ABSTRACT
We have previously shown that the lactic acid bacterium Lactobacillus brevis 174A, isolated from Citrus iyo fruit, produces a bacteriocin designated brevicin 174A, which is comprised of two antibacterial polypeptides (designated brevicins 174A-β and 174A-γ). We have also found a gene cluster, composed of eight open reading frames (ORFs), that contains genes for the biosynthesis of brevicin 174A, self-resistance to its own bacteriocin, and two transcriptional regulatory proteins. Some lactic acid bacterial strains have a system to start the production of bacteriocin at an adequate stage of growth. Generally, the system consists of a membrane-bound histidine protein kinase (HPK) that senses a specific environmental stimulus and a corresponding response regulator (RR) that mediates the cellular response. We have previously shown that although the HPK- and RR-encoding genes are not found on the brevicin 174A biosynthetic gene cluster in the 174A strain, two putative regulatory genes, designated breD and breG, are in the gene cluster. In the present study, we demonstrate that the expression of brevicin 174A production and self-resistance is positively controlled by two transcriptional regulatory proteins, designated BreD and BreG. BreD is expressed together with BreE as the self-resistance determinant of L. brevis 174A. DNase I footprinting analysis and a promoter assay demonstrated that BreD binds to the breED promoter as a positive autoregulator. The present study also demonstrates that BreG, carrying a transmembrane domain, binds to the common promoter of breB and breC, encoding brevicins 174A-β and 174A-γ, respectively, for positive regulation.
IMPORTANCE The problem of the appearance of bacteria that are resistant to practical antibiotics and the increasing demand for safe foods have increased interest in replacing conventional antibiotics with bacteriocin produced by the lactic acid bacteria. This antibacterial substance can inhibit the growth of pathogenic bacteria without side effects on the human body. The bacteriocin that is produced by a Citrus iyo-derived Lactobacillus brevis strain inhibits the growth of pathogenic bacteria such as Listeria monocytogenes, Staphylococcus aureus, and Streptococcus mutans. In general, lactic acid bacterial strains have a system to start the production of bacteriocin at an adequate stage of growth, which is called a quorum-sensing system. The system consists of a membrane-bound histidine protein kinase that senses a specific environmental stimulus and a corresponding response regulator that mediates the cellular response. The present study demonstrates that the expression of the genes encoding bacteriocin biosynthesis and the self-resistance determinant is positively controlled by two transcriptional regulatory proteins.
KEYWORDS: bacteriocin, lactic acid bacteria, transcriptional regulatory protein
INTRODUCTION
The term “lactic acid bacteria” (LAB) (bacteria that are known as probiotics) is a generic name given to bacteria that produce large amounts of lactic acid from various sugars and other carbohydrates. Bacteriocins, which are antibacterial polypeptides produced by some LAB strains, inhibit the growth of their related LAB strain and several pathogenic microorganisms (1, 2). Therefore, the bacteriocin-producing LAB strains can be recognized as biopreservatives that produce an antibacterial substance without side effects (3, 4). In fact, a bacteriocin, nisin A, is used worldwide (5, 6).
In a previous study, we isolated the bacteriocin-producing Lactobacillus brevis strain 174A from Citrus iyo fruit (7). The bacteriocin designated brevicin 174A consists of two polypeptides, named brevicins 174A-β and 174A-γ. Brevicin 174A inhibits the growth of several LAB strains and pathogenic bacteria such as Listeria monocytogenes, Staphylococcus aureus, and Streptococcus mutans. We have also showed in previous work that the bacteriocin biosynthetic gene cluster of L. brevis 174A is identical to that of L. brevis 925A isolated from kimuchi (7, 8). The study demonstrated that the gene clusters of both strains contain two genes encoding the deduced transcriptional regulators, designated breD and breG. Protein motif analysis using the SOSUI program (http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html) (9) showed that the BreD protein (molecular mass, 7.4 kDa) has a helix-turn-helix (HTH) motif at the N-terminal region. On the other hand, the BreG protein (24 kDa) has the HTH motif at the N-terminal region together with four-transmembrane-spanning domains at the C-terminal region.
Bacteriocins are classified into two classes designated classes I and II (4, 10, 11). The class I bacteriocins, which are also called lantibiotics, contain a typical intrapeptide cyclic structure composed of thioether-bound amino acids, lanthionine, and methyllanthionine. The class II bacteriocins are further classified into four subclasses, class IIa (active to Listeria and pediocin-like), IIb (two peptide), IIc (cyclic), and IId (nonpediocin single linear). In general, the production of bacteriocins classified into classes I and II (12–17) is controlled by a two-component regulatory system consisting of a histidine protein kinase (HPK) and a response regulator (RR) in the presence of the specific autoinducer peptide (AIP). We have proposed in the previous study that the regulatory mechanism to express the two-polypeptide bacteriocin brevicin 174A produced by the 174A strain is different from that for the two-component system (7). To clarify the hypothesis, in the present study, we determined the binding site of the deduced positive regulator BreD, which is present near the promoter region of breE, encoding an immunity protein as the self-resistance determinant in L. brevis 174A. We also demonstrate that another positive regulator, BreG, binds upstream of the common promoter of breB and breC to express brevicins 174A-β and 174A-γ, respectively.
In the present study, we show a new regulatory mechanism for the expression of two-polypeptide bacteriocin.
RESULTS
Determination of DNA-binding sites of BreD and BreG.
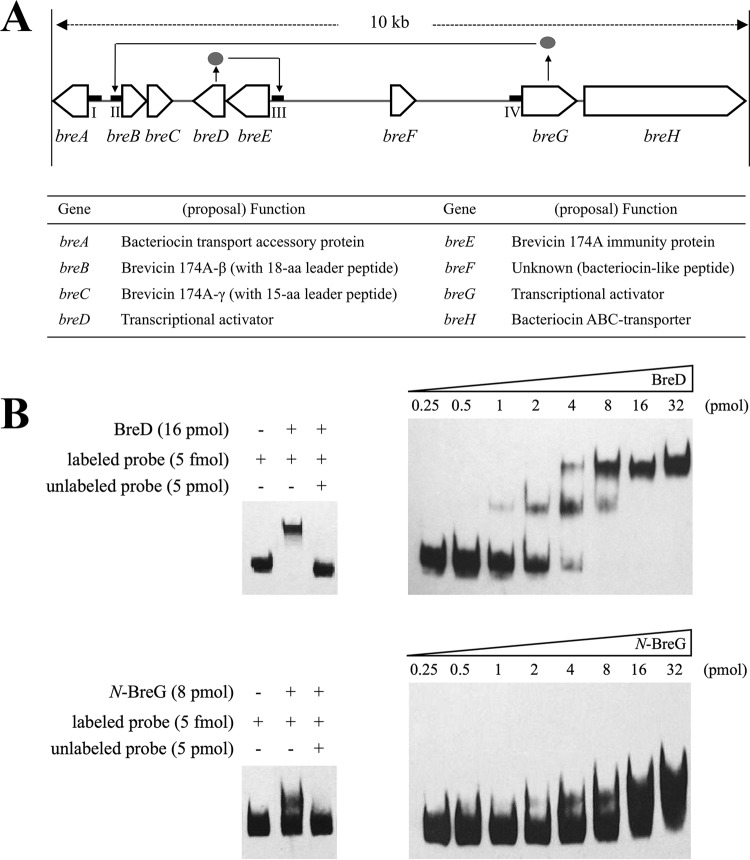
To determine the DNA-binding sites of BreD and BreG, each protein was expressed using an Escherichia coli host-vector system and purified. DNA fragments named regions I to IV, which include the predicted promoter region found in the brevicin 174A biosynthetic gene cluster (Fig. 1A), were amplified to assay the binding affinities of BreD and BreG using the electrophoretic mobility shift assay (EMSA) method. Figure 1B shows that BreD and BreG bind to region III and region II, respectively. The former and latter regions contain the breED promoter (PbreED) and breBC promoter (PbreBC), respectively, and the binding intensity increased in a protein concentration-dependent manner.
FIG 1.
Determination of the promoter region for the binding of BreD and BreG proteins. (A) Gene organization and predicted functions of ORFs in the brevicin 174A biosynthetic gene cluster composed of breA to breH. The promoter regions used for the electrophoretic mobility shift assay (EMSA) are indicated as four thick bars and named regions I, II, III, and IV. The solid line with an arrowhead (→) indicates the binding between the breD- or breG-encoded protein (•) and the promoter region as the target. (B) Profiles of the EMSA due to the concentration-dependent addition of BreD or N-BreG together with the labeled probes (right). When the unlabeled probe was added in excess, the specific band shift was inhibited (left). In this experiment, DNA fragments identical to regions III and II were used to detect BreD and N-BreG proteins, respectively.
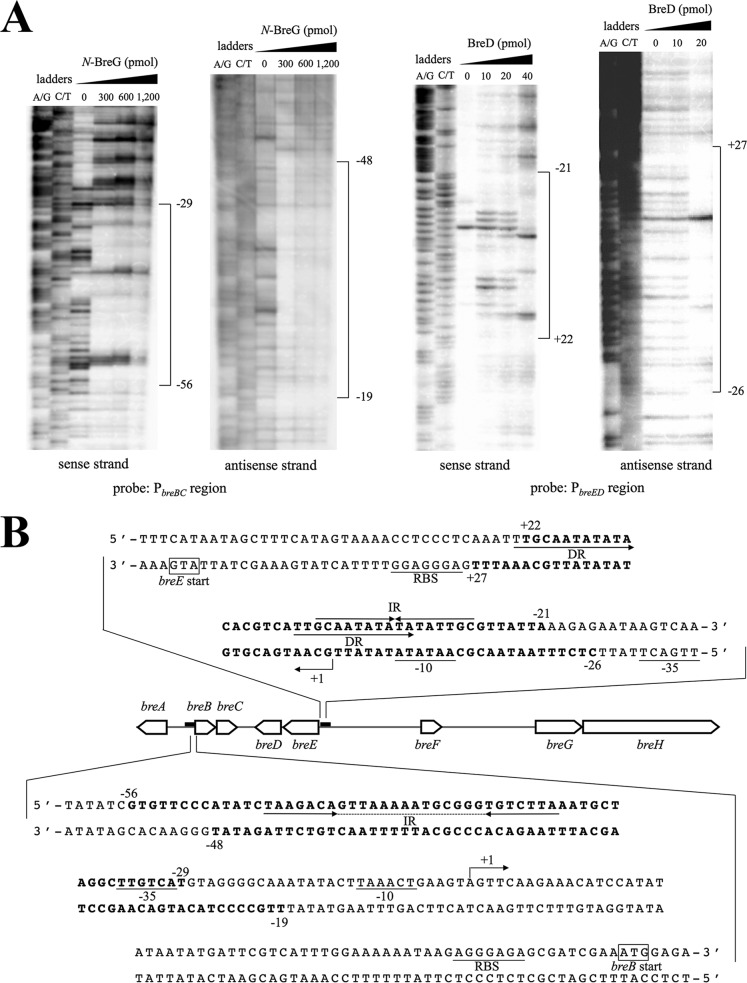
To obtain more information on the binding sites of both proteins, a DNase I footprinting assay was performed (Fig. 2A). Figure 2B shows that the binding site of BreD contains the 16-bp inverted-repeat (IR) sequence 5′-GCAATATATATATTGC-3′ located between the ribosome-binding site (RBS) and the −10 region of PbreED. The twice-repeated 12-bp direct-repeat (DR) sequences are also found in the same region. With regard to the nucleotide sequence of the BreG-binding site, two possible IR sequences, 5′-TAAGACA-N14-TGTCTTA-3′ (28 bp) and 5′-GACA-N8-TGTC-3′ (16 bp), are found upstream of the PbreBC −35 sequence (Fig. 2B). The transcriptional start sites of breB and breE were also determined by the primer extension analysis (data not shown) and are shown in Fig. 2B as +1.
FIG 2.
Binding sites of BreD and BreG to the promoter region of breED and breBC, respectively. (A) DNase I footprinting for the determination of BreD and BreG binding sites upstream of breED and breBC, respectively. The amounts of BreD (10 to 40 pmol) and N-BreG (300 to 1,200 pmol) used for the assay are indicated at the top of each lane. A probe amplified with a 5′-end-biotinylated primer was used. The nucleotide corresponding to the first nucleotide of the transcribed RNA is numbered +1. (B) Nucleotide sequences of the predicted binding site of BreD or BreG. The numbers indicate positions from the transcription starting point, beginning with +1. The putative RBS, −10, and −35 regions are underlined. The regions protected by both regulators are in bold. The arrows indicate direct repeats (DR) and inverted repeats (IR). The dotted line indicates spacer sequences. The transcriptional start point is marked with a bent arrow.
Promoter assay.
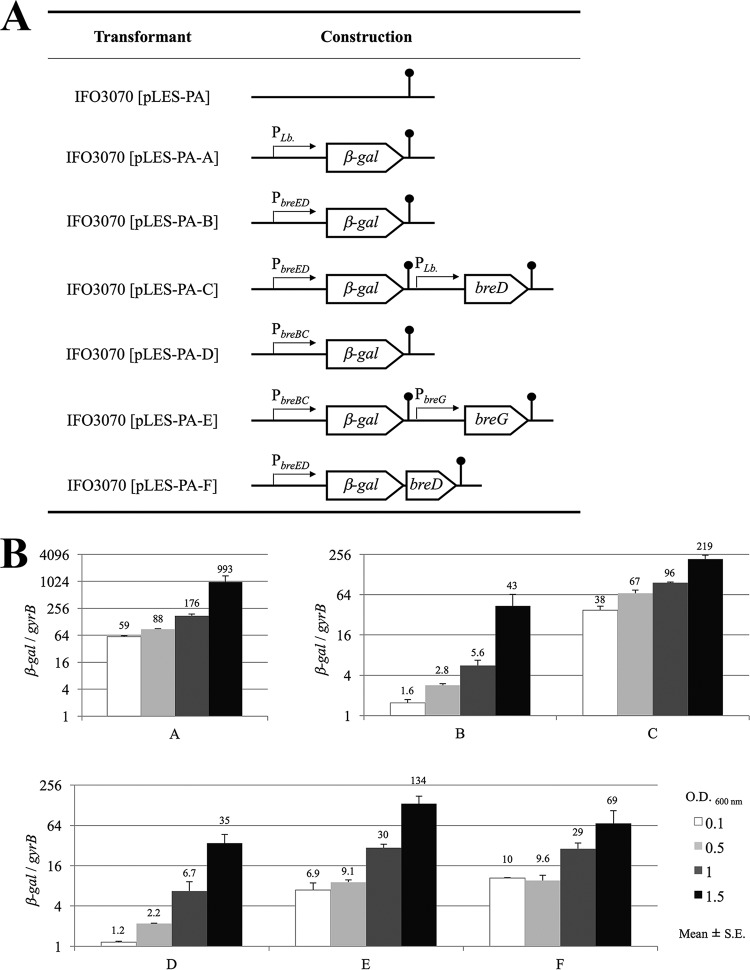
To elucidate whether BreD and BreG function as a positive or negative regulators, a promoter assay using the quantitative reverse transcription-PCR (qRT-PCR) method was performed using the β-galactosidase (β-gal) structural gene as a reporter gene. The β-gal gene was located under the control of PbreBC or PbreED instead of PLb, which is the synthetic promoter for constitutive gene expression in Lactobacillus cells (18), and the transcriptional activities in the presence and absence of the corresponding regulator were compared (Fig. 3A). Interestingly, Fig. 3B shows that the transcriptional activities of PbreBC and PbreED were enhanced in the presence of the breG (pLES-PA-D versus pLES-PA-E) and breD (pLES-PA-B versus pLES-PA-C) genes, respectively. The result indicates that BreD and BreG function as transcriptional activators to produce brevicin 174A and the immunity protein for expression of self-resistance, respectively. The enhancement was even higher at the early stage of cell growth and gradually decreased toward the stationary stage. When both the β-gal and breD genes were under the control of PbreED as a polycistronic transcriptional unit, like for the breED transcription (pLES-PA-F), the expression levels of the β-gal gene were almost the same as those in pLES-PA-E. The regulations of β-gal gene expression in pLES-PA-E and pLES-PA-F are regarded as the same as those of breBC and breED expression in the brevicin 174A biosynthetic gene cluster, respectively. The result shows that the productivities of bacteriocin and immunity protein in L. brevis 174A have balanced with each other.
FIG 3.
β-Gal gene expression of L. plantarum IFO3070 cells transformed with the indicated plasmids. (A) Plasmids constructed for the promoter assay. Four kinds of promoters were used for the expression of the β-gal gene, breD, or breG. The terminator is marked with a lollipop. (B) Ratio of expression of β-gal mRNA to gyrB mRNA. The host L. plantarum IFO3070 was transformed with each expression plasmid. Each of the resulting transformants was grown until it reached the indicated cell concentration (optical density at 600 nm).
Genotypic and phenotypic changes in breD or breG mutants.
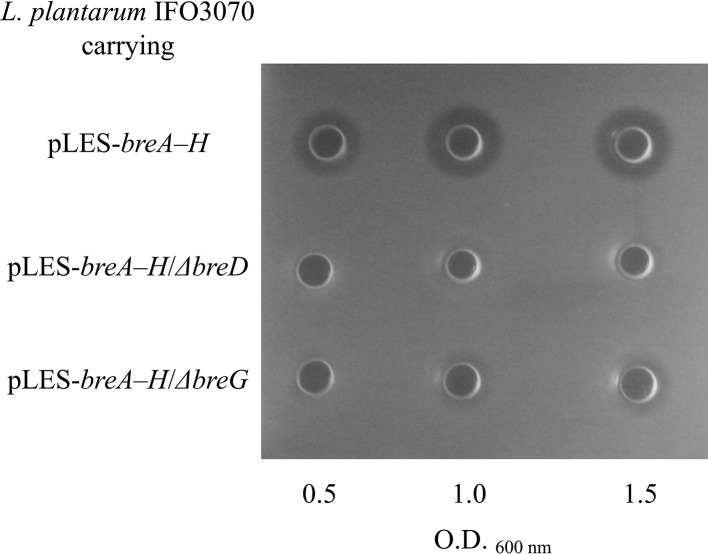
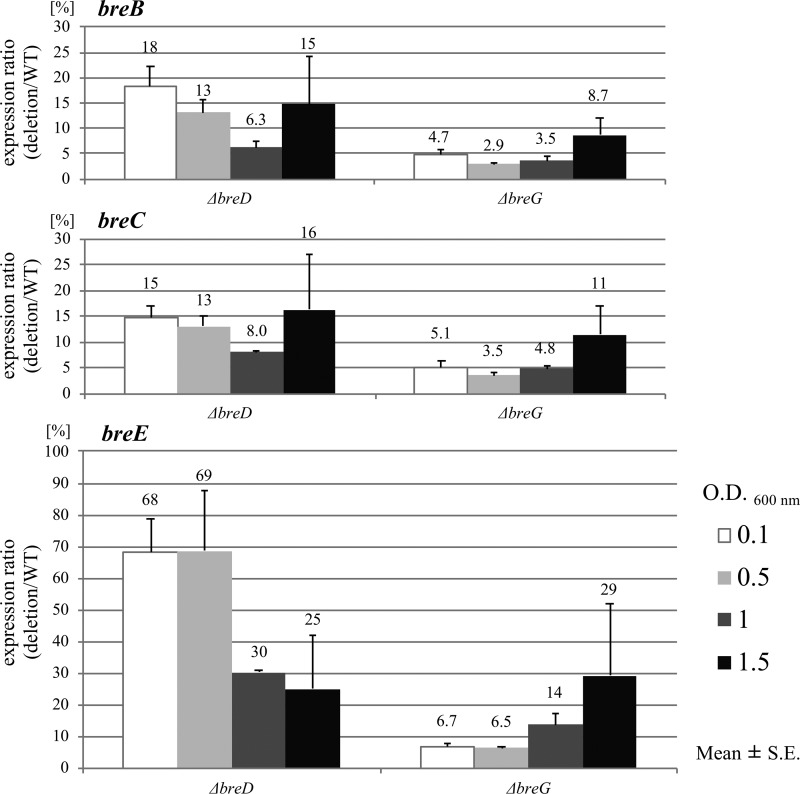
A plasmid-curing experiment and the Southern blot analysis showed that the brevicin 174A biosynthetic gene cluster is present in the largest plasmid harbored in L. brevis 174A (data not shown). To clarify the roles of breD and breG, which were deduced to be the transcriptional regulators, a brevicin 174A biosynthetic gene vector carrying a nonsense mutation of breD or breG was constructed. In the experiment, the vector designated pLES-breA–H carries the wild-type brevicin 174A biosynthetic gene cluster. Figure 4 shows that Lactobacillus plantarum IFO3070 transformed with pLES-breA–H definitely produces brevicin 174A. In contrast, the same host harboring the plasmid pLES-breA–H/ΔbreG (named the ΔbreG mutant) displayed slight antibacterial activity. This phenomenon corresponds with the result obtained via qRT-PCR that the expression of breB and breC was scarcely detected in host cells carrying the ΔbreG mutation (Fig. 5). This observation corresponds with the result that BreG promoted the transcription of breB and breC (Fig. 3). However, the antibacterial activity was scarcely present in the culture supernatant of the host transformed with the mutant plasmid pLES-breA–H/ΔbreD (named the ΔbreD mutant) (Fig. 4). In addition, the expression of both breB and breC was decreased in the ΔbreD mutant (Fig. 5).
FIG 4.
Brevicin 174A productivity of L. plantarum IFO3070 transformed with the plasmid pLES-breA–H carrying the breD- or breG-disrupted gene. The bacteriocin activity detected in the culture supernatant from the host cells transformed with each mutant plasmid was confirmed by the agar well diffusion method.
FIG 5.
Expression ratio of the deletion mutant to the wild-type (deletion/WT) breB, breC, and breE genes in L. plantarum IFO3070 transformed with pLES-breA–H/ΔbreD (ΔbreD mutant) and pLES-breA–H/ΔbreG (ΔbreG mutant) to the same host harboring pLES-breA–H. The relative quantification of each target gene in the ΔbreD and ΔbreG mutants in comparison to that observed in the host harboring pLES-breA–H was measured. The relative expression ratio of the target gene is shown. The expression of each gene was measured at the indicated growth phase of each transformant.
DISCUSSION
In the present study, we show that the expression of bacteriocin produced by L. brevis 174A is regulated by a system different from the two-component regulatory system with HPK and RR. As evidence, a BLAST search suggests that the BreD and BreG proteins encoded by ORFs found in the brevicin 174A biosynthetic gene cluster are homologous to several Lactobacillus DNA-binding proteins, with about 60% and 40% identities on average, respectively, and these proteins have an HTH motif (19). The motif in the BreD and BreG proteins was similar to the cro/C1-type HTH motif, and the DNA-binding domain found in the motif consists of five α-helices. The second and third helices in the DNA-binding domain are especially necessary to recognize the DNA-binding sequence of the promoter (20, 21). The transcriptional regulators, which are grouped as DNA-binding protein, roughly divided into two types, transcriptional repressor and activator. In general, the repressor protein binds to the operator sequence and inhibits the binding of RNA polymerase. On the other hand, the activator protein binds to the specific binding sequence and facilitates the binding of RNA polymerase to the promoter. Several DNA-binding proteins, such as the bacteriophage 434-derived cro/C1-type HTH domain (20), the Xre family transcriptional regulator from the Bacillus subtilis prophage PBSX (22), and CopR, which is a two-component system-response regulator for copper resistance of the broad-host-range plasmid pIP501 (23), have been reported to be repressor proteins.
On the other hand, it has been reported that the bacitracin resistance in Enterococcus faecalis is mediated by an ATP-binding cassette (ABC) transporter and is positively regulated by a novel protein (24). The regulatory protein designated BcrR, which carries the cro/C1-type HTH motif for DNA binding at the N terminus, functions as an activator to express the bacitracin resistance (24, 25). The BcrR protein has a four-transmembrane-spanning domain at the C terminus, as does BreG. In the presence of bacitracin, the BcrR protein positively regulates the transcription of the bcrABD operon, encoding an ABC transporter that confers resistance to the bacitracin. It has been confirmed that the bcrABD operon is not transcribed in the bcrR deletion mutant (22). The function of BreG in the expression of breBC is similar to that of BcrR. Highly virulent strains of E. faecalis express a pore-forming exotoxin called cytolysin, which lyses both bacterial and eukaryotic cells in response to quorum-sensing signals (26). The expression of cytolysin in the bacterium has been reported to be regulated by the CylR1/CylR2 system (27, 28). This membrane-associated regulatory system does not depend on HPK/RR phosphorylation signaling (25, 26). In the regulatory model, a membrane-bound protein, CylR1, recognizes a signal peptide. When the peptide has accumulated to a threshold to allow the transcription of cytolysin, repression against the cytolysin promoter is cancelled by the binding of CylR2 to CylR1. Thus, CylR and BcrR recognize cytolysin and bacitracin, respectively (24, 25, 27, 28).
Based on those reports, it is likely that some signal molecules are produced in the culture supernatant from the brevicin 174A biosynthetic gene cluster. Therefore, in the present study, we measured the transcriptional activities for the promoters PbreBC and PbreED in the presence or absence of the supernatant from the culture broth of L. plantarum IFO3070 harboring the brevicin 174A biosynthetic gene cluster. However, the transcriptional activities of PbreBC and PbreED did not change even in the presence of the bacteriocin (data not shown). The breF gene product found in the brevicin 174A biosynthetic gene cluster, which is annotated as a hypothetical protein, has a double-glycine motif followed by polylysine residues, like BreB and BreC. Therefore, we also investigated in this study whether the BreF peptide molecule exhibits an antibacterial activity, as do brevicins 174A-β and 174A-γ, or functions as a signal peptide (data not shown). The productivity of brevicin 174A and resistance to the bacteriocin in L. plantarum IFO3070 carrying pLES-breA–H/ΔbreF were almost the same as those in the same host carrying pLES-breA–H.
Furthermore, the mature form of BreF predicted from the amino acid sequence did not exhibit antibacterial activity. In addition, the peptide molecule did not enhance the antibacterial activity of brevicins 174A-β and 174A-γ. At this time, we cannot identify the signal molecule that regulates the brevicin 174A biosynthesis.
In the regulatory system to produce brevicin 174A, the transcription of breD and breE is positively autoregulated by BreD. It has been shown that two-thirds of transcriptional regulators seem to be autoregulated in the bacterium (29). In many autoregulation circuits, the negative regulatory system has been reported to reduce the time to reach the required steady-state concentration and provide stability for homeostasis (30, 31). On the other hand, although it needs more time to reach steady-state expression, the bistability network is provided by the positive autoregulation (32–34). Furthermore, the bistability is sometimes associated with hysteresis. Once upregulated, the activated transcription is maintained even when the stimulating factors are no longer present in the hysteresis state (35, 36). Since BreD is too small a protein to function as a receptor carrying the HTH motif, it is not reasonable to speculate that the function of BreD bound with some signal molecules is altered. Even without the selective pressure of bacteriocin, a sufficient amount of the immunity protein might be produced by the regulatory system, suggesting that the system is meaningful to maintain the self-resistance of brevicin 174A.
In this study, we showed that when breD was inactivated, the positive autoregulation was lost, followed by decreased expression of breE (Fig. 5). The ΔbreD mutant may manage to survive the lethal effect of its own produced brevicin 174A by maintaining the breE expression level. For example, the mutant might increase the number of replicated plasmids. In fact, the expression of the rep gene, which is necessary for plasmid replication, was approximately three times higher than that in the same host harboring pLES-breA–H at the early stage of growth (data not shown). In the ΔbreD mutant, even if BreG is intact, the strain cannot produce enough brevicin 174A, because when an excess amount of brevicin 174A is present, the ΔbreD mutant might be killed due to poor expression of the immunity protein. Therefore, the production of brevicin 174A in the ΔbreD mutant is likely to be controlled at a lower level during cultivation (Fig. 4). On the other hand, expression of both breB and breC was decreased in the ΔbreG mutant, and the breE level was also repressed during cultivation (Fig. 5). Unlike the case for the ΔbreD mutant, the production of brevicin 174A in the ΔbreG mutant is not lethal. As shown in Fig. 4, the bacteriocin is detected in the supernatant of the ΔbreG mutant culture. The results obtained with these two mutants show that brevicin 174A is significantly productive only when the bacteriocin and immunity protein are adequately supplied.
It has been shown that transcriptional regulatory proteins binding to the promoter region are predicted to be repressors but not activators. However, a research group has reported that a transcriptional activator protein belonging to the Streptomyces antibiotic-regulatory protein (SARP) family, named AfsR, binds a 9-bp direct repeat located only 8 bp from the −10 region of the target gene promoter (37). Although the promoter sequences of target genes of BreD and BreG do not have a similarity to those of SARP, the present study demonstrates that both BreD and BreG act as activators to regulate brevicin 174A biosynthesis and self-resistance. The production of two-polypeptide bacteriocins, in general, is regulated by the two-component regulatory system with HPK and RR (11). For example, the production of ABP-118 by Lactobacillus salivarius subsp. salivarius UCC118 (38), of plantaricin E/F and J/K by L. plantarum C11 (39–41), and of plantaricin NC8 by L. plantarum NC8 (42) is controlled by the system. Although determination of the precise activation mechanisms of both the BreD and BreG regulators is still in progress, in the present study, we have found a novel regulatory system to produce the two-polypeptide bacteriocin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. L. brevis 174A and L. plantarum IFO3070 were grown at 28°C in de Man, Rogosa, and Sharpe (MRS) broth (Merck). E. coli was grown at 37°C in LB medium (43). When required, ampicillin (100 μg/ml), kanamycin (50 μg/ml), erythromycin (300 μg/ml for E. coli and 100 μg/ml for LAB strains), and 1.5% (wt/vol) agar were supplemented. E. coli BL21(DE3) and the pET-28a(+) plasmid (Novagen) were used for the overexpression of BreD and BreG gene products. An E. coli-LAB shuttle vector, pLES003-b, which carries the AatII fragment reversed from pLES003 (DDBJ accession no. AB370338) (8) as the replication origin, was used for the promoter assay and the reconstruction of the brevicin 174A biosynthetic gene cluster. E. coli DH5α and the pUC19, pGEM-T (Promega), and pTA2 (Toyobo) plasmids were used for DNA cloning and sequencing. E. coli HST04 was used for the preparation of a Dam and/or Dcm methylation-free plasmid. L. plantarum IFO3070 was used as a host for the promoter assay and reconstruction of the bacteriocin biosynthetic gene cluster.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| L. plantarum strains | ||
| IFO3070 | ||
| IFO3070(pLES-PA-A) | IFO3070 transformed with pLES-PA-A | This study |
| IFO3070(pLES-PA-B) | IFO3070 transformed with pLES-PA-B | This study |
| IFO3070(pLES-PA-C) | IFO3070 transformed with pLES-PA-C | This study |
| IFO3070(pLES-PA-D) | IFO3070 transformed with pLES-PA-D | This study |
| IFO3070(pLES-PA-E) | IFO3070 transformed with pLES-PA-E | This study |
| IFO3070(pLES-PA-F) | IFO3070 transformed with pLES-PA-F | This study |
| IFO3070(pLES-breA–H) | IFO3070 transformed with pLES-breA–H | This study |
| IFO3070(pLES-breA–HΔbreD) | IFO3070 transformed with pLES-breA–HΔbreD | This study |
| IFO3070(pLES-breA–HΔbreG) | IFO3070 transformed with pLES-breA–HΔbreG | This study |
| Plasmids | ||
| pET-28a(+) | E. coli expression vector; Kmr | Novagen |
| pET-28a(+)/breD | pET-28a(+) harboring 0.2-kb NdeI-XhoI fragment containing entire breD with N-terminal His6-tag | This study |
| pET-28a(+)/N-breG | pET-28a(+) harboring 0.3-kb NdeI-XhoI fragment containing N-terminal region of breD with N-terminal His6-tag | This study |
| pLES003 | E. coli-LAB shuttle vector; Ampr Ermr | 8 |
| pLES003-b | The replication origin region for LAB (AatII fragment) of this plasmid is reversed from that of pLES003 | This study |
| pLES-PA | pLES003-b lacking lacZα, CAP-binding site, and lac operator | This study |
| pLES-PA-A | pLES-PA containing β-gal gene (PLb) | This study |
| pLES-PA-B | pLES-PA containing β-gal gene (PbreED) | This study |
| pLES-PA-C | pLES-PA containing β-gal gene (PbreED) and breD (PLb) | This study |
| pLES-PA-D | pLES-PA containing β-gal gene (PbreBC) | This study |
| pLES-PA-E | pLES-PA containing β-gal gene (PbreBC) and breG (PbreG) | This study |
| pLES-PA-F | pLES-PA containing β-gal gene and breD (PbreBC, polycistronic) | This study |
| pLES-breA–H | pLES003-b harboring 10-kb fragment containing entire brevicin 174A synthetic gene cluster | This study |
| pLES-breA–HΔbreD | pLES-breA–H containing 3-bp substitution mutation (positions +13 to +15 relative to breD start codon) | This study |
| pLES-breA–HΔbreG | pLES-breA–H containing 3-bp substitution mutation (positions +16 to +18 relative to breG start codon) | This study |
DNA preparation, manipulation, and sequencing.
A plasmid from E. coli was isolated using the Wizard Plus Minipreps DNA purification system (Promega). Chromosomal DNA and plasmids from Lactobacillus strains were isolated according to a method described previously (8). Genes and nucleotide fragments located on the brevicin 174A-biosynthesizing gene cluster (DDBJ accession no. LC062087) were PCR amplified using the plasmid isolated from the L. brevis 174A cells. Nucleotide sequencing was performed on an ABI Prism 310 genetic analyzer using the BigDye Terminator v1.1 cycle sequencing kit as described in the manufacturer's protocol (Applied Biosystems). Genetic analysis was performed using ATGC and GENETYX software (Genetyx Corporation). All of the PCRs were carried out using PrimeSTAR GXL DNA polymerase (TaKaRa) for plasmid construction.
Expression in E. coli and purification of His-tagged BreD and N-BreG.
The BreG protein was not expressed in a soluble fraction when overexpressed as a recombinant protein using an E. coli host vector system, because the C-terminal region of BreG was predicted to have a four-transmembrane-spanning domain. Therefore, we expressed only the N-terminal region of BreG (N-BreG, amino acids [aa]1 to 81). The breD and N-terminal parts of breG genes were amplified by PCR using the Ex-breD and Ex-breG primer sets, respectively (Table 2). The amplified DNA fragments were digested with NdeI and XhoI and inserted into the same site of pET-28a(+) to yield pET-28a(+)/breD and pET-28a(+)/N-breG, respectively. E. coli BL21(DE3) harboring the expression vector was grown in LB medium at 28°C. At the exponential phase of growth (optical density at 600 nm [OD600] = 0.6), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at a concentration of 1 mM to induce the expression of objective proteins. After an additional incubation for 6 h, the cells were harvested by centrifugation, washed, and resuspended in binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 5 mM imidazole). After the cell mass was disrupted by sonication, the cell debris was removed by centrifugation. The resulting cell extract was subjected to Ni(II)-chelate affinity chromatography using HisBind resin (Novagen) to purify the N-terminal histidine-tagged proteins as described in the supplier's instruction manual: the column was washed with a wash buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 60 mM imidazole), and the protein was eluted with a linear gradient of 60 to 1,000 mM imidazole. The fractions having objective proteins were pooled and concentrated by using an ultracentrifuge unit (Amicon Ultra; Millipore). The protein concentrations of the purified BreD and N-BreG were determined using a Bio-Rad protein assay kit. The purified protein was dialyzed against EMSA buffer (20 mM triethanolamine-HCl [pH 7.0], 20% [vol/vol] glycerol, 2 mM 2-mercaptoethanol, 1 mM dithiothreitol [DTT], and 100 mM KCl) and stored at 4°C until use.
TABLE 2.
Primers used in this study
| Use | Name | Sequence (5′→3′) | Restriction site | Target | Comment |
|---|---|---|---|---|---|
| Protein expression | Ex-breD-F | CAACATATGGACAATAAAATTCGTGAATATC | NdeI | breD | |
| Ex-breD-R | AAACTCGAGTTAGAAAGACTTGGGATTAAA | XhoI | breD | ||
| Ex-breG-F | CACCATATGAATATTTCTCAGAAACTTAAAC | NdeI | breG | ||
| Ex-breG-R | CTCGAGCTAAAGTTTATTATCATTTTTATTC | XhoI | breG | ||
| EMSA | EMSA-breA-F | Biotin-TCACGCTCAGCTGCTGTTTT | Region I | ||
| EMSA-breA-R | TATTTTGGTCGCGGGTACCT | Region I | |||
| EMSA-breB-F | Biotin-CAGTTTAGAAAGGTTGTTAAATCGG | Region II | |||
| EMSA-breB-R | GAATTTCTCCATTTCGATCGCTC | Region II | |||
| EMSA-breE-F | Biotin-AGCTTTCATAGTAAAACCTCCCTC | Region III | |||
| EMSA-breE-R | GTGCAATTGATAAACTATGAATCGACG | Region III | |||
| EMSA-breG-F | Biotin-ATACGTTTGTCCAAGCAGTTGTC | Region IV | |||
| DNase I footprinting assay and primer extension | FP-breBC-F | TCTGTCCTTTAAAGACCAAAAAATT | Upstream of breB | ||
| FP-breBC-R | GAATTTCTCCATTTCGATCGCTCTC | Upstream of breB | |||
| Biotin-FP-breBC-F | Biotin-TCTGTCCTTTAAAGACCAAAAAATT | Upstream of breB | |||
| Biotin-FP-breBC-R | Biotin-GAATTTCTCCATTTCGATCGCTCTC | Upstream of breB | |||
| FP-breED-F | GTAATCATTGCTACGCCAAGTAATT | Upstream of breE | |||
| FP-breED-R | TAAACTATGAATCGACGATTCCGTT | Upstream of breE | |||
| Biotin-FP-breED-F | Biotin-GTAATCATTGCTACGCCAAGTAATT | Upstream of breE | |||
| Biotin-FP-breED-R | Biotin-TAAACTATGAATCGACGATTCCGTT | Upstream of breE | |||
| Vector construction for promoter assay | PA01 | TATACTGCAGAAAACGAGGCAATTTGCCTCGTTTTCTTTTTCACTGCCCGCTTTCCAGTC | PstI | For constructing pre-pLES-PA | |
| PA02 | GCATGCCTGCAGGTCGACTC | PstI | For constructing pre-pLES-PA | ||
| PA03 | CAGTGAATTCGAGCTCGGTACCCGGGGATCCTCT | EcoRI | For constructing pLES-PA | ||
| PA04 | TATAGAATTCCATATGATAGACGGTTTTTC | EcoRI | For constructing pLES-PA | ||
| PA-β-gal-F1 | TATAGGTACCAGTTGTTGACAGAATGGACATACTATGATATATTGTTGCTATAGCGTCACACAGGAAACAGCTATG | KpnI | β-Gal gene | Containing PLb sequence | |
| PA-β-gal-R1 | TATAGGATCCAAAAGAAAACGAGGCAAATTGCCTCGTTTTTAATGGATTTCCTTACGCGA | BamHI | β-Gal gene | Containing terminator sequence | |
| PA-β-gal-F2 | TATAGGTACCTCACACAGGAAACAGCTATG | KpnI | β-Gal gene | ||
| PA-β-gal-R2 | TATAGGATCCGCCCGGTTATTATTATTTTTG | BamHI | β-Gal gene | ||
| PA-PBreBC-F | TATAGGTACCAGACCAAAAAATTATAAATATATGG | KpnI | PBreBC | For pLES-PA-D and E | |
| PA-PBreBC-R | TATAGGTACCTTATTTTTTCCAAATGACGAATCAT | KpnI | PBreBC | For pLES-PA-D and E | |
| PA-PBreED-F | GAGCGGTACCAACTTTTAGATTTTTCATAATTAAT | KpnI | PBreED | For pLES-PA-B, C, and F | |
| PA-PBreED-R | TATAGGTACCAAATTTGCAATATATACACGTCATT | KpnI | PBreED | For pLES-PA-B, C, and F | |
| PA-BreD-F1 | TATAGGATCCGAGGCGAATCCAATGGACAATAAAA | BamHI | breD | For pLES-PA-C | |
| PA-BreD-F2 | AAAAGGATCCAGTTGTTGACAGAATGGACA | BamHI | breD | For pLES-PA-F | |
| PA-BreD-R | GCGCCTGCAGTTAGAAAGACTTGGGATTAAATAGA | PstI | breD | For pLES-PA-C and F | |
| PA-BreG-F | TTTTGGATCCATTAGAGGCATTCATGAACCATGAT | BamHI | breG | For pLES-PA-E | |
| PA-BreG-R | TATACTGCAGAGAATATACTTTTAAAGCCAGAATA | PstI | breG | For pLES-PA-E | |
| PA-β-gal-deletion-F | AACAGCTATGGACAATAAAATTCGT | For pLES-PA-F | |||
| PA-β-gal-deletion-R | TTGTCCATAGCTGTTTCCTGTGTGAC | For pLES-PA-F | |||
| Brevicin 174A biosynthetic gene cluster construction | IF_breA–H_insert-F | GGTACCCGGGGATCCAGTTATGGATTTGCACCAGAACCTA | Gene cluster | ||
| IF_breA–H_insert-R | CTTGCATGCCTGCAGTTAAACTACCAGACGCCAATTGAGG | Gene cluster | |||
| IF_breA–H_vector-F | CTGCAGGCATGCAAGCTTGG | pLES003-b | |||
| IF_breA–H_vector-R | GGATCCCCGGGTACCGAGCT | pLES003-b | |||
| breD_mutation-F | GAGGCGAATCCAATGGACAATAAATAGCGTGAATATCGGAAG | For breD mutation | |||
| breD_mutation-R | CTTCCGATATTCACGCTATTTATTGTCCATTGGATTCGCCTC | For breD mutation | |||
| breG_mutation-F | GATATGAATATTTCTCAGTAGCTTAAACAATGTCGTAGTGCC | For breG mutation | |||
| breG_mutation-R | GGCACTACGACATTGTTTAAGCTACTGAGAAATATTCATATC | For breG mutation | |||
| qRT-PCR | qPCR-breB-F | GAAATATACCGGACCAAACTACCG | breB | ||
| qPCR-breB-R | ACCACCAACGATACCACCAAC | breB | |||
| qPCR-breC-F | GGGGAAATGCAGCAACAG | breC | |||
| qPCR-breC-R | ATTGTGAAACGCCCCAGATAG | breC | |||
| qPCR-breE-F | ACCGAGAAGCCATAAAACACCT | breE | |||
| qPCR-breE-R | CGGCAACAAACGATCCAA | breE | |||
| qPCR-rep-F | GGCAGTCTAACACCGACCA | rep | |||
| qPCR-rep-R | CTTTGCGTTCAGGTTTCCA | rep | |||
| qPCR-gyrB-F | TGAATACCGAAAAGGCCAAA | gyrB | |||
| qPCR-gyrB-R | ACCAACACCCGCACCAA | gyrB | |||
| qPCR-β-gal-F | CAGGTAGCAGAGCGGGTAAA | β-Gal gene | |||
| qPCR-β-gal-R | ATCCCAGCGGTCAAAACA | β-Gal gene |
EMSA.
Biotin-labeled 200-bp DNA fragments which were designed to obtain the breA, breB, breE, and breG promoter regions located about 190 bases upstream from each start codon were amplified by PCR using the appropriate electrophoretic mobility shift assay (EMSA) gene primer sets (Table 2). Nonbiotinylated primers were used for the preparation of nonlabeled DNA fragments. An appropriate amount of purified proteins (0.25 to 256 pmol) and 5 fmol of biotinylated DNA fragments were incubated in EMSA buffer containing 50 μg/ml poly(dI-dC) in a volume of 20 μl at 25°C for 15 min. When necessary, 5 pmol of nonlabeled DNA was added to the reaction mixture. After the addition of 5 μl of loading dye (20% [vol/vol] glycerol, 0.1% [wt/vol] xylene cyanol, and 1% [wt/vol] bromophenol blue), the mixture was subjected to 6% native polyacrylamide gel electrophoresis in TBE buffer (90 mM Tris-borate [pH 8.3] and 2 mM EDTA) at 100 V for 3 h. The DNA fragments were transferred on a Hybond-N+ membrane (GE Healthcare) with a 0.5× TBE buffer using a Trans-Blot SD semidry transfer cell (Bio-Rad) at 380 mA for 30 min. After the electroblotting procedure, the DNA fragments were fixed on the membrane via heat treatment at 80°C for 2 h. DNA detection was performed using a chemiluminescent nucleic acid detection module (Pierce) and an ImageQuant LAS 4000 biomolecular imager (GE Healthcare) according to the manufacturer's instruction manual.
DNase I footprinting assay.
Each of 291-bp DNA fragments which were designed with the breE and breB promoter regions, including 193 and 279 bases upstream of each start codon, respectively, was amplified by PCR using the FP-breED and FP-breBC primer sets (Table 2). If necessary, 5′-end-biotinylated primer was used to prepare a biotin-labeled DNA fragment. A reaction mixture containing 0.6 pmol of the target DNA fragment and appropriate amounts of purified BreD or N-BreG (10 to 40 pmol or 300 to 1,200 pmol, respectively) was prepared in 200 μl of EMSA buffer. After incubation at 25°C for 30 min, a 30-μl portion of DNase I solution (0.04 U/μl DNase I [TaKaRa], 400 mM Tris-HCl [pH 7.5], 80 mM MgCl2, and 50 mM DTT) was added to the mixture, followed by further incubation for 4 min on ice. The reaction was stopped by the addition of 270 μl of a solution composed of 100 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1% (wt/vol) sodium N-lauroyl sarcosinate, 10 mM EDTA-NaOH (pH 8.0), and 25 μl/ml of salmon sperm DNA. After phenol-chloroform extraction and ethanol precipitation, 4 μl of nuclease-free water and 1 μl of a formamide-dye mixture (44) were added to the pellet, and the resulting DNA sample was subjected to 6% polyacrylamide–7 M urea gel electrophoresis at 25 W for 2 h. DNA (A+G and C+T) ladders were prepared by the Maxam-Gilbert chemical cleavage method (45). The separated fragments were visualized using the chemiluminescent nucleic acid detection module and ImageQuant LAS 4000 biomolecular imager according to the manufacturer's instruction manual.
RNA extraction and primer extension analysis.
Total RNA was extracted from each LAB cell using NucleoSpin RNA II (Macherey-Nagel) according to the manufacturer's instruction manual. After extracting the RNA, residual DNA fragments were digested with DNase I (TaKaRa) in reaction buffer (40 mM Tris-HCl [pH 7.5], 0.8 U/μl RNase inhibitor [TaKaRa], 8 mM MgCl2, 1 mM CaCl2, 1 mM MnCl2, and 5 mM DTT). A portion (15 μg) of the total RNA was used to synthesize cDNA using the PrimeScript RT-PCR kit (TaKaRa) according to the manufacturer's instruction manual. The 5′-biotinylated biotin-FP-breBC-R and biotin-FP-breED-F primers (Table 2), designed with an antisense sequence that pairs with nucleotides −13 to +12 and +86 to +110 of breB and breE, respectively, were used for the primer extension analysis. The reaction sample was subjected to 6% polyacrylamide–7 M urea gel electrophoresis, and detected was as described in the previous section.
Transformation of LAB.
The preparation of competent cells derived from the LAB strain was conducted according to methods described previously (46, 47). Competent cells (50 μl) mixed with the given plasmid vector were placed into a 0.1-cm electroporation cuvette (Bio-Rad) and held on ice for 5 min. After pulsing under specific conditions (field strength, 7.5 kV/cm; capacitance, 25 μF; resistance, 400 Ω) using a Gene Pulser XCell (Bio-Rad), the cell suspension was immediately diluted with 1 ml of MRS broth containing 0.5 M sucrose and 0.1 M MgCl2 and then incubated at 28°C for 6 h. A portion of the cell suspension (100 μl) was placed on selective medium and incubated anaerobically at 28°C for 1 to 2 days. Before the transformation, all plasmid constructs were prepared as the methylation-free plasmid from E. coli HST04.
Promoter assay.
All primers used for constructing the vectors are listed in Table 2. A 6.0-kb DNA fragment was amplified from pLES003-b by PCR using the primers PA01 and PA02. The amplified DNA was digested with PstI and self-ligated to yield pre-pLES-PA. A 5.8-kb DNA fragment was amplified to form pre-pLES-PA by PCR using the primers PA03 and PA04. The amplified DNA was digested with EcoRI and self-ligated to yield pLES-PA, which was designed to have a terminator sequence and to lose a lacZα gene, CAP binding site, and lac operator sequence. To construct plasmid pLES-PA-A, a KpnI/BamHI-digested 3.2-kb fragment, which corresponds to the β-gal gene originating from an E. coli K-12 W3110 genome sequence (DDBJ accession no. AP009048) (48, 49) with PLb and terminator sequence, was amplified by using the PA-β-gal-F1 and PA-β-gal-R1 primers and inserted into the same site of pLES-PA. Additionally, plasmids pLES-PA-B to -F were constructed by insertion of the appropriate fragments (Fig. 3) by PCR amplification using the primer sets (Table 2). The L. plantarum IFO 3070 strain was transformed with the constructed plasmid. To measure β-gal gene transcription, qRT-PCR analyses were performed. The gyrB gene was used as a housekeeping gene. Both the gyrB and β-gal genes were amplified using the gene-specific primer sets listed in Table 2.
Construction of plasmids carrying the brevicin 174A biosynthetic gene cluster.
The brevicin 174A biosynthetic gene cluster was PCR amplified using the IF_breA–H_insert primer sets and the whole region of pLES003-b except for the multicloning site by PCR using the IF_breA–H_vector primer sets. The two amplified DNA fragments were ligated using an In-Fusion HD cloning kit (Clontech) to construct the pLES-breA–H plasmids.
We constructed two mutants of the biosynthetic gene cluster. The mutation was performed using PrimeSTAR Max DNA polymerase according to the manufacturer's instruction manual. To construct each mutant plasmid having the nonsense breD or breG mutations, respectively, the breD or breG_mutation primer sets were used. The strain L. plantarum IFO 3070, which has been confirmed to have neither the brevicin 174A-biosynthesizing gene cluster nor the brevicin 174A resistance gene, was transformed with each resulting mutant plasmid. The transcriptional assay of each gene was done by qRT-PCR. For the qRT-PCR analysis, gyrB and breB, breC, and breE were amplified using each cDNA preparation as a template and the gene-specific primer sets listed in Table 2.
Assay of antibacterial activity.
The antibacterial activity of brevicin 174A was confirmed by the agar well diffusion method described previously (2, 8, 50).
qRT-PCR analysis.
Total RNA, which was extracted and purified from each construct at several growth points (OD600 = 0.1, 0.5, 1.0, and 1.5), was converted to cDNA using the ReverTra Ace qPCR RT master mix with gDNA remover (Toyobo) according to the manufacturer's instruction manual. qRT-PCR was performed on the PikoReal real-time PCR system (Thermo Fisher Scientific) using the Kapa SYBR Fast qPCR master mix (Kapa Biosystems). The PCR was performed as follows: an initial 30 s at 95°C followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. The relative transcriptions of the target genes were normalized to a housekeeping gene (gyrB) by the ΔΔCT method. In this experiment, a proportional relationship was observed between the dilution factors of the template and the signal intensity.
ACKNOWLEDGMENTS
This work was financially supported by a JSPS Grant-in-Aid for Scientific Research (grant number 20790065).
We are grateful to the Research Center for Molecular Medicine, the Faculty of Medicine, and the Analysis Center of Life Science, Hiroshima University, for the use of their facilities.
REFERENCES
- 1.Tagg JR, Dajani AS, Wannamaker LW. 1976. Bacteriocins of gram-positive bacteria. Bacteriol Rev 40:722–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klaenhammer TR. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 3.McAuliffe O, Ross RP, Hill C. 2001. Lantibiotics: structure and mode of action. FEMS Microbiol Rev 25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 5.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholts J. 1996. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 6.Delves-Broughton J. 2005. Nisin as a food preservative. Food Australia 57:525–527. [DOI] [PubMed] [Google Scholar]
- 7.Noda M, Miyauchi R, Danshiitsoodol N, Higashikawa F, Kumagai T, Matoba Y, Sugiyama M. 2015. Characterization and mutational analysis of a two-polypeptide bacteriocin produced by citrus iyo-derived Lactobacillus brevis 174A. Biol Pharm Bull 38:1902–1909. doi: 10.1248/bpb.b15-00505. [DOI] [PubMed] [Google Scholar]
- 8.Wada T, Noda M, Kashiwabara F, Jeon HJ, Shirakawa A, Yabu H, Matoba Y, Kumagai T, Sugiyama M. 2009. Characterization of four plasmids harboured in a Lactobacillus brevis strain encoding a novel bacteriocin, brevicin 925A, and construction of a shuttle vector for lactic acid bacteria and Escherichia coli. Microbiology 155:1726–1737. doi: 10.1099/mic.0.022871-0. [DOI] [PubMed] [Google Scholar]
- 9.Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 10.Oppegård C, Rogne P, Emanuelsen L, Kristiansen PE, Fimland G, Nissen-Meyer J. 2007. The two-polypeptide class II bacteriocins: structure, production, and mode of action. J Mol Microbiol Biotechnol 13:210–219. doi: 10.1159/000104750. [DOI] [PubMed] [Google Scholar]
- 11.Nissen-Meyer J, Oppegård C, Rogne P, Haugen HS, Kristiansen PE. 2010. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob Proteins 2:52–60. doi: 10.1007/s12602-009-9021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 13.de Vos WM, Kulpers OP, van der Meer JR, Slezen RJ. 1995. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol 17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 14.Nilsen T, Nes IF, Holo H. 1998. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J Bacteriol 180:1948–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brurberg MB, Nes IF, Eijsink VG. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol 26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 16.Diep DB, Johnsborg O, Risøen PA, Nes IF. 2001. Evidence for dual functionality of the operon plnABCD in the regulation of bacteriocin production in Lactobacillus plantarum. Mol Microbiol 41:633–644. doi: 10.1046/j.1365-2958.2001.02533.x. [DOI] [PubMed] [Google Scholar]
- 17.Kleerebezem M, Kuipers OP, de Vos WM, Stiles ME, Quadri LEN. 2001. A two-component signal-transduction cascade in Carnobacterium piscicola LV17B: two signaling peptides and one sensor-transmitter. Peptides 22:1597–1601. doi: 10.1016/S0196-9781(01)00494-6. [DOI] [PubMed] [Google Scholar]
- 18.Rud I, Jensen PR, Naterstad K, Axelsson L. 2006. A synthetic promoter library for constitutive gene expression in Lactobacillus plantarum. Microbiology 152:1011–1019. doi: 10.1099/mic.0.28599-0. [DOI] [PubMed] [Google Scholar]
- 19.Brennan RG, Matthews BW. 1989. The helix-turn-helix DNA binding motif. J Biol Chem 264:1903–1906. [PubMed] [Google Scholar]
- 20.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. 1988. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 21.Steinmetzer K, Behlke J, Brantl S, Lorenz M. 2002. CopR binds and bends its target DNA: a footprinting and fluorescence resonance energy transfer study. Nucleic Acids Res 30:2052–2060. doi: 10.1093/nar/30.9.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonnell GE, McConnell DJ. 1994. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J Bacteriol 176:5831–5834. doi: 10.1128/jb.176.18.5831-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brantl S, Wagner EG. 1997. Dual function of the copR gene product of plasmid pIP501. J Bacteriol 179:7016–7024. doi: 10.1128/jb.179.22.7016-7024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manson JM, Keis S, Smith JM, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother 48:3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauntlett JC, Gebhard S, Keis S, Manson JM, Pos KM, Cook GM. 2008. Molecular analysis of BcrR, a membrane-bound bacitracin sensor and DNA-binding protein from Enterococcus faecalis. J Biol Chem 283:8591–8600. doi: 10.1074/jbc.M709503200. [DOI] [PubMed] [Google Scholar]
- 26.van Tyne D, Martin MJ, Gilmore MS. 2013. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 29:895–911. doi: 10.3390/toxins5050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas W, Shepard BD, Gilmore MS. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 28.Rumpel S, Razeto A, Pillar CM, Vijayan V, Taylor A, Giller K, Gilmore MS, Becker S, Zweckstetter M. 2004. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J 23:3632–3642. doi: 10.1038/sj.emboj.7600367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen-Orr SS, Milo R, Mangan S, Alon U. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 30.Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld N, Elowitz MB, Alon U. 2002. Negative autoregulation speeds the response times of transcription networks. J Mol Biol 323:785–793. doi: 10.1016/S0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 32.Pinho R, Garcia V, Irimia M, Feldman MW. 2014. Stability depends on positive autoregulation in boolean gene regulatory networks. PLoS Comput Biol 10:e1003916. doi: 10.1371/journal.pcbi.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maamar H, Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams JW, Cui X, Levchenko A, Stevens AM. 2008. Robust and sensitive control of a quorum-sensing circuit by two interlocked feedback loops. Mol Syst Biol 4:234. doi: 10.1038/msb.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sha W, Moore J, Chen K, Lassaletta AD, Yi CS, Tyson JJ, Sible JC. 2003. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci U S A 100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagowski CP, Ferrell JE Jr. 2001. Bistability in the JNK cascade. Curr Biol 11:1176–1182. doi: 10.1016/S0960-9822(01)00330-X. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. 2007. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333. doi: 10.1016/j.jmb.2007.02.096. [DOI] [PubMed] [Google Scholar]
- 38.Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 39.Diep DB, Håvarstein LS, Nes IF. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol Microbiol 18:631–639. [DOI] [PubMed] [Google Scholar]
- 40.Diep DB, Håvarstein LS, Nes IF. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol 178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diep DB, Myhre R, Johnsborg O, Aakra A, Nes IF. 2003. Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol Microbiol 47:483–494. doi: 10.1046/j.1365-2958.2003.03310.x. [DOI] [PubMed] [Google Scholar]
- 42.Maldonado A, Jiménez-Díaz R, Ruiz-Barba JL. 2004. Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific gram-positive bacteria is mediated by an autoinduction mechanism. J Bacteriol 186:1556–1564. doi: 10.1128/JB.186.5.1556-1564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 44.Maxam AM, Gillbert W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavage. Methods Enzymol 65:499–560. doi: 10.1016/S0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 45.Maxam AM, Gilbert W. 1977. A new method for sequencing DNA. Proc Natl Acad Sci U S A 74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alegre MT, Rodriguez MC, Mesas JM. 2004. Transformation of Lactobacillus plantarum by electroporation with in vitro modified plasmid DNA. FEMS Microbiol Lett 241:73–77. doi: 10.1016/j.femsle.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Dunny GM, Lee LN, LeBlanc DJ. 1991. Improved electroporation and cloning vector system for Gram-positive bacteria. Appl Environ Microbiol 57:1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Reenen CA, Dicks LM, Chikindas ML. 1998. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol 84:1131–1137. doi: 10.1046/j.1365-2672.1998.00451.x. [DOI] [PubMed] [Google Scholar]