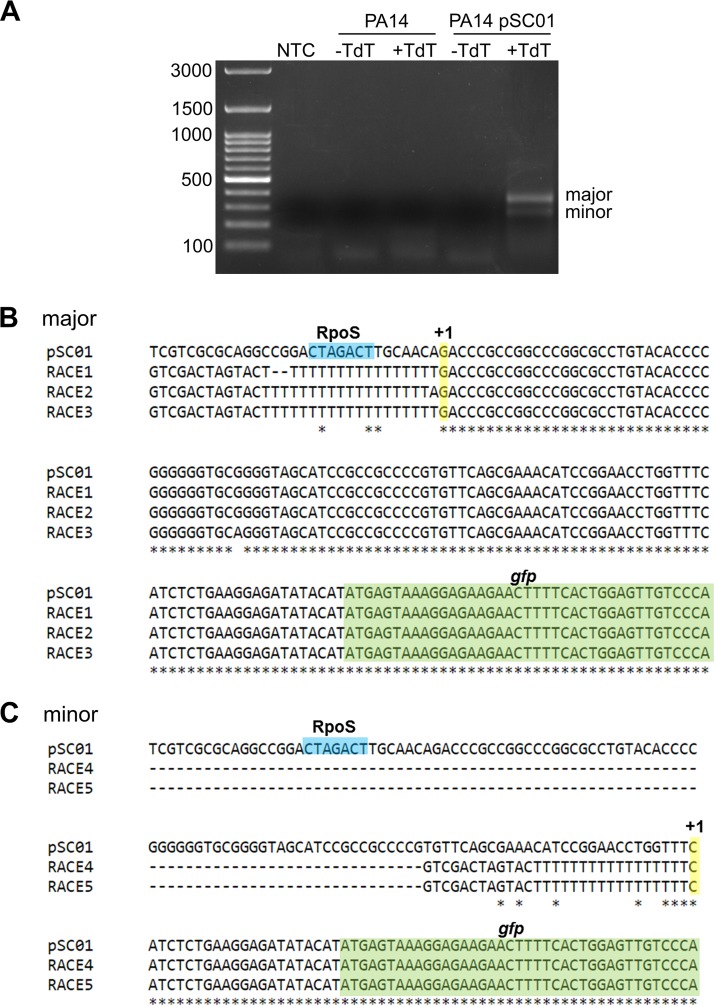
If the CTAGACT sequence in the ndvB promoter serves as the −10 element for RpoS binding, then transcription of ndvB should be initiated approximately 10 bp downstream of this sequence. Wurtzel et al. (40) mapped the transcriptional start sites (TSS) for about half of all protein-coding genes in P. aeruginosa using RNA-seq. Unfortunately, however, their work did not identify the ndvB TSS in spite of the fact that RNA from early stationary-phase cells was used for the analysis. In order to map the TSS of ndvB, 5′ rapid amplification of cDNA ends (RACE) was performed with RNA from PA14/pSC01 and gfp-specific primers (Fig. 8A). Sequencing of the major 5′ RACE product revealed a TSS located 8 bp downstream of the CTAGACT sequence, which is consistent with this motif serving as the −10 element for RpoS binding in the ndvB promoter (Fig. 8B). A smaller, minor 5′ RACE product was also obtained, and sequencing revealed a second potential TSS that would be located 7 bp upstream of the annotated translational start site of ndvB (Fig. 8C). However, we reasoned that the promoter of this second TSS is not likely to play a significant role in ndvB expression under our conditions because activity of the ndvB promoter was insignificant when the major, RpoS-directed promoter was mutated (Fig. 7C).
FIG 8.
Identification of the transcriptional start of ndvB by 5′ RACE. (A) RNA was extracted from PA14 or PA14/pSC01, and first-strand cDNA synthesis was performed with a gfp-specific primer. First-strand cDNA was incubated with dATP without or with TdT (−TdT and +TdT, respectively). The dA-tailed cDNA was subsequently amplified by two rounds of PCR as described in Materials and Methods. The 5′ RACE products were analyzed by electrophoresis on a 1.2% agarose gel. Two 5′ RACE products (major and minor) were obtained. Note that 5′ RACE products were observed only for the reaction that used RNA from PA14/pSC01 and that had been incubated with TdT, demonstrating the specificity of the gfp gene-specific primers and the dependence of the obtained the products on the poly(dA) tail added by TdT. NTC, no-template control. (B) Three pUC18 plasmids carrying the major 5′ RACE product (RACE1, -2, and -3) were sequenced using the M13R primer (StemCore Laboratories, Ottawa Hospital Research Institute), and the obtained sequences were aligned to that of pSC01 using Clustal Omega (80). Sequence identity is shown by asterisks. The GTCGACTAGTAC(T)17 sequence is derived from the 3′ RACE adapter primer. The −10 RpoS element is shown in blue, the TSS is shown in yellow with a +1, and the gfp coding sequence is in green. (C) Two pUC18 plasmids carrying the minor 5′ RACE product (RACE4 and -5) were analyzed as described for the major product.
Together, the presence of a putative −10 RpoS binding site in the ndvB promoter, the necessity of this motif for ndvB promoter activity, and the presence of a TSS 8 bp downstream of this sequence support the hypothesis that RpoS directly regulates transcription of ndvB. The sequences of the RpoS regulatory motif and the transcriptional start site of ndvB within the upstream regulatory region of ndvB are shown in their genomic context in Fig. 9.
FIG 9.
The ndvB promoter region. The divergently transcribed PA14_49350 and ndvB genes are shown schematically. The sequence of the 210-bp intergenic region between PA14_49350 and ndvB is presented below the schematic. Relevant features are annotated on the sequence.
ndvB does not appear to contribute to stationary-phase tolerance to tobramycin.
Previous data from our group and others have implicated ndvB in biofilm resistance to tobramycin (18, 24, 25). Since we discovered in the current study that ndvB is additionally expressed in stationary-phase cells, we hypothesized that the ΔndvB mutant would be more susceptible to tobramycin than wild-type PA14 in stationary phase. To address this hypothesis, overnight stationary-phase cultures of the wild type and the ΔndvB mutant were washed and subsequently exposed to various concentrations of tobramycin for 8 h at 37°C (see Materials and Methods). Survival posttreatment was assessed by colony counts. To our surprise, the susceptibility of the ΔndvB mutant did not differ from that of wild-type PA14 for any concentration of tobramycin tested (Fig. 10A).
FIG 10.
Concentration-dependent tobramycin killing of washed stationary-phase cells. (A) Washed stationary-phase cultures of wild-type PA14 and ΔndvB and ΔrpoS strains were exposed to various tobramycin (TOB) concentrations (0 to 500 μg/ml) for 8 h at 37°C. Cell survival following antibiotic treatment was assessed by colony counts using the drop plate method. Data are presented as means ± SEMs for three independent experiments. In some cases, error bars are smaller than the symbols. Statistical significance was determined by two-way ANOVA with Dunnett's post hoc multiple-comparison test for each concentration of tobramycin (PA14 was used as the comparison control). (B) Log10 reduction of PA14, ΔndvB, and ΔrpoS stationary-phase cells exposed to 200 μg/ml of tobramycin (calculated as the difference between survival at 0 μg/ml and survival at 200 μg/ml from panel A). Data are means + SEMs for three experiments. Statistical significance was determined by one-way ANOVA with Dunnett's multiple-comparison test (PA14 was used as the comparison control). (C) Washed stationary-phase PA14 or ΔrpoS cells carrying pUCP19 or pUCP19::rpoS+ were exposed to 0 or 200 μg/ml of tobramycin. Data are means + SEMs for four independent experiments. Statistical significance was determined by two-way ANOVA with Tukey's multiple-comparison test. (D) Log10 reduction of PA14 and ΔrpoS strains carrying pUCP19 or pUCP19::rpoS+ when exposed to 200 μg/ml of tobramycin (calculated as the difference between survival at 0 μg/ml and survival at 200 μg/ml from panel C). Data are means + SEMs for four experiments. Statistical significance was determined by one-way ANOVA with Tukey's multiple-comparison test.
While NdvB-derived cyclic glucans have traditionally been thought to be located in the periplasm (18), it has been demonstrated that the cyclic glucans produced by NdvB can be released into the growth medium and are a major carbohydrate component of the extracellular matrix in PA14 biofilms (24, 41). The relative contributions of periplasmic and extracellular glucan to tobramycin resistance have not been determined. We hypothesized that a phenotype for the ΔndvB mutant was not observed in our stationary-phase susceptibility assay because extracellular glucan that can interact with tobramycin was being removed during washing of the stationary-phase cells. To address this methodological concern, unwashed overnight, stationary-phase cells were used for the tobramycin susceptibility assay instead of washed cells. Since cyclic glucans can sequester aminoglycosides (18, 24), it was expected that unwashed PA14 would be less susceptible to tobramycin than the unwashed ΔndvB strain. However, no statistically significant difference in susceptibility was observed between wild-type PA14 and the ΔndvB strain under these conditions (data not shown).
RpoS contributes to stationary-phase tolerance to tobramycin.
P. aeruginosa rpoS contributes to stationary-phase tolerance to carbapenems and fluoroquinolones (42). To our knowledge, rpoS has not been previously implicated in stationary-phase tobramycin tolerance. In fact, work by another group suggests that deletion of rpoS in the wild-type PAO1 background does not impact stationary-phase tobramycin tolerance in P. aeruginosa (43). To address the role of RpoS in stationary-phase tobramycin tolerance under our conditions, washed overnight cultures of the ΔrpoS mutant were exposed to increasing concentrations of tobramycin as described above. Surprisingly, the ΔrpoS mutant was found to be more susceptible to tobramycin than the wild-type PA14 strain during stationary-phase (Fig. 10A). For instance, when treated with 200 μg/ml of tobramycin for 8 h, survival of the ΔrpoS mutant was approximately 2 log10 units lower than that of the wild-type strain (Fig. 10B). Complementation of the ΔrpoS mutant with the pUCP19::rpoS+ plasmid restored survival of the mutant to wild-type levels when treated with 200 μg/ml of tobramycin (Fig. 10C and D).
DISCUSSION
The ability of bacteria to survive antibiotic exposure depends, to a large extent, on their intrinsic resistance functions. Thus, understanding the requirements for expression of intrinsic resistance functions can provide insights for the development of therapies aimed at weakening bacterial defenses. In this study, we investigate the environmental and genetic regulation of ndvB, a gene that contributes to antibiotic tolerance in P. aeruginosa biofilms. We developed a fluorescent transcriptional reporter to examine the expression of ndvB in cells at different growth phases. The use of an unstable variant of GFP was necessary to accurately represent the downregulation of ndvB transcription that occurs during planktonic growth and upregulation in biofilms, described previously by our group (18, 25).
A novel finding of this study was that ndvB is also upregulated in stationary-phase cells. We subsequently demonstrated that induction of ndvB in stationary-phase planktonic cells and in biofilms depends on the stationary-phase sigma factor RpoS. This report provides indirect, but compelling, evidence that RpoS binds directly to the ndvB promoter to induce ndvB transcription. Mature biofilms and stationary-phase planktonic cultures are thought to exhibit similar physiological states, characterized by high cell density, nutrient depletion, and buildup of waste products (44). Likewise, protein levels of RpoS have been shown to be similar between stationary-phase and biofilm cells (45), although the distribution of rpoS transcripts is heterogeneous among different biofilm subpopulations (46–48). This similarity is supported by global transcriptome comparisons between planktonic and biofilm cultures of P. aeruginosa, which indicate that gene expression profiles of stationary-phase planktonic cultures are more similar to those of mature biofilms than exponential-phase planktonic cultures (49–52). It has been traditionally thought that biofilm cells existing in a stationary-phase-like state are slow growing (17, 53, 54) and that the reduction in growth rate is, at least partially, responsible for the reduced antibiotic susceptibility of biofilm cells relative to that of rapidly growing, exponential-phase cells. Our data suggest that a stationary-phase-like state also contributes to biofilm recalcitrance through the RpoS-mediated expression of ndvB and, potentially, of other resistance and/or tolerance determinants. Interestingly, the psl locus, which encodes the biosynthetic enzymes necessary for Psl polysaccharide synthesis, has also been shown to be regulated by RpoS (55). Psl plays an important structural role in the biofilm matrix of some P. aeruginosa strains, and it has a proposed role in biofilm antibiotic resistance (56).
The contribution of ndvB to biofilm resistance to tobramycin has been well documented (18, 24, 25). It was, therefore, surprising that ndvB did not contribute to tobramycin tolerance in stationary-phase cells given that we have demonstrated in this report that ndvB is also expressed in stationary phase. Differing levels of cyclic glucan in stationary-phase and biofilm cells could account for this result. For instance, Sadovskaya et al. (24) compared relative cyclic glucan contents of planktonic and biofilm PA14 cells. The biofilms were grown in M63 (with Casamino Acids) statically at 30°C for 64 h, while the planktonic cultures were grown under the exact same conditions except with shaking. Given the extended incubation time, the planktonic cultures were presumably in stationary phase. The glucan content per gram of cells was 50% higher in biofilms than in the planktonic cells. Therefore, absolute differences in cyclic glucan content between biofilms and stationary-phase cells may explain the importance of these molecules for biofilm, but not stationary-phase, resistance to tobramycin. Indeed, the enzymatic activity of some NdvB homologues is regulated posttranslationally in other bacterial species (57, 58), but whether or how NdvB protein activity is regulated in P. aeruginosa and if this regulation is different in stationary-phase and biofilm cells constitute an open area for investigation. Furthermore, it is relevant to note that while biofilm communities are grossly stationary-phase-like, biofilms consist of heterogeneous subpopulations of cells that experience different environments (59, 60). In contrast, stationary-phase cells experience a relatively homogenous environment due to mixing of the liquid culture. It is possible that cyclic glucans protect a specific biofilm subpopulation that is not represented in stationary-phase cultures. Further work will be required to better pinpoint where cyclic glucans are present within biofilms and what cells are protected from antibiotic treatment by the glucans.
In P. aeruginosa, RpoS has been implicated in protection against a variety of stressors, including hyperosmolar or acidic growth conditions, exposure to hydrogen peroxide or ethanol, carbon starvation when grown in minimal media with glucose, and heat shock (61, 62). Interestingly, a role for RpoS in mediating antibiotic tolerance of stationary-phase cells to ofloxacin, biapenem, and imipenem has been demonstrated using time- and concentration-dependent killing assays (42). While a recent report indicated that deletion of rpoS in the wild-type PAO1 background does not impact stationary-phase tobramycin tolerance (43), our results suggest that rpoS does play a role in stationary-phase tolerance of PA14 to tobramycin. The discrepancy in results may be due to strain differences or variations in the experimental methods. Highlighting the potential impact of differences in experimental approach, Viducic et al. (43) observed significant killing of PAO1 after 8 h of incubation with 32 μg/ml of tobramycin, while we observed no killing of PA14 after 8 h of incubation with 50 μg/ml of tobramycin. Our stationary-phase cultures were also at a higher density (approximately 5 × 109 CFU/ml) than in the study by Viducic et al. (1 × 108 CFU/ml), which might contribute to the differences in results since stationary-phase cell density is known to affect antibiotic susceptibility (63). Overall, the contribution of RpoS to stationary-phase tobramycin tolerance appears to be highly dependent on the experimental conditions.
While not addressed in this paper due to the focus on stationary-phase cells, the contribution of RpoS to antimicrobial resistance or tolerance in P. aeruginosa biofilms has been somewhat controversial. While biofilms formed by an rpoS mutant have been described as thicker and more resistant to tobramycin than wild-type biofilms (64, 65), Stewart et al. (66) demonstrated that RpoS did not play a role in biofilm tolerance to tobramycin. RpoS did, however, contribute to ciprofloxacin tolerance in P. aeruginosa biofilms (66). Somewhat paradoxically, we have demonstrated that RpoS regulates the expression of a known mechanism of aminoglycoside resistance in biofilms (18). These findings can be reconciled by the fact that RpoS is a pleiotropic regulator with many target genes that may have opposing effects on antimicrobial susceptibility.
ndvB was initially described as a biofilm-specific antibiotic resistance gene due to its in vitro phenotype when comparing exponential-phase planktonic cells and biofilms (18). By more precisely mapping the activity of the ndvB promoter, we have refined our understanding of the expression pattern of this gene. Determining that ndvB expression is induced in both stationary-phase planktonic cultures and biofilm cells allowed us to identify RpoS as the sigma factor responsible for ndvB promoter activity. Thus, ndvB can be situated within a broader response to stresses encountered by stationary-phase and biofilm cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Tables 1 and 2. All P. aeruginosa strains were derived from the burn wound isolate UCBPP-PA14 (67). The unmarked P. aeruginosa ΔrpoS deletion mutant was generated by two-step allelic exchange with the pEX18Gm suicide vector as previously described (68, 69). Bacteria were grown aerobically at 37°C with LB medium (70) or M63 medium supplemented with 0.4% arginine and 1 mM MgSO4 (25). Pseudomonas isolation agar (PIA; Difco) or LB with 0.1 μg/ml of ciprofloxacin or 20 μg/ml of nalidixic acid was used for counterselection of E. coli following E. coli-Pseudomonas conjugation. Saccharomyces cerevisiae was grown aerobically at 30°C in yeast extract-peptone-dextrose medium (1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose). Yeast selections were plated on synthetic, defined uracil drop-out medium, which contained (per liter) 6.7 g of yeast nitrogen base without amino acids (Wisent), 2 g of uracil drop-out supplement, and 20 g of glucose. Plasmids were transformed into E. coli by the calcium chloride or Inoue method (70) and into P. aeruginosa by electroporation (71), conjugation with E. coli S17-1, or triparental mating with the pRK2013 helper plasmid (72). Antibiotics were used at the following concentrations: 100 μg/ml for ampicillin (Ap), 25 μg/ml for kanamycin (Km), and 15 μg/ml for gentamicin (Gm) with E. coli; 250 μg/ml for carbenicillin (Cb) and 50 to 100 μg/ml for gentamicin (plasmids); or 20 μg/ml for gentamicin (chromosomal insertions) with P. aeruginosa.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide name | Sequence (5′–3′)a | Function |
|---|---|---|
| ndvBp-F-2b | cttctcctttactcatatgtatatctccttcAGAGATGAAACCAGGTTCCGG | PndvB-gfp fusion |
| ndvBp-R-2b | caaaaacgcgtaacaaaagtgtctataatcaACCCGGCAGGCCGGGTTG | PndvB-gfp fusion |
| AH-GFP-Tag-Fb | tcatccgccaaaacagccaagcttgcatgcctgcagactagtGGAGTCCAAGCTCAGCTAATTAAGC | GFP stability tag |
| AH-GFP-Tag-Rb | acagctgctgggattacacatggcatggatgaactatacaaaAGGCCTGCAGCAAACGACGAA | GFP stability tag |
| rpoSdel1 | ATCCATGGATCCTAGTCTGATCGGCCGTTTTG | rpoS deletion |
| rpoSdel2 | ATCATCGTGGTCAAACTCCG | rpoS deletion |
| rpoSdel3 | CGGAGTTTGACCACGATGATGACAAGCAGCGTGAGGTGGT | rpoS deletion |
| rpoSdel4 | ATCCATCTGCAGTGCCAGAAAACCCAGCGAAG | rpoS deletion |
| rpoScompF | AATAATGGTACCCTGCGAGCGGTAGTCTGATC | rpoS complementation |
| rpoScompR | CATCATAAGCTTATCTACTTAGGCTCACACGC | rpoS complementation |
| ndvBp-lacZ for | ATCCATCTGCAGGGACTAGACTTGCAACAGAC | PndvB-lacZ fusion |
| ndvBp-mut-lacZ forc | ATCCATCTGCAGGGAAGAGACTTGCAACAGAC | Mutant PndvB-lacZ fusion |
| ndvBp-lacZ rev | ATCCATAAGCTTAGAGATGAAACCAGGTTCCG | PndvB-lacZ fusion |
| Tn7R | CACAGCATAACTGGACTGATTTC | Tn7 insertion verification |
| glmS-down | GCACATCGGCGACGTGCTCTC | Tn7 insertion verification |
| ndvB-F | ACAAGGGCTTCTTCCACATC | ndvB qPCR |
| ndvB-R | TCTTCGGTGATGCACCATTC | ndvB qPCR |
| rpoD-F | TCCATCGCCAAGAAGTACAC | rpoD qPCR |
| rpoD-R | TTGTAGCCACGACGGTATTC | rpoD qPCR |
| rpoS-F | CGAACCTTCACCCGAAGAAA | rpoS qPCR |
| rpoS-R | AAGAGAGACGTCTACCGAAGT | rpoS qPCR |
| gfp-gsp1 | AGCACGTGTCTTGTAGTTCC | 5′ RACE |
| gfp-gsp2 | ATCTGGGTATCTCGCAAAGC | 5′ RACE |
| gfp-gsp3 | CTACAGGATCCGAAAGTAGTGACAAGTGTTGG | 5′ RACE |
| 3′ RACE adapter primer | GGCCACGCGTCGACTAGTAC(T)17 | 5′ RACE |
| AUAP | GGCCACGCGTCGACTAGTAC | 5′ RACE |
| M13R | TCACACAGGAAACAGCTATGAC | Sequencing |
Restriction sites are indicated in bold.
Primer used for yeast-recombination cloning. Uppercase in the sequence indicates sequences targeted for PCR amplification. Lowercase in the sequence indicates sequences homologous to the vector used for recombination.
Primer contains two changes (underlined) to the predicted ndvB promoter element.
Construction of gfp reporter vectors by yeast recombination.
A yeast recombination gap repair method was used to generate the gfp reporter vectors as described by Shanks et al. (33). In brief, a yeast-bacterium shuttle vector was digested with a restriction enzyme, creating a gap. A PCR amplicon with ends homologous to vector sequences on both sides of the gap was cotransformed with the linearized vector into yeast cells. Recombination between the vector and amplicon by endogenous yeast functions generated recircularized vectors containing the inserted sequences. Yeast cells carrying circularized vectors were selected using a vector-encoded marker.
The base vector of the gfp reporters was the yeast-E. coli-Pseudomonas shuttle vector pMQ80 (33), which carries the bright gfp variant (gfpmut3) downstream of the arabinose-inducible promoter system (araC-PBAD). Selectable markers on the vector backbone include the aacC1 gentamicin resistance gene (bacteria) and the URA3 gene, which complements uracil auxotrophy (yeast) (33). To generate the PndvB-gfp reporter (pSC01), the 500-bp region upstream of the ndvB start codon was PCR amplified from PA14 chromosomal DNA with primers ndvBp-F-2 and ndvBp-R-2 (Table 2). The primers included 31-bp tails homologous to pMQ80 vector sequences, which targeted the amplicon for recombination upstream of the Shine-Dalgarno sequence of gfpmut3, effectively replacing the araC-PBAD sequences with the ndvB promoter. The purified PCR product was cotransformed with BamHI-digested pMQ80 into yeast cells auxotrophic for uracil, using the “Lazy Bones” protocol (73). Transformants were selected on uracil drop-out medium and streaked for isolation. Plasmids were isolated from yeast cultures (74) and PCR screened for presence of the insert.
The reporters with unstable gfp variants were similarly generated using pSC01 as the base vector. The variants were amplified from four vectors (pJBA110 to pJBA113 [Table 1]) that carry gfp with 11-amino-acid C-terminal protease tags (AANDENYALAA), with each vector encoding a different final triplet (underlined) (34). The primers AH-GFP-Tag-F and AH-GFP-Tag-R (Table 2) were used to amplify 150-bp fragments that included the final 81 nucleotides of the tagged gfp genes. The inserts were recombined in yeast with pSC01 digested with SpeI, which cleaved close to the gfp stop codon. The resulting PndvB-gfp reporters differed in the final three amino acids of the protease tag as follows: pAH05, LAA; pAH06, LVA; pAH07, AAV; and pAH08, ASV. An arabinose-inducible gfp reporter encoding the ASV tag (pAH04) was constructed in the same manner using the pMQ80 base vector. The yeast-derived vectors were transformed into E. coli DH5α, verified by Sanger sequencing (StemCore Laboratories, Ottawa Hospital Research Institute), and mobilized into P. aeruginosa by triparental mating with selection on PIA containing 100 μg/ml of gentamicin.
Biofilm and planktonic growth assays and microscopy.
Biofilms were grown in M63-arginine by two methods: the air-liquid interface coverslip assay and the colony biofilm assay (36). For the air-liquid interface coverslip assay, exponential-phase LB cultures of PA14 carrying the reporter plasmids were diluted 1:100 in 4 ml of M63-arginine with 30 μg/ml of gentamicin in flat-bottom six-well plates. A flame-sterilized glass coverslip was half-submerged in each well, and plates were incubated for 19 h at 37°C to allow biofilms to form on the coverslip. The coverslips were gently rinsed with saline to remove nonadherent cells, and the biofilms that formed at the air-liquid interface of the coverslip were observed by phase-contrast and fluorescence microscopy. The colony biofilms were prepared by diluting exponential-phase LB cultures to an optical density at 600 nm (OD600) of 0.05 in M63-arginine and spotting 5 μl on semipermeable membranes (0.22-μm pore size) on M63-arginine agar plates containing 30 μg/ml of gentamicin. Following 20 h of incubation at 37°C, the membranes were removed, and the cells were gently stamped on glass slides and examined by fluorescence microscopy.
Planktonic growth curves were inoculated by washing 19-h LB cultures in M63-arginine and diluting them in 500 ml M63-arginine containing 30 μg/ml of gentamicin. For LB growth curves, 19-h LB cultures were diluted (1:100) in 50 ml of LB containing 30 μg/ml of gentamicin. Cultures were incubated at 37°C in Erlenmeyer flasks with agitation at 250 rpm, and samples were removed periodically for OD600 measurements, RNA isolation, and microscopy.
Images of planktonic and biofilm cultures were taken at a magnification of ×630 using a Leica AF6000 microscope with a Leica DFC350 FX camera. Fluorescence images were obtained using the BrightLine GFP-3035B filter set (Semrock) and Leica application suite advanced fluorescence software. Exposure times for phase-contrast and fluorescence images were 1.5 s and 8 s, respectively.
Quantification of GFP stability.
Green fluorescence was quantified in PA14 cells expressing arabinose-inducible stable and ASV-tagged GFP from vectors pMQ80 and pAH04 (Table 1). Exponential cultures were grown in LB broth supplemented with gentamicin, and GFP expression was induced by adding arabinose at 10 mM (pMQ80) or 100 mM (pAH04). After 4 h of induction, the cells were pelleted, washed, and resuspended in M63-arginine lacking arabinose to stop GFP induction. The cultures were transferred to a black 96-well clear-bottom plate (Corning) and incubated at 37°C in a plate reader (Tecan Infinite F200 PRO) that measured cell density (OD595) and green fluorescence (excitation wavelength, 500 nm; emission wavelength, 535 nm) at 15-min intervals. The fluorescent signal specific to GFP was determined by subtracting the background signal of PA14 cells not expressing GFP from the total signal. The GFP-specific fluorescence measured immediately after the shift was set at 100%, and fluorescence measured over time was plotted as the percentage of this initial measurement. The data were fitted with exponential regression lines to estimate the decay constants (λ) of the equation N(t) = N(i)eλt, where N(i) is the initial percent fluorescence and N(t) is the percent fluorescence at time t. The GFP half-lives (t1/2) were then determined by the equation t1/2 = −ln(2)/λ.
RNA extraction and qPCR.
Overnight cultures of wild-type PA14 and the ΔrpoS mutant were diluted 1:1,000 into LB and grown on a culture roller at 37°C to exponential phase (OD600 = 0.5) or stationary phase (OD600 = 2). Biofilms were grown as colony biofilms as described above. Cultures were pelleted, and RNA was extracted from the cells using TRIzol reagent and the PureLink RNA minikit (Ambion). RNA was digested with DNase as necessary, and complete removal of genomic DNA from the samples was confirmed by PCR using the rpoD-F and rpoD-R primers. The iScript cDNA synthesis kit (Bio-Rad) was used for first-strand cDNA synthesis from 2 μg of RNA in a volume of 40 μl as recommended by the manufacturer.
qPCR experiments were performed using a MyiQ single-color detection system (Bio-Rad) with 20-μl reaction mixtures containing SYBR green Supermix (Bio-Rad), 500 nM (each) forward and reverse gene-specific primers, and 2 μl of cDNA. The thermocycler program was 95°C for 90 s followed by 45 cycles of 60 s at 95°C, 30 s at 56°C, and 30 s at 72°C. Melt curve analysis was performed to ensure that amplification yielded single, specific products. Relative gene expression was calculated using the 2−ΔΔCT method, and fold changes were expressed as 2−ΔΔCT. Expression of the housekeeping gene rpoD was used for normalization. For each condition, qPCR experiments were performed in triplicate for at least three biological replicate samples.
Construction of pUCP19::rpoS+.
To enable complementation of the ΔrpoS mutant, a 1.57-kb fragment containing the rpoS promoter, coding, and terminator sequences (75, 76) was amplified by PCR using PA14 genomic DNA as the template and primers rpoScompF and rpoScompR. The fragment was ligated into the KpnI/HindIII sites of pUCP19 to create pUCP19::rpoS+. The pUCP19 and pUCP19::rpoS+ plasmids were mobilized into P. aeruginosa strains by electroporation.
Construction of PndvB-lacZ reporters and Miller assays.
A fragment encompassing part of the region immediately upstream of ndvB (−112 to −1 relative to the ndvB translational start site) was PCR amplified from PA14 genomic DNA using primers ndvBp-lacZ-for/ndvBp-lacZ-rev and subsequently cloned into the PstI/HindIII sites of the pUC18-mini-Tn7T-Gm-lacZ vector (77). Sequence-verified plasmids (StemCore Laboratories, Ottawa Hospital Research Institute) were introduced into PA14 by electroporation with the pTNS2 helper plasmid, as previously described (78). Transformants were selected on LB plates with 20 μg/ml of gentamicin and screened for successful chromosomal insertion of the mini-Tn7 reporter construct by PCR using the glmS-down/Tn7R primer pair. To generate an ndvBp-lacZ fusion with a mutated RpoS binding site, the ndvB promoter was amplified using the primer pair ndvBp-mut-lacZ for/ndvBp-lacZ rev, and the insert was cloned into the PstI/HindIII restriction sites of pUC18-mini-Tn7T-Gm-lacZ as described above. This promoter fusion mutated the putative −10 RpoS binding site in the ndvB promoter from CTAGACT to AGAGACT (mutated nucleotides are underlined). Miller assays were performed with stationary-phase cells carrying the lacZ reporters as described previously (79).
TSS mapping.
The 5′ RACE system for rapid amplification of cDNA ends (version 2.0; Invitrogen) was used per the manufacturer's recommendations for templates with high GC content. Briefly, RNA was isolated from PA14 or PA14/pSC01 stationary-phase cells as described above. Reverse transcription was performed using the gfp-gsp1 primer at 50°C for 50 min. Column-purified first-strand cDNA was incubated with dATP in the presence or absence of TdT (+TdT or −TdT, respectively). The resulting dA-tailed cDNA was subjected to PCR with gfp-gsp2 and the 3′ RACE adapter primer followed by nested PCR with gfp-gsp3 and AUAP. 5′ RACE products were run on a 1% agarose gel and visualized with ethidium bromide staining. The major and minor 5′ RACE products from PA14/pSC01 +TdT were gel purified and ligated into the SalI/BamHI sites of pUC18. Plasmids were sequenced, and the ndvB transcriptional start site (TSS) was localized by alignment of the plasmid sequencing results with the ndvB promoter region.
Stationary-phase susceptibility assays.
The stationary-phase assay was adapted from previous studies, with some modifications (42, 43, 63). Overnight (18-h) stationary-phase cultures of each strain were grown in LB at 37°C. For strains carrying pUCP19 or pUCP19::rpoS+, the overnight cultures were grown in the presence of 250 μg/ml of carbenicillin to maintain plasmid selection. The stationary-phase cultures were washed twice in fresh LB, after which the washed cells were resuspended at the original concentration in LB medium. One-milliliter aliquots of the washed stationary-phase cells were dispensed into 1.5-ml Eppendorf tubes, and various concentrations of tobramycin in water were added in a fixed volume of 50 μl to obtain final tobramycin concentrations of 0, 50, 100, 200, 300, 400, and 500 μg/ml. Cells were incubated for 8 h at 37°C with rotation on a tube revolver/rotator (Thermo Scientific; speed, 40 rpm). Following antibiotic treatment, 200-μl aliquots of treated cells were washed twice in phosphate-buffered saline (PBS, pH 7.4) and resuspended at the original concentration in PBS. Washed cells were 10-fold serially diluted in PBS, and survival was assessed by colony counting using the drop plate method. Survival was expressed as log10 CFU per milliliter. Log10 reductions were calculated as the difference between survival at 0 μg/ml of tobramycin and the survival at 200 μg/ml.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 7 software. The statistical tests that were used are specified in the figure legends when appropriate. Statistical significance is indicated as follows: ns, not significant (P > 0.05); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001.
ACKNOWLEDGMENTS
We thank Robert Shanks (University of Pittsburgh) for providing the yeast shuttle vector and for assistance in the design of the reporter fusion. We also thank Alexandre Poulain (University of Ottawa) for the use of the plate reader and members of Kristin Baetz's laboratory (University of Ottawa) for the yeast strain, reagents, and technical assistance. We thank Antonius T. M. Van Kessel for assistance with the Miller assays.
This research was supported by grants to T.-F.M. from Natural Sciences and Engineering Research Council of Canada and Cystic Fibrosis Canada. C.W.H. was supported by a Vanier Canada Graduate Scholarship.
T.-F.M., A.J.H., C.W.H., and L.Z. designed the experiments, A.J.H., C.W.H., L.G., L.Z., J.-P.N., S.C., and B.S. performed the experiments, and T.-F.M., A.J.H., and C.W.H. wrote the manuscript.
REFERENCES
- 1.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox G, Wright GD. 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol 303:287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, Singh PK, Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci U S A 106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 12.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 13.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 14.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Mah T-F. 2012. Biofilm-specific antibiotic resistance. Future Microbiol 7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 16.Conlon BP, Rowe SE, Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- 17.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Mah T-F. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Fritsch M, Hammond L, Landreville R, Slatculescu C, Colavita A, Mah T-F. 2013. Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One 8:e61625. doi: 10.1371/journal.pone.0061625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao J, Schurr MJ, Sauer K. 2013. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol 195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson M, Rybtke M, Givskov M, Høiby N, Twetman S, Tolker-Nielsen T. 2016. The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int J Antimicrob Agents 48:298–304. doi: 10.1016/j.ijantimicag.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Hall CW, Mah T-F. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 24.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated β-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 25.Beaudoin T, Zhang L, Hinz AJ, Parr CJ, Mah T-F. 2012. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J Bacteriol 194:3128–3136. doi: 10.1128/JB.06178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naylor LH. 1999. Reporter gene technology: the future looks bright. Biochem Pharmacol 58:749–757. doi: 10.1016/S0006-2952(99)00096-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Campbell RE, Ting AY, Tsien RY. 2002. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 28.Sternberg C, Christensen BB, Johansen T, Toftgaard Nielsen A, Andersen JB, Givskov M, Molin S. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol 65:4108–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Southward CM, Surette MG. 2002. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol Microbiol 45:1191–1196. doi: 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- 31.Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Høiby N. 2004. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 48:1168–1174. doi: 10.1128/AAC.48.4.1168-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 33.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 36.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K, Takahashi H. 1994. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene 150:81–85. [DOI] [PubMed] [Google Scholar]
- 38.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 39.Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dötsch A, Hornischer K, Bruchmann S, Düvel J, Häussler S. 2015. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog 11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coulon C, Vinogradov E, Filloux A, Sadovskaya I. 2010. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One 5:e14220. doi: 10.1371/journal.pone.0014220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami K, Ono T, Viducic D, Kayama S, Mori M, Hirota K, Nemoto K, Miyake Y. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol Lett 242:161–167. doi: 10.1016/j.femsle.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. 2017. RpoN promotes Pseudomonas aeruginosa survival in the presence of tobramycin. Front Microbiol 8:839. doi: 10.3389/fmicb.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 45.Xu KD, Franklin MJ, Park C-H, McFeters GA, Stewart PS. 2001. Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously-fed Pseudomonas aeruginosa biofilms. FEMS Microbiol Lett 199:67–71. doi: 10.1111/j.1574-6968.2001.tb10652.x. [DOI] [PubMed] [Google Scholar]
- 46.Babin BM, Atangcho L, van Eldijk MB, Sweredoski MJ, Moradian A, Hess S, Tolker-Nielsen T, Newman DK, Tirrell DA. 2017. Selective proteomic analysis of antibiotic-tolerant cellular subpopulations in Pseudomonas aeruginosa biofilms. mBio 8:e01593-. doi: 10.1128/mBio.01593-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Osorio AC, Williamson KS, Franklin MJ. 2010. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J Bacteriol 192:2991–3000. doi: 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heacock-Kang Y, Sun Z, Zarzycki-Siek J, McMillan IA, Norris MH, Bluhm AP, Cabanas D, Fogen D, Vo H, Donachie SP, Borlee BR, Sibley CD, Lewenza S, Schurr MJ, Schweizer HP, Hoang TT. 2017. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol Microbiol 106:976–985. doi: 10.1111/mmi.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol 187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dötsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, Geffers R, Häussler S. 2012. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One 7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, Mazurie A, Stewart PS. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol 10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, Curtis MA. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderl JN, Zahller J, Roe F, Stewart PS. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 47:1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown MR, Allison DG, Gilbert P. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother 22:777–780. [DOI] [PubMed] [Google Scholar]
- 55.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Billings N, Ramirez Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breedveld MW, Miller KJ. 1994. Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol Rev 58:145–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingram-Smith C, Miller KJ. 1998. Effects of ionic and osmotic strength on the glucosyltransferase of Rhizobium meliloti responsible for cyclic β-(1,2)-glucan biosynthesis. Appl Environ Microbiol 64:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 60.Wimpenny J, Manz W, Szewzyk U. 2000. Heterogeneity in biofilms. FEMS Microbiol Rev 24:661–671. doi: 10.1111/j.1574-6976.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 61.Jørgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart GSAB. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835–844. [DOI] [PubMed] [Google Scholar]
- 62.Suh S-J, Silo-Suh L, Woods DE, Hassett DJ, West SEH, Ohman DE. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol 181:3890–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heydorn A, Ersbøll BK, Hentzer M, Parsek MR, Givskov M, Molin S. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- 65.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 66.Stewart PS, Franklin MJ, Williamson KS, Folsom JP, Boegli L, James GA. 2015. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 59:3838–3847. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. [DOI] [PubMed] [Google Scholar]
- 68.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- 69.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 71.Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 72.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shanks RMQ, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. [DOI] [PubMed] [Google Scholar]
- 75.Fujita M, Tanaka K, Takahashi H, Amemural A. 1994. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol Microbiol 13:1071–1077. [DOI] [PubMed] [Google Scholar]
- 76.Kang Y, Lunin VV, Skarina T, Savchenko A, Schurr MJ, Hoang TT. 2009. The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA-operon in Pseudomonas aeruginosa. Mol Microbiol 73:120–136. doi: 10.1111/j.1365-2958.2009.06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 78.Choi K-H, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 79.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 80.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. [DOI] [PubMed] [Google Scholar]
- 82.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. [Google Scholar]
- 83.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]
- 85.Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–112. [DOI] [PubMed] [Google Scholar]