ABSTRACT
Pseudomonas aeruginosa exhibits flagellum-mediated swimming in liquid and swarming on hydrated surfaces under diverse nutrient conditions. Prior studies have implicated a phosphodiesterase, DipA, in regulating these flagellum-mediated motilities, but collectively, the necessity for DipA was unclear. In this study, we find that the medium composition conditionally constrains the influence of DipA on flagellar motility. We show that DipA exhibits more influence on minimal medium supplemented with glutamate or glucose, where flagellar motility was negated for the dipA mutant. Conversely, a dipA-deficient mutant exhibits flagellar motility when growing with LB Lennox broth and minimal medium supplemented with Casamino Acids. Swarming under these rich medium conditions occurs under elevated levels of c-di-GMP. We also demonstrate that the influence of DipA upon swimming often differs from that upon swarming, and we conclude that a direct comparison of the motility phenotypes is generally valid only when characterizing motility assay results from the same medium composition. Our results are consistent with the explanation that DipA is one of several phosphodiesterases responding to the nutrient environment sensed by P. aeruginosa. On minimal medium with glutamate or glucose, DipA is dominant; however, on rich medium, the necessity of DipA is fully negated.
IMPORTANCE Motile and ubiquitous bacteria such as Pseudomonas aeruginosa can quickly colonize surfaces and form biofilms in numerous environments such as water distribution systems, soil, and the human lung. To effectively disrupt bacterial colonization, it is imperative to understand how bacteria regulate motility in these different growth environments. Here, we show that the phosphodiesterase DipA is not required for flagellar motility under all nutrient conditions. Thus, the maintenance of intracellular c-di-GMP levels to promote flagellar motility or biofilm development must be conditionally regulated by differing phosphodiesterases in variation with select nutrient cues.
KEYWORDS: Pseudomonas aeruginosa, cyclic di-GMP, flagellar motility, media, swarming
INTRODUCTION
Pseudomonas aeruginosa is a widespread environmental bacterium and opportunistic pathogen (1). To enable growth in a wide array of environments, P. aeruginosa boasts a robust metabolism, including an ability to metabolize a variety of sugars and proteins, though amino acids are a preferred carbon source (2). P. aeruginosa is also a motile organism that expresses a single polar flagellum, which enables both swimming and swarming motilities (3, 4). Swimming is the cell-based motility of individual cells through a liquid medium. Swarming is the community-based movement of cell groups through a thin liquid layer on a surface. Swimming and swarming are both considered to offer competitive advantages over nonmotile microbes, which include an ability of flagellum-motile bacteria to seek out favorable growth environments through the exploration and movement toward nutrients or away from toxins and the self-assemblage of enduring biofilm communities (5).
Swarming in P. aeruginosa can be affected by many factors. Previous studies of P. aeruginosa swarming have noted the role of rhamnolipid (6, 7), a surfactant produced by P. aeruginosa that lowers the surface tension and viscosity, thus improving surface motility. Rhamnolipid production can be regulated by the acyl homoserine lactone-regulated Rhl quorum-sensing regulon (8, 9). However, rhamnolipid production is not solely controlled by population density and previously established quorum-sensing regulatory mechanisms, as nutrient effects (for specific carbon and nitrogen compounds and iron limitation) are well known for the pseudomonads (10–12). Other studies have also shown that supplementation with specific amino acids and carboxylic acids (13, 14) or limiting phosphate (15) or iron (7) all appear to promote and improve swarming. The regulatory link(s) between these nutrient cues and swarming is still not clear. It is likely that many of these environmental nutrients are signals that cue changes in intracellular concentrations of bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP). For example, Pseudomonas fluorescens acts to interpret extracellular phosphate concentrations through LapD (16), but a comparable cascade has not been demonstrated in P. aeruginosa. Although specific nutrients have not been connected to c-di-GMP regulation in P. aeruginosa, it has been hypothesized that P. aeruginosa regulates its surface behavior in a hierarchical cascade, where cAMP signals from PilY1 govern downstream c-di-GMP regulation (17).
It has been shown that low levels of c-di-GMP promote P. aeruginosa motility, while high levels promote sessile (biofilm) lifestyles (3, 18). c-di-GMP is synthesized by diguanylate cyclases containing a GG(D/E)EF domain; phosphodiesterases containing an EAL domain cleave c-di-GMP (18). P. aeruginosa harbors genes for the production of many phosphodiesterases which could be involved in controlling motility, including the phosphodiesterase dipA (dispersion induced phosphodiesterase A [PA5017]) (19). When DipA is not produced, c-di-GMP levels in the cell have been shown to increase, promoting a nonmotile phenotype (20). This sessile lifestyle could also result from an interaction between flagella and chemotaxis machinery, partially mediated by DipA (21).
The results from previous studies are somewhat varied on which flagellum-mediated motilities are affected by dipA, citing either strain differences or medium differences for the reported observations (20). Here, we show that dipA confers swarming and flagellar motility in many, but not all, growth media. We find more generally that medium selection for motility assays greatly impacts the effects of select specific nutrients and the effect of dipA on swimming and swarming motilities. Additionally, we show that increasing the concentration of nutrients in media can actually stimulate motility to different degrees, highlighting the importance of medium uniformity on the interpretation of results from motility assays. Lastly, we report Difco Lennox broth and M8 medium supplemented with 0.2% glucose and 0.5% Casamino Acids promote robust swarming during conditions when cellular levels of c-di-GMP are elevated.
RESULTS
Effect of dipA on flagellum-mediated motility is nutritionally conditional.
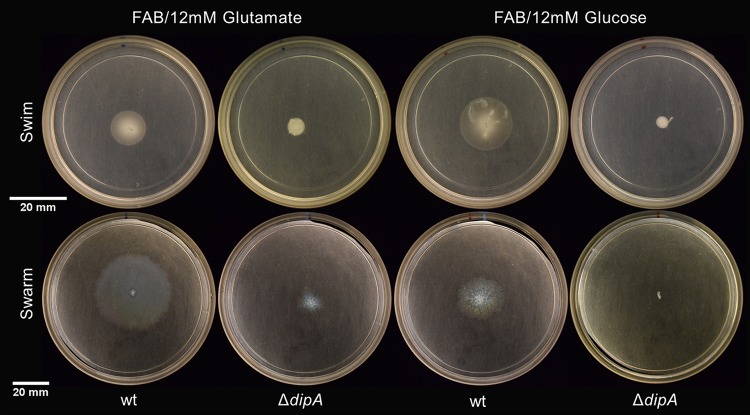
The predominant description of P. aeruginosa swarm motility depends upon an active polar flagellum and a sufficient quorum-sensing active population to produce the biosurfactant rhamnolipid (12, 13, 22). However, some previous studies have shown varied phenotypes of P. aeruginosa swarming that depend upon the presence of select amino acids. The requirement for rhamnolipid production can be negated when growing P. aeruginosa under these “select” conditions (14). We were interested to identify the regulatory elements that specifically influence P. aeruginosa swarming in a nutrient-dependent manner. In our initial experiments, we screened for reduced swarming in an rhlAB-deficient parent that still exhibited swim motility from a library of roughly 3,000 mariner transposon mutants. One of the transposon mutants that showed consistent behaviors in subsequent experiments was disrupted in the phosphodiesterase dipA (Fig. 1). On minimal medium containing either 12 mM glutamate or 12 mM glucose as the carbon source, a deletion mutant of dipA constructed in an isogenic background showed little to no swarming or swimming (Fig. 2). In trans complementation of dipA restored the swarming phenotype (see Fig. S1 in the supplemental material).
FIG 1.
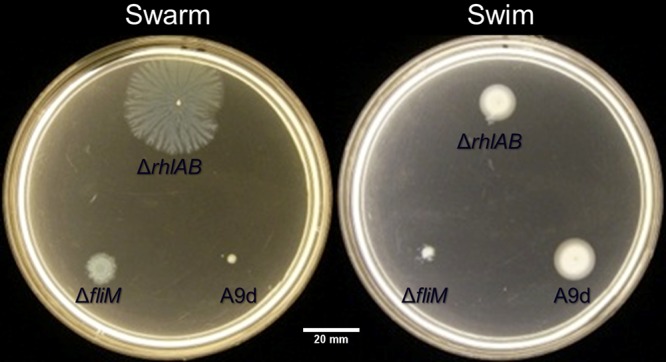
Transposon mutant PA_A9d (bottom right on plates) is a swarming-negative and swim-positive mutant of the parent rhamnolipid-deficient strain. A flagellar mutant (ΔfliM) strain is included as negative controls at the bottom left of the plates. The ΔrhlAB parent strain is shown at the top. Assays were conducted on FAB-glutamate. Swim assay contained 0.3% (wt/vol) agar; swarm assay contained 0.45% (wt/vol) Noble agar.
FIG 2.
Variations in flagellum-mediated motility are apparent on minimal media. The wt swims and swarms to a lesser extent on minimal medium with glutamate or glucose as the sole carbon source. The ΔdipA mutant does not swim, nor does it swarm.
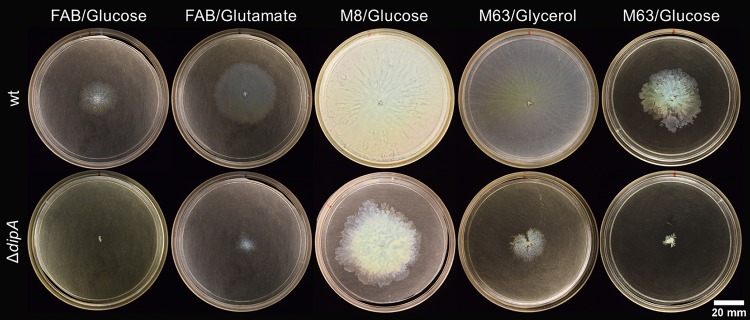
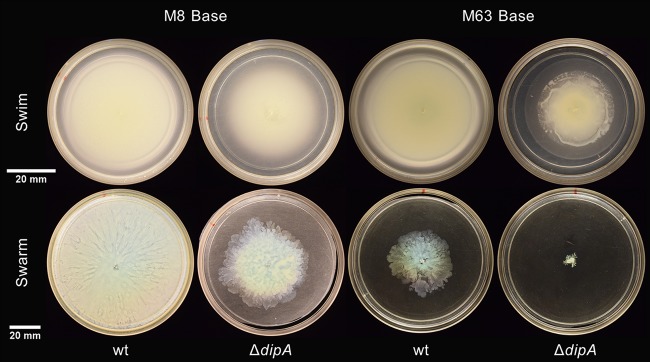
Previous studies have examined P. aeruginosa swarming using M8 (19) or M63 (20) minimal medium supplemented with 0.2% glucose and 0.5% Casamino Acids and have yielded varied results. We hypothesized that this variation was not just due to differing lab protocols or strain backgrounds, and we examined the swimming and swarming of both wild-type (wt) and ΔdipA strains on M8 and M63 media in our laboratory (Fig. 3). The ΔdipA mutant exhibited both swimming and swarming motility on M8 medium. On all media examined, the motility of the ΔdipA strain was some degree less than the motility of the wt strain. Interestingly, all flagellar motility was substantially less when growing on M63 medium compared to that on other media (Fig. 3). When M63 medium was supplemented with 0.2% glycerol (a manufacturer-suggested alternative to glucose) and 0.5% Casamino Acids, the swarming of the wt was much more robust (Fig. 4). A substitution of glycerol for glucose in M63 medium marginally improved swarming in the ΔdipA mutant. The swim motility of the ΔdipA mutant was also somewhat impaired on M63 medium; furthermore, the ΔdipA mutant exhibited some amount of surface motility on these 0.2% (wt/vol) agar assays (Fig. 5).
FIG 3.
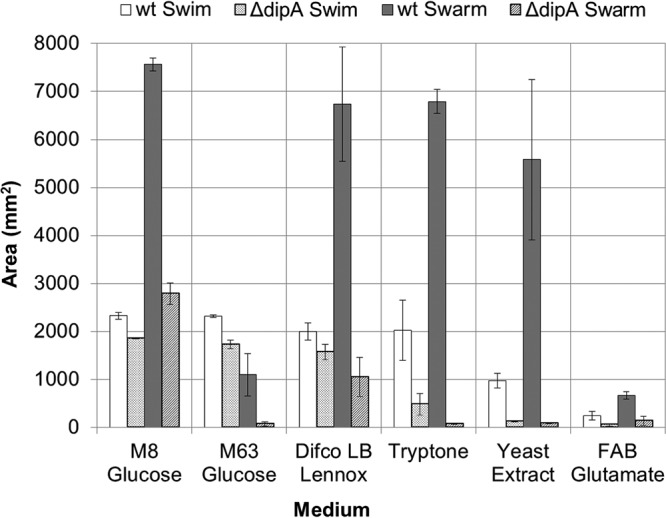
Comparison of swim and swarm areas for wt and dipA deficient mutant strains on tested media. Average swarm area for wt is consistent, unless using M63 medium with glucose or FAB-glutamate, for which all strains show decreased motility. The wt swim area varies more than wt swarming, but not as much as the ΔdipA mutant. The ΔdipA mutant swarms only on M8 medium with glucose and LB Lennox broth. All assays were performed in triplicates. Error bars indicate standard deviations. Maximum swarm area is 7,853 mm2, and maximum swim area is 2,827 mm2 (based on plate size).
FIG 4.
Swarming of the wt (top) on different media compared to that of a ΔdipA deficient mutant (bottom). FAB was supplemented only with 12 mM carbon source. M8 and M63 media were supplemented with either 0.2% glucose or 0.2% glycerol and 0.5% Casamino Acids.
FIG 5.
Variations in flagellum-mediated motility on M8 (left) and M63 (right) media supplemented with 0.2% glucose and 0.5% Casamino Acids. The wt swims (top) and swarms (bottom) abundantly on M8 medium, but swarming is reduced on M63 medium. The swimming of the ΔdipA mutant is uniform on both media, though the mutant swarms more on M8 than on M63.
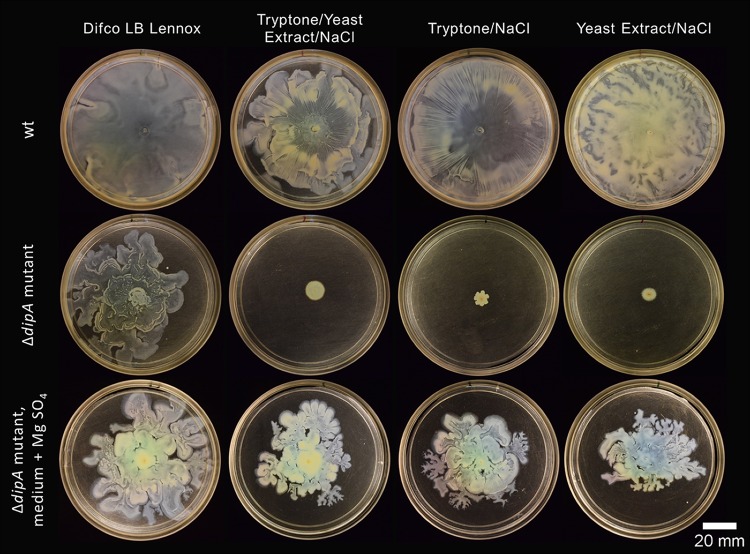
We expanded our investigation of nutrient influences to include other rich media commonly used by P. aeruginosa researchers to examine if the differences in the swim- and swarm-impaired ΔdipA mutant phenotypes were due to a lack of available nutrients. Motility assays were conducted using LB Lennox broth, as well as with tryptone and yeast extract—the organic constituents of LB (we did not focus on NaCl). All rich media supported wt swarming and swimming, as expected (Fig. 3). There was no significant difference between wt swarm areas on supplemented or rich media (assessed using a single-factor analysis of variance [ANOVA]); wt swarm zones were significantly smaller on fastidious anaerobe broth (FAB)-glutamate. Again, the flagellar motility exhibited by the ΔdipA mutant neither equaled nor paralleled that of the wt. LB Lennox broth clearly supported the swim and swarm motility of the ΔdipA mutant. However, the ΔdipA mutant was significantly impaired for flagellar motility on both tryptone and yeast extract (Fig. 6). Unexpectedly, recombining tryptone and yeast extract in the same ratio as that found in LB Lennox broth did not restore the swarming phenotype in the ΔdipA mutant (Fig. 7). We additionally observed that the motilities with LB Lennox broth compared to those with tryptone plus yeast extract plus NaCl were not uniformly equivalent in these experiments. The motility differences we observed for both wt and ΔdipA mutant strains did not correlate with their planktonic growth rates. As expected, minimal media supported less cell growth than rich or supplemented media (see Fig. S2). However, the most robust planktonic growth was seen on a medium that did not support swarming in the ΔdipA mutant.
FIG 6.
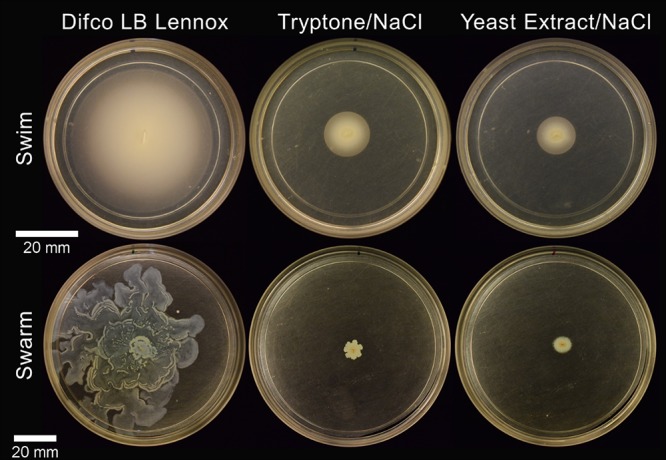
ΔdipA mutant flagellum-mediated motility assays on rich media. The dipA deficient mutant readily swims (top) and swarms (bottom) on LB Lennox broth (left). However, when the medium contains only tryptone (middle) or yeast extract (right) with NaCl, the strong motility phenotype is lost.
FIG 7.
The wt (top) swarms easily on rich media, while the ΔdipA mutant (middle) only swarms on LB Lennox broth. Addition of 1 mM MgSO4 (bottom) enables the ΔdipA mutant to swarm, restoring a robust swarming phenotype on the remaining rich media tested.
As tryptone can be magnesium deficient (23), magnesium sulfate was added to tryptone and yeast extract media to a concentration of 1 mM. This addition restored swarming in the ΔdipA mutant to near the amount observed on LB Lennox broth (Fig. 7). However, magnesium limitation did not fully explain the lack of swarming in the ΔdipA mutant on minimal media, as M8, M63, and FAB minimal media all had abundant (≈1 mM) levels of magnesium.
To investigate the possibility that these variations in medium compositions were acting through DipA to promote changes in rhamnolipid production (or some possible alternative surfactant), we performed drop collapse assays (6, 10, 24–26). No differences were apparent between the wt and the ΔdipA mutant (compared to the ΔrhlAB mutant control), indicating that these swarming differences do not stem from differences in surfactant production (see Fig. S3).
Doubling rich medium components, but not glucose or glutamate, increases flagellum-mediated motility.
As strains grown on supplemented and rich media generally swarmed and grew more under planktonic conditions than on minimal media, we examined whether reduced swarming or swimming ΔdipA phenotypes were due to a lack of available nutrients. To test this, medium conditions that did not support flagellum-mediated motility in the ΔdipA mutant, primarily tryptone and yeast extract, were doubled and the motility areas were measured as before.
The doubling of tryptone and yeast extract generally increased the motility 1.5- to 3-fold. The strongest response came on 2× tryptone swarm plates, where the area covered by the wt swarm was 2.7 times greater and that covered by the ΔdipA swarm was 3.2 times greater than that in the 1× condition. Yeast extract swarm plates were the only tested condition where the doubling of the amount in the medium had no significant effect (Fig. 8).
FIG 8.
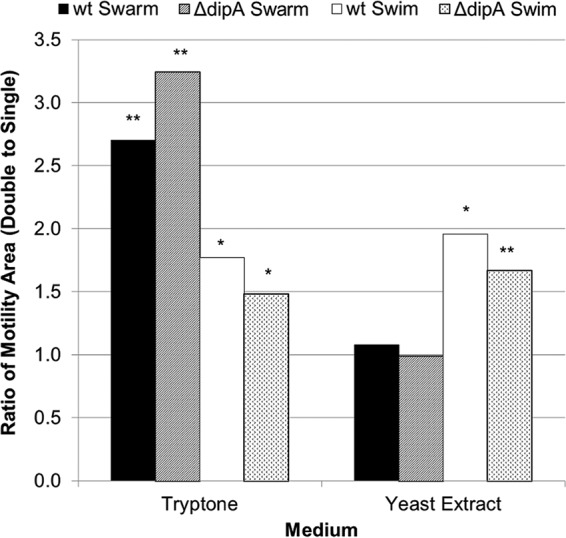
Effect of doubling the concentration of tryptone or yeast extract on motility. Average areas of motility were measured for each concentration. The double-concentration areas were then divided by the single-concentration areas to get the ratios shown. Doubling the tryptone had a positive impact on flagellum-mediated motility, significantly increasing both wt and ΔdipA strain swimming and swarming. However, doubling the yeast extract only increased swimming motility and had no impact on swarming in either strain. All assays were performed in triplicates. *, P value of <0.05 compared to the 1× concentration; **, P value of <0.01 compared to the 1× concentration.
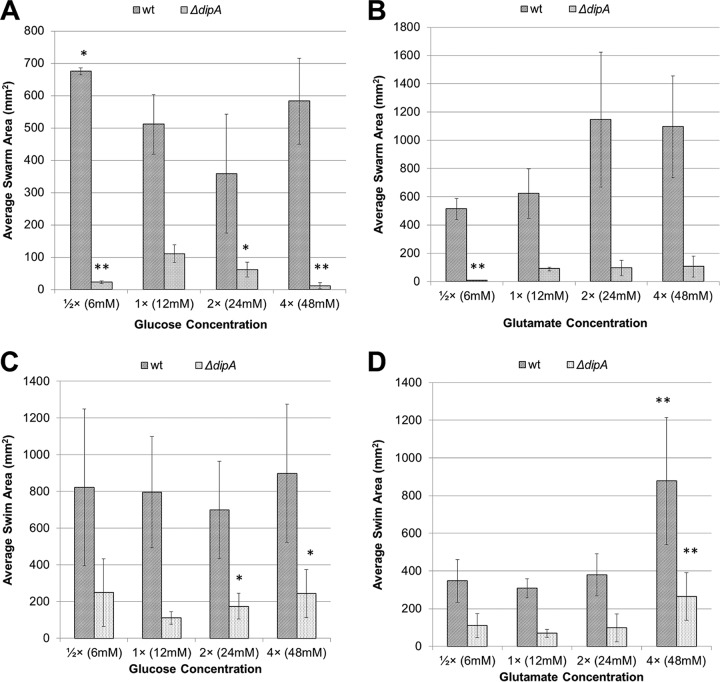
Because doubling these rich medium components generally improved the motility, we hypothesized that similar results would be observed in defined minimal media. However, the swarming of the wt on glucose actually decreased when the concentration was increased to the 2× condition (Fig. 9). Furthermore, decreasing the glucose concentration ([1/2]×) promoted increased swarming. The highest (4×) glucose condition yielded a swarm area comparable to that from the [1/2]× condition. Using glutamate, the trend was for more swarming, as expected, as 2× glutamate increased swarming. However, further increases did not additionally increase the wt swarm area.
FIG 9.
Effects of carbon source concentration on average swarm and swim areas in wt and the ΔdipA mutant. (A) Lowering the glucose concentration significantly improved wt swarming. Raising the glucose concentration appeared to inhibit wt swarming, though not to a degree that was statistically significant. Both raising and lowering the glucose concentrations significantly decreased swarm areas in the ΔdipA mutant. (B) Raising the glutamate concentration generally increased swarming in the wt, though not significantly according to Student's t tests. Lowering the glutamate concentration did significantly impair swarming in the ΔdipA mutant. Raising the glutamate concentration had no effect on the ΔdipA mutant. Swimming was less affected by changes in concentration than swarming. (C) On glucose, wt swimming did not differ significantly. However, the ΔdipA mutant showed significant increases in swimming motility under the 2× (24 mM) and 4× (48 mM) conditions. (D) On glutamate, only the 4× (48 mM) concentration showed significant increases compared to the 1× concentration. This held for both the wt and the ΔdipA mutant. All plates used the FAB base medium. Error bars indicate standard deviations. *, P < 0.05 compared to wt growing under the 1× (12 mM) condition. **, P ≤ 0.01 compared to ΔdipA mutant growing under the 1× (12 mM) condition.
The swarm area trends of the ΔdipA mutant did not mirror those of the wt (Fig. 9). Reducing the concentration of glucose or glutamate by 50% resulted in decreased swarming for the ΔdipA mutant. Increasing the concentration of glucose also decreased the swarm area. Increasing the concentration of glutamate in the swarm assay had no effect on ΔdipA mutant swarming.
Changing the carbon source concentration had much less of an effect on swimming motility, and only minimal differences were apparent (Fig. 9). The wt swim areas did not change with changes in glucose concentration; however, the ΔdipA mutant did show increases under the 2× and 4× conditions compared to that under the 1× condition. The only glutamate condition that resulted in a significant difference in swim area for either strain was the 4× condition. This indicated that the ΔdipA deficient strain was more sensitive to changes in the carbon source, as it showed significant changes more often on both swim and swarm assays.
ΔdipA deficient mutant swarms despite elevated levels of c-di-GMP.
Generally, low levels of c-di-GMP correlate with increased bacterial motility, while high levels of c-di-GMP correlate with nonmotile lifestyles (18). After detailing the divergence in ΔdipA mutant swarm phenotypes on several medium conditions, we were curious whether the mutant was simply producing less c-di-GMP on rich media and more c-di-GMP on minimal media. We utilized a PcdrA::lux reporter (27) to assess c-di-GMP production levels on LB Lennox broth, lab-mixed LB (tryptone, yeast extract, and NaCl), and M8-glucose-Casamino Acids medium and FAB-glutamate. This c-di-GMP-responsive reporter is a transcriptional fusion of the cdrA promoter with the gene encoding luciferase and enables real-time evaluation of c-di-GMP-regulated activities through the quantification of luminescence. c-di-GMP reporters utilizing this transcriptional fusion-based approach were previously published and validated (27–30). In a swarming assay, the wt harboring PcdrA::lux showed minimal luminescence 3 h after inoculation on surfaces (i.e., after sufficient time for the transition to a swarming mode) for all media tested, which is expected for motile bacteria (Fig. 10). The ΔdipA mutant consistently exhibited much more luminescence than the wt by the PcdrA::lux reporter on each given medium, indicating the mutant harbors elevated levels of c-di-GMP, as would be expected from a cell missing a phosphodiesterase.
FIG 10.
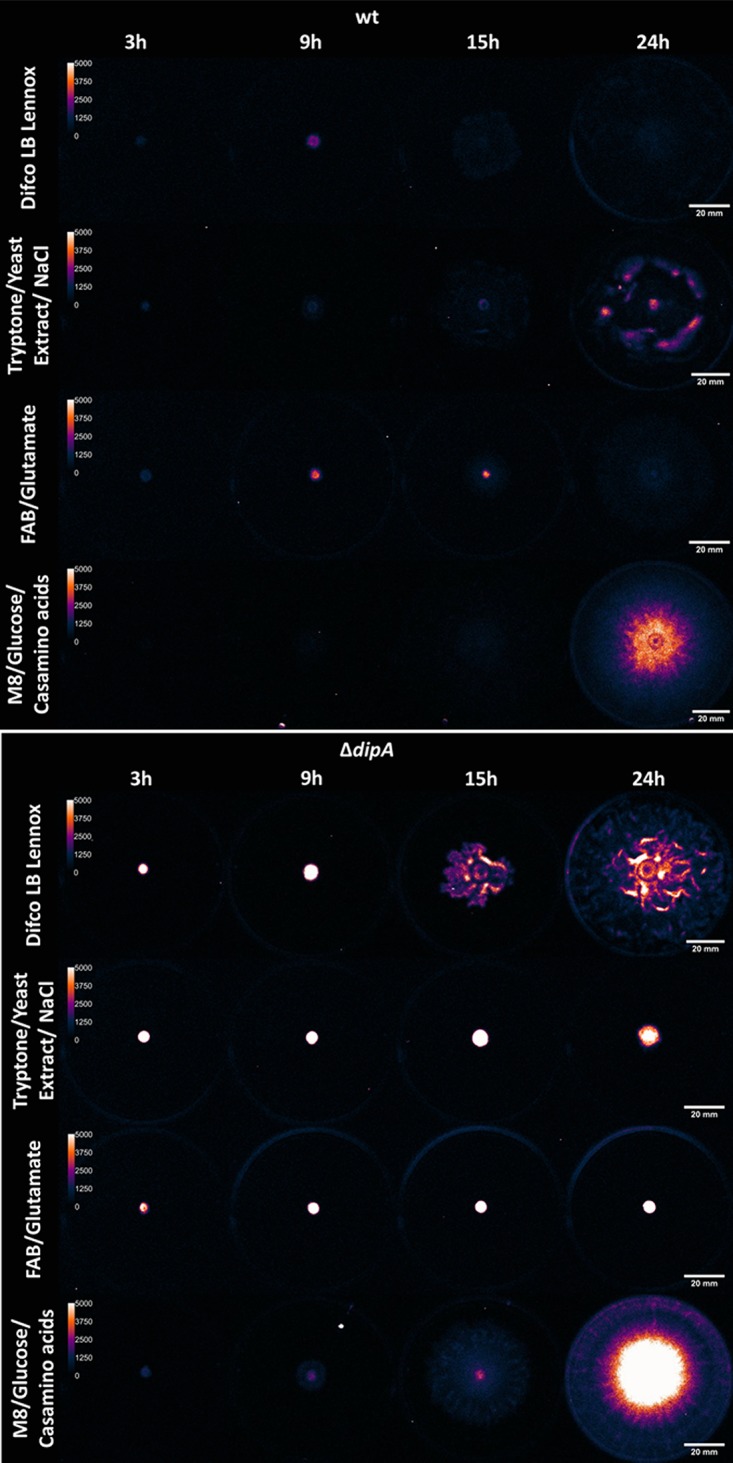
In vivo luminescence images show the variation of c-di-GMP throughout the swarming event on different media. After a 3-h incubation, the c-di-GMP PcdrA::lux reporter emits a weaker signal in the wt (top) than in the ΔdipA mutant (bottom), as would be expected when a phosphodiesterase is removed. There is no significant difference between the reporter signals in the wt on the media tested. Contrastingly, there is differential reporter activity in the ΔdipA mutant, further indicating that the role of DipA is nutritionally conditional. As would be expected, the nonswarming ΔdipA mutant on tryptone, yeast extract, and NaCl emits a very high signal compared to when it swarms on LB Lennox broth. However, the most striking result is the robust swarming combined with high levels of c-di-GMP seen when cultivated on M8 medium with glucose and Casamino Acids. Each plate has a 2-μl drop of broth, normalized to an OD600 of ≈0.5 before inoculation and incubation. Pictures were taken after 3, 9, 15, and 24 h of incubation at 30°C. Brightness maximum was set to 5,000 photons/s/mm2. Luminescence images are falsely colored for contrast.
Intensity of the PcdrA::lux signal varied between the different media tested in both the wt and ΔdipA backgrounds. Any differences became greatly accentuated after 15 h (see Table S1). The luminescence of the wt was routinely consistent at the 3-h and 9-h time points: a single-factor ANOVA indicates no significant difference (for α ≤ 0.01) in corrected total swarm luminescence between the 4 medium formulations tested. However, after 24 h, the wt luminescent signal was clearly much higher on M8 medium, in addition to regions of increased intensity (“hot spots”) within the swarm on tryptone, yeast extract, and NaCl. These hot spots could be explained by pooling cells commonly seen in many published swarm images; indeed, digital single-lens reflex (DSLR) images of our plates show denser areas of cells within the swarm (see Fig. S4). The ΔdipA mutant shows increased variation in signal intensity 3 or more h after inoculation. The differences in intensity are much less apparent after 24 h, when the nonswarming mutant on tryptone, yeast extract, and NaCl and FAB-glutamate and the swarming mutant on LB Lennox broth and M8-glucose-Casamino Acids medium all produce intense luminescent signals.
DISCUSSION
We conclude that DipA is the dominant phosphodiesterase in regulating P. aeruginosa motility behavior in environments that equate with minimal media. Overall, we find the necessity for DipA to regulate intracellular levels of c-di-GMP is nutritionally dependent, as we show a ΔdipA mutant swarms on select medium conditions. DipA was previously linked to both swimming and swarming but not on every medium examined (19, 20). Our results consistently show significant reductions in swimming or swarming by a ΔdipA strain compared to that of the wt and demonstrate that the medium used determines the extent of this difference. On minimal M8 medium supplemented with 0.2% glucose and 0.5% Casamino Acids, the ΔdipA mutant produced swim and swarm areas larger than those seen on rich LB Lennox broth. Changing the base medium from M8 to M63 while keeping the supplements constant supported swimming to a similar extent but severely decreased swarm motility. Thus, the maintenance of low intracellular c-di-GMP levels that should promote flagellar motility must be conditionally regulated by differing phosphodiesterases in variation with select nutrient cues. We speculate there is substantial redundancy by P. aeruginosa phosphodiesterases to appropriately regulate responses from the nutrient environment to increase or decrease flagellar motility. Our results are consistent with an explanation that DipA is most influential in responding to changes of certain carbon compounds, such as glutamate and glucose. Such a nutrient-sensing behavior bears similarity to those for the range of compounds sensed by some methyl-accepting chemotaxis proteins essential for classical chemotaxis (31). Collectively, the importance of DipA to flagellar motility is very apparent on some undefined rich medium components, such as tryptone. This is particularly the case for swarming, perhaps indicating the DipA-specific control of surface motility and not all flagellar motility. Our findings also suggest that the maximum nutrient concentrations that boost expansion are distinct for swarming and swimming, further illustrating the difficulties with directly comparing different flagellum-mediated motility assays.
In addition to a possible new surface-sensing function of DipA, this work shows P. aeruginosa swarming in the presence of elevated levels of c-di-GMP. The ΔdipA mutant consistently expressed higher levels of c-di-GMP via the PcdrA::lux reporter and yet, under a few conditions, was able to swarm quite well. This is most apparent on M8 medium supplemented with 0.2% glucose and 0.5% Casamino Acids and on LB Lennox broth. Although the ΔdipA mutant on M8 medium emits the lowest signal, comparatively, at the beginning, its lux signal increases as the swarm progresses. On LB Lennox broth, the ΔdipA mutant starts at an elevated c-di-GMP level, similar to those seen under nonswarming conditions, but is able to swarm at a still comparatively high level after 15 h. This illustrates that c-di-GMP levels may not always govern when P. aeruginosa switches between a motile and sessile lifestyle. Such a notion has already been proposed by Yan et al., who found “no apparent correlation” between c-di-GMP, biofilm formation, and swarming levels across a number of P. aeruginosa clinical isolates (32). Although they found that the deletion of dipA resulted in a total loss of swarming, they also proposed a model of c-di-GMP networking that could account for swarming under higher levels of c-di-GMP. Understanding which factors are important under different environmental conditions will be paramount as we continue to unravel the complexities of bacterial motility.
Within our overall assessment of the conditional influence of DipA on flagellar motility, several specific aspects of our results are interesting and can be used to reconcile prior studies. For example, although the base recipes for both M8 and M63 media contain potassium phosphate, M8 contains disodium phosphate, sodium chloride, and no inherent nitrogen source, while M63 contains ammonium sulfate, iron sulfate, and potassium hydroxide (Table 1). Those differences in iron and ammonium concentrations could account for differences in motility between these minimal media, as iron limitation has been shown to increase rhamnolipid production (7), while ammonium has been shown to inhibit rhamnolipid production (33). However, using M63 as the base and supplementing with 0.2% glycerol instead of glucose supported much more robust wt swarming. The resultant swarming differences thus appear to come from a combination of the nutrients available from the base medium and all supplements rather than just from inherent differences in the base media or the presence of one single constituent.
TABLE 1.
Recipes of media used in this study
| Medium | Ingredient | Concn (per liter) | Reference or source |
|---|---|---|---|
| Lennox broth | Tryptone, yeast extract, NaCl mix | 20 g | BD Difco (VWR) |
| Lab-mixed LB | Tryptone | 10 ga | Sigma |
| Yeast Extract | 5 ga | Fluka Biochemika | |
| NaCl | 5 ga | BDH Chemicals (VWR) | |
| M8 base | Na2HPO4 · 7H2O | 64 g | 13 |
| KH2PO4 | 15 g | ||
| NaCl | 2.5 g | ||
| M63 base | (NH4)2SO4 | 2 g | Amresco |
| KH2PO4 | 13.6 g | ||
| FeSO4 · 7H2O | 0.5 mg | ||
| KOHb | Thermo Fisher Scientific | ||
| FABc base | (NH4)2SO4 | 2 g | 37 |
| Na2HPO4 · 7H2O | 9 g | ||
| KH2PO4 | 3 g | ||
| NaCl | 3 g | ||
| MgCl2 · 6H2O | 198 mg | ||
| CaCl2 · H2O | 14 mg | ||
| CaSO4 · 2H2O | 0.2 mg | ||
| FeSO4 · 7H2O | 0.2 mg | ||
| MnSO4 · H2O | 20 μg | ||
| CuSO4 · 5H2O | 20 μg | ||
| ZnSO4 · 7H2O | 20 μg | ||
| CoSO4 · 7H2O | 10 μg | ||
| NaMoO4 · H2O | 10 μg | ||
| H3BO3 | 5 μg | ||
| Casamino Acids | Casein hydrolysate | Unspecified | 46, Amresco |
| MgSO4 | |||
| Beta-alanine | |||
| Nicotinic acid | |||
| Pimelic acid | |||
| CuSO4 | |||
| ZnSO4 | |||
| MnCl2 | |||
| HCl | |||
| H2O | |||
| Cystine | |||
| Maltose | |||
| CuCl2 | |||
| FeSO4 |
Concentration based on approximate recipe of BD Difco LB Lennox broth.
To pH 7.
FAB, fastidious anaerobe broth.
Tryptone and yeast extract also supported robust swimming and swarming motility in the wt, but the ΔdipA mutant was severely impaired for both motilities, particularly swarming. Combining tryptone and yeast extract did not restore the swarming phenotype in the ΔdipA mutant, indicating that any negative impact on motility is not simply due to differences in total available nutrients. There is likely a difference in chemical composition due to variations in manufacturing, highlighting the need for consistent medium use. Further study into these differences on a chemical level is required. One possible difference in chemical composition could be Mg2+, as adding MgSO4 to tryptone and yeast extract restored the swarming phenotype in the ΔdipA mutant to levels seen on LB Lennox broth. Mg2+ has been reported to be required for or enhance the catalytic activity of the phosphodiesterase enzyme in a number of bacteria (34–36). It could be that additional Mg2+ in the medium enables other phosphodiesterases in P. aeruginosa to hydrolyze c-di-GMP more efficiently, partially negating the absence of DipA and reducing c-di-GMP levels just enough to enable cells to start swarming.
We also examined the possibility of a nutrient threshold to support swimming and swarming. Below such a threshold, no motility would be possible due to the lack of required nutrients; above such a threshold, there would be no increase in movement due to the lack of pressure to colonize and seek out nutrients. In testing this, we found positive correlations between tryptone and both swimming and swarming for both the wt and the ΔdipA mutant. However, yeast extract contains or lacks a compound that specifically affects surface motility, as increasing the amount of yeast extract increased only the swimming motility. In minimal medium containing glucose or glutamate, we observed mixed trends between motility and carbon source concentration. While the glucose concentration had no effect on swimming, it had a negative correlation to wt swarming. In contrast, wt swarm areas increased with increasing glutamate concentrations, plateauing above the 2× concentration. Overall, these differences we detailed in wt motility suggest that the low motility observed in any particular strain could be attributed to using a nonoptimal concentration of the carbon source.
These results neither confirm nor negate the “threshold hypothesis” as we originally imagined it, as changing the nutrient concentrations did have an effect, but the trends were not consistent. These data do suggest that DipA is more important for sensing amino acids than sugars. We were able to discern some of this behavior directly by comparing the swarming of the wt and the ΔdipA mutant on glutamate versus glucose. Interestingly, this DipA influence is more pronounced during swarming than during swimming. A seemingly related role for DipA is observed when growing P. aeruginosa on different rich media.
MATERIALS AND METHODS
Growth conditions.
A list of all media used can be found in Table 1. Minimal medium FAB (37) broth cultures were supplemented with 30 mM glucose or glutamate as the sole carbon source. M8 and M63 media were supplemented with 0.2% glucose, 0.5% Casamino Acids, and 1 mM MgSO4, as described by other research groups (13, 19, 20, 38). Difco LB Lennox broth was prepared as suggested by the manufacturer (20 g/liter of approximately 10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl). Tryptone medium was made with 10 g/liter Sigma tryptone and 5 g/liter NaCl. Yeast extract medium was made with 5 g/liter Fluka yeast extract and 5 g/liter NaCl. All media had a neutral pH, between 6 and 7. Planktonic cultures were grown in 6 ml of medium with shaking at 240 rpm at 37°C for 18 to 24 h.
Growth curves were measured using a BioTek Powerwave ×340 microplate reader, courtesy of NDnano Center for Nanoscience and Technology. Cells were grown in LB for 18 h, washed twice with 1× phosphate-buffered saline, and resuspended in nuclease-free water to a starting optical density at 600 nm (OD600) of ≈0.05. (Due to the use of multiple spectrophotometers, OD600 measurements were approximately 4× higher when measured with the plate reader.) Twenty-five microliters of suspended cells was added to 225 μl of medium in a flat-bottom 96-well microplate. Each condition was completed in quadruplicates. Cells were grown at 37°C with shaking for 18 h.
Swarm assay preparation.
Swarm assays were made with 0.1% (wt/vol) Gelzan agar substitute (Sigma) added to the prepared base medium and autoclaved for 22 min at 121°C. The bottles were allowed to cool on stir plates for 15 min. For minimal media, the carbon source was added from a 1.2 M stock solution to a final concentration of 12 mM (unless otherwise indicated) (14). Medium (25 ml) was pipetted into a 100-mm-diameter petri dish (VWR) within 30 min of taking the medium out of the autoclave. The plates were cured, covered, on the bench overnight and were dried, uncovered, in a fume hood for 5 h prior to point inoculation of 18- to 24-h planktonic cultures using a platinum wire (except where indicated). The plates were incubated inverted at 30°C, and pictures of the developed swarms were taken between 20 and 24 h. Measurements were made using ImageJ 1.47v (39). All assays were completed in triplicates. For experiments using various carbon source concentrations in minimal media, 12 mM was used as “1×,” 6 mM for “[1/2]×,” 24 mM for “2×,”and 48 mM for “4×.”
Swim assay preparation.
Swim assays were made with 0.2% (wt/vol) Noble agar (Sigma) added to the respective prepared base media and autoclaved for 22 min at 121°C. The bottles were allowed to cool to 50 to 55°C on stir plates. For minimal media, the carbon source was added to a 12 mM final concentration (unless otherwise indicated) (14). Fifteen milliliters of medium was pipetted into 60-mm-diameter petri dishes (VWR). The plates were cured, covered, on the bench overnight prior to point inoculation with 18- to 24-h broth cultures. The plates were incubated at 30°C, and pictures of the developed swim zones were taken between 20 and 24 h. All assays were completed in triplicates. For accurate comparisons between rich media, additional pictures were taken before the swim zones reached the edges of the plates, between 9 and 10 h. Area measurements were made from images of whole plates using ImageJ 1.47v.
Deletion of dipA.
A list of the strains and plasmids used can be found in Table 2; primer sequences can be found in Table 3. Traditional amplification methods were employed, but ligation into the pEX18Gm plasmid (40) was according to SLiCE protocols (41). Successful recombinant plasmids were electroporated into Escherichia coli DH10B (Thermo Fisher Scientific) and screened for correct deletion sequences. Electrocompetent E. coli SM10 cells were prepared as previously described (42). E. coli SM10 colonies were screened using LB supplemented with 15 μg/ml gentamicin. Conjugational mating was used for plasmid transfer to P. aeruginosa PAO1C. Colonies were screened on VBE plates supplemented with 40% citrate and 100 μg/ml gentamicin. Recombination was completed by screening the colonies on LB plates without NaCl supplemented with 10% sucrose. Potential clean-deletion mutant colonies were screened by colony PCR using outside primers (A9dPCR1-F and A9dPCR2-R) (Table 3). Clean-deletion mutants were sequenced at the Genomics & Bioinformatics Core Facility at the University of Notre Dame, using sequencing primers internal to and flanking dipA.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Select characteristics | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH10B | Electromax, F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG | Thermo Fisher Scientific |
| SM10 | Conjugational competent donor, thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | 14, 47 |
| Pseudomonas aeruginosa strains | ||
| PAO1C | Wild-type laboratory strain, ATCC collection strain 15692 | 14, 47 |
| PAO1C ΔdipA | Dispersion-induced phosphodiesterase A-deficient mutant of PAO1C | This study |
| Plasmids | ||
| pEX18Gm | 5.8-kb plasmid, Gmr | 40 |
| PcdrA::lux | cdrA reporter in pMS402 luxCDABE-based plasmid, Kmr | 27 |
| pJN-dipA-V5/6× His | C-terminal V5/6× His-tagged dipA cloned into pJN105, Gmr | 20 |
TABLE 3.
Primers created for and used in this study
| Primer | Sequence |
|---|---|
| Pseudomonas aeruginosa PAO1C | |
| A9dPCR1-F | CA GGA AAC AGC TAT GAC CAT GAT TAC GAA TTC GCG ACC CTG CTC CTG TAA TAC |
| A9dPCR1-R | AAG CCT GGG CGA TCA GTG ATG ACT TTT CAT GCG AGG CT |
| A9dPCR2-F | AGC CTC GCA TGA AAA GTC AT CAC TGA TCG CCC AGG CTT |
| A9dPCR2-R | GA CGT TGT AAA ACG ACG GCC AGT GCC AAG CTT TAG TAG AAG GGC GGG ACG T |
| A9dSeq-F | TTC ACC ATC TCC AGT CTC |
| A9dSeq-R1 | TCAG GTA GAC CTT GTA GAG |
| A9dSeq-R2 | CAG CTC AAG CGT TTC TAC |
| Escherichia coli DH10B | |
| lacZ-F | GCTCGTATGTTGTGTGGAATTGTG |
| lacZ-R | CGAAGGCGATTAAGTTGGGTAAC |
lux reporter imaging.
A cdrA lux reporter was electroporated into the wt and the ΔdipA mutant strains according to previously described methods (43). Transformed P. aeruginosa colonies were selected on LB supplemented with 500 μg/ml kanamycin. Relative c-di-GMP levels were assessed by imaging cells growing on swarm agar containing LB Lennox both, medium with tryptone, yeast extract, and NaCl, M8 medium with 0.2% glucose and 0.5% Casamino Acids, and FAB with 12 mM glutamate, where 2 μl of planktonic culture, normalized to an OD600 of 0.5, was inoculated on swarm plates made as described above. The plates were incubated at 30°C for 3 h, at which point there were no apparent variations in population sizes between the medium conditions but there had been ample time for attachment and initial stages of swarming. Images were taken at this initial 3-h stage, during swarm formation, at 9 and 15 h, and after swarms were fully developed, at 24 h. Images were taken using a Bruker In Vivo Extreme at the Notre Dame Integrated Imaging Facility, as previously described (44). In vivo swarm images were processed in ImageJ 1.47v as previously described (44), though adjusted to a uniform maximum brightness of 5,000 photons/s/mm2. Luminescence was measured in ImageJ 1.47v by using a method adapted from McCloy et al. (45). Briefly, integrated densities and areas were measured for luminescent swarms in each image. These measurements were also taken from nonluminescent portions of the corresponding image. The corrected total swarm luminescence was calculated by subtracting the average background internal density (based on the mean gray value and area) from the measured internal density of the swarm area.
Drop collapse assays.
Drop collapse assays were completed with broths grown at 37°C with shaking for 48 h. Ten microliters of cell culture was dropped onto a glass slide covered in Parafilm to ensure a consistently hydrophobic surface. Each condition was repeated in triplicates. The contact angles were measured using ImageJ 1.47v and the Contact_angle plug-in.
Statistical analysis.
Student's t tests and single-factor ANOVAs were completed for all conditions tested using a P value of 0.01.
Supplementary Material
ACKNOWLEDGMENTS
Plasmids were generously provided by Herbert Schweizer (University of Florida) and Karin Sauer (Binghamton University). Assistance with some assays was provided by Devin North. Primers to verify lacZ clones were designed by Nydia Morales-Soto. We thank Alan Wolfe (Loyola University Chicago) for input and helpful suggestions.
Support for this work is provided by NIH (NIAID) grant no. R21AI109417.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02847-17.
[This article was published on 19 March 2018 without Nydia Morales-Soto's name in the byline. The byline was updated in the version posted on 18 July 2018.]
REFERENCES
- 1.Costa D, Bousseau A, Thevenot S, Dufour X, Laland C, Burucoa C, Castel O. 2015. Nosocomial outbreak of Pseudomonas aeruginosa associated with a drinking water fountain. J Hosp Infect 91:271–274. doi: 10.1016/j.jhin.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz T, Armant O, Bretschneider N, Hahn A, Kirchen S, Seifert M, Dotsch A. 2015. Whole genome and transcriptome analyses of environmental antibiotic sensitive and multi-resistant Pseudomonas aeruginosa isolates exposed to waste water and tap water. Microb Biotechnol 8:116–130. doi: 10.1111/1751-7915.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarter LL, Gomelsky M. 2015. Fifty ways to inhibit motility via cyclic di-GMP: the emerging Pseudomonas aeruginosa swarming story. J Bacteriol 197:406–409. doi: 10.1128/JB.02483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, Wang XL, Liu JF, Nememan I, Singh AH, Weiss H, Levin BR. 2011. The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility. Proc Natl Acad Sci U S A 108:4047–4052. doi: 10.1073/pnas.1013499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deziel E, Lepine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 7.Glick R, Gilmour C, Tremblay J, Satanower S, Avidan O, Déziel E, Greenberg E, Poole K, Banin E. 2010. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochsner UA, Reiser J. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Mawgoud AM, Lepine F, Déziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soberón-Chávez G, Lépine F, Déziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 13.Köhler T, Curty L, Barja F, van Delden C, Pechère J. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrout J, Chopp D, Just C, Hentzer M, Givskov M, Parsek M. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 15.Bains M, Fernandez L, Hancock REW. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 19.Li YL, Xia HM, Bai F, Xu HJ, Yang L, Yao HM, Zhang L, Zhang XM, Bai YL, Saris PEJ, Tolker-Nielsen T, Qiao MQ. 2007. Identification of a new gene PA5017 involved in flagella-mediated motility, chemotaxis and biofilm formation in Pseudomonas aeruginosa. FEMS Microbiol Lett 272:188–195. doi: 10.1111/j.1574-6968.2007.00753.x. [DOI] [PubMed] [Google Scholar]
- 20.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge JD, Harshey RM. 2013. Swarming: flexible roaming plans. J Bacteriol 195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen DG, Orr JS, Rao CV, Wolfe AJ. 2017. Increasing growth yield and decreasing acetylation in Escherichia coli by optimizing the carbon-to-magnesium ratio in peptide-based media. Appl Environ Microbiol 83:e03034-16. doi: 10.1128/AEM.03034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain DK, Collins-Thompson DL, Lee H, Trevors JT. 1991. A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiol Methods 13:271–279. doi: 10.1016/0167-7012(91)90064-W. [DOI] [Google Scholar]
- 25.Bodour AA, Miller-Maier RM. 1998. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J Microbiol Methods 32:273–280. doi: 10.1016/S0167-7012(98)00031-1. [DOI] [Google Scholar]
- 26.Tugrul T, Cansunar E. 2005. Detecting surfactant-producing microorganisms by the drop-collapse test. World J Microbiol Biotechnol 21:851–853. doi: 10.1007/s11274-004-5958-y. [DOI] [Google Scholar]
- 27.Borlee BR, Borlee GI, Martin KH, Irie Y. 2017. Cyclic di-GMP-responsive transcriptional reporter bioassays in Pseudomonas aeruginosa. Methods Mol Biol 1657:99–110. doi: 10.1007/978-1-4939-7240-1_9. [DOI] [PubMed] [Google Scholar]
- 28.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5060–5069. doi: 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawar SV, Messina M, Rinaldo S, Cutruzzola F, Kaever V, Rampioni G, Leoni L. 2016. Novel genetic tools to tackle c-di-GMP-dependent signalling in Pseudomonas aeruginosa. J Appl Microbiol 120:205–217. doi: 10.1111/jam.12984. [DOI] [PubMed] [Google Scholar]
- 30.Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortega Á, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, Deforet M, Boyle KE, Rahman R, Liang R, Okegbe C, Dietrich LEP, Qiu W, Xavier JB. 2017. Bow-tie signaling in c-di-GMP: machine learning in a simple biochemical network. PLoS Comput Biol 13:e1005677. doi: 10.1371/journal.pcbi.1005677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulligan C, Gibbs B. 1989. Correlation of nitrogen metabolism with biosurfactant production by Pseudomonas aeruginosa. Appl Environ Microbiol 55:3016–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 38.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O'Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YW, Werling U, Edelmann W. 2012. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res 40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook JR, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 43.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Morales-Soto N, Anyan ME, Mattingly AE, Madukoma CS, Harvey CW, Alber M, Deziel E, Kearns DB, Shrout JD. 7 April 2015. Preparation, imaging, and quantification of bacterial surface motility assays. J Vis Exp doi: 10.3791/52338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller JH, Miller PA. 1941. Production of diphtheric toxin of high potency (100 Lf) on a reproducible medium. J Immun 40:21–32. [Google Scholar]
- 47.Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, Shrout JD. 2014. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 111:18013–18018. doi: 10.1073/pnas.1414661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.