ABSTRACT
Gut metagenomic sequences provide a rich source of microbial genes, the majority of which are annotated by homology or unknown. Genes and gene pathways that encode enzymes catalyzing biotransformation of host bile acids are important to identify in gut metagenomic sequences due to the importance of bile acids in gut microbiome structure and host physiology. Hydroxysteroid dehydrogenases (HSDHs) are pyridine nucleotide-dependent enzymes with stereospecificity and regiospecificity for bile acid and steroid hydroxyl groups. HSDHs have been identified in several protein families, including medium-chain and short-chain dehydrogenase/reductase families as well as the aldo-keto reductase family. These protein families are large and contain diverse functionalities, making prediction of HSDH-encoding genes difficult and necessitating biochemical characterization. We located a gene cluster in Eggerthella sp. CAG:298 predicted to encode three HSDHs (CDD59473, CDD59474, and CDD59475) and synthesized the genes for heterologous expression in Escherichia coli. We then screened bile acid substrates against the purified recombinant enzymes. CDD59475 is a novel 12α-HSDH, and we determined that CDD59474 (3α-HSDH) and CDD59473 (3β-HSDH) constitute novel enzymes in an iso-bile acid pathway. Phylogenetic analysis of these HSDHs with other gut bacterial HSDHs and closest homologues in the database revealed predictable clustering of HSDHs by function and identified several likely HSDH sequences from bacteria isolated or sequenced from diverse mammalian and avian gut samples.
IMPORTANCE Bacterial HSDHs have the potential to significantly alter the physicochemical properties of bile acids, with implications for increased/decreased toxicity for gut bacteria and the host. The generation of oxo-bile acids is known to inhibit host enzymes involved in glucocorticoid metabolism and may alter signaling through nuclear receptors such as farnesoid X receptor and G-protein-coupled receptor TGR5. Biochemical or similar approaches are required to fill in many gaps in our ability to link a particular enzymatic function with a nucleic acid or amino acid sequence. In this regard, we have identified a novel 12α-HSDH and a novel set of genes encoding an iso-bile acid pathway (3α-HSDH and 3β-HSDH) involved in epimerization and detoxification of harmful secondary bile acids.
KEYWORDS: bile acid, Eggerthella, metagenome, hydroxysteroid dehydrogenase, targeted gene synthesis
INTRODUCTION
Metagenomic sequencing of microbial DNA in environmental samples, including human stool, has provided a massive gene catalogue to the microbiologist, only an estimated 50% of which has been annotated; many of these annotations are homology based, and a miniscule number have been characterized biochemically (1). The Human Microbiome Project (2) and Metagenomics of the Human Intestinal Tract (MetaHIT) have provided a “second human genome” to decode (3). One set of functional pathways within this “second human genome” is the gut sterolbiome (4), which participates in the endocrine function of the host as well as, in the case of bile acids, regulation of microbiome structure and functions. Gut bacterial metabolism of bile acids is known to affect signaling through the farnesoid X receptor (5), G-protein-coupled protein receptor TGR5 (6), muscarinic receptor (7), and vitamin D receptor (8). In addition, the formation of oxo-bile acids has been shown to inhibit the enzyme 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1), which has been reported to alter the gut microbiome community structure (9).
The oxidation of bile acid hydroxyl groups by gut bacteria also allows epimerization by the concerted action of two regiospecific enzymes differing in stereospecificity such that the conversion of, for instance, 7α-hydroxylated chenodeoxycholic acid (CDCA) to 7β-hydroxylated ursodeoxycholic acid (UDCA) bile proceeds through a stable 7-oxolithocholic acid intermediate (10). Epimerization of the 3α-hydroxy results in formation of so-called iso-bile acids (3β-hydroxyl) (11) that are more hydrophilic and less toxic than the 3α-hydroxyl host-derived bile acid (12, 13), as is also the case with UDCA, which is used therapeutically in hepatobiliary diseases (14). The least-studied pathway is the epimerization of the 12α-hydroxyl→12-oxo→12β-hydroxyl of cholic acid derivatives (15, 16).
Eggerthella lenta (formerly Eubacterium lentum) is a Gram-positive, non-spore-forming, nonmotile, obligate anaerobic bacterium whose growth is stimulated by arginine (17, 18). E. lenta is a particularly important participant in the gut sterolbiome, various strains of which have the potential to encode enzymes that metabolize cardiac glycosides (19, 20) and 21-dehydroxylate corticosteroids (21), hydrolyze conjugated bile acids (22), and oxidize bile acid hydroxyl groups (23, 24). E. lenta strains have previously been grouped according to their pattern of hydroxysteroid dehydrogenase (HSDH) activities, with some strains having no HSDH activity (group V) and others expressing 3α-HSDH, 3β-HSDH, 7α-HSDH, and 12α-HSDH (group III) (24). E. lenta and other members of the Coriobacteriaceae family have been correlated with changes in the host liver metabolome, specifically through reduction in serum triglycerides, cholesterol excretion, and metabolism of xenobiotics (25).
The E. lenta sterolbiome offers a potential link between associations with strains of this species and changes in host liver metabolome and physiology. Alteration of steroid structure and absorption of E. lenta steroid metabolites (and derivatives from additional microbial biotransformations) from the colon and transport to the liver via the portal circulation may alter nuclear receptor and G-protein-coupled receptor activation, as well as affect liver enzyme activities, leading to altered lipid and steroid metabolism. Currently, the sterolbiome of E. lenta is poorly characterized. Genes encoding HSDHs, particularly 7α-HSDH and 12α-HSDH, have yet to be identified. It may be expected that diverse isoforms of currently identified 3α-HSDH and 3β-HSDH may exist, requiring functional characterization of genes whose protein products are predicted to encode these enzymes, namely, those within the family of short-chain dehydrogenases/reductases (SDRs) (26). A major barrier to such mechanistic study is the need to identify and characterize much of the E. lenta sterolbiome.
In the current study, we queried the nonredundant database with the amino acid sequence from the only characterized 12α-HSDH to date, identified in Clostridium sp. strain ATCC 29733/VPI C48-50 (15). We located an open reading frame (ORF) in a gut metagenome of Eggerthella sp. CAG:298, a sequence from Metagenomics of the Human Intestinal Tract (MetaHIT), that is part of a gene cluster with additional NAD(P)H-dependent dehydrogenases in the SDR and aldo-keto reductase protein families. Utilizing targeted gene synthesis, we identified and cloned these three synthesized genes for heterologous expression in Escherichia coli. We report the characterization of a gene encoding a novel 12α-HSDH as well as two genes encoding an iso-bile acid pathway (3α-HSDH and 3β-HSDH), distinct from those previously reported (13).
RESULTS
Identification of a gene cluster predicted to encode multiple HSDHs in a metagenomic sequence.
12α-HSDHs (EC 1.1.1.176) have been reported in gut bacteria such as Clostridium perfringens (27), Clostridium leptum (28), E. lenta (23), and Clostridium sp. strain ATCC 29733/VPI C48-50 (15). To date, the only reported 12α-HSDH whose gene sequence is known is that of Clostridium sp. strain ATCC 29733/VPI C48-50 (29). Previously, a phylogenetic analysis of the 12α-HSDH amino acid sequence (ERJ00208.1) was performed, suggesting that this function may be widespread in gut microbiota (30); however, biochemical characterization has yet to confirm this. An updated query of the nonredundant database identified numerous gut microbial phyla, particularly Firmicutes and Actinobacteria. One particular sequence, CDD59475, belonging to Eggerthella sp. CAG:298 and not reported previously (30), shared 71% identity (E value = 1e−134) with ERJ00208.1. This predicted 266-amino-acid protein in the 3-ketoacyl-(acyl-carrier protein) reductase/SDR family was unique among gene products that populated the BLAST query in that it is encoded by part of a three-gene cluster encoding two additional putative NAD(P)(H)-dependent oxidoreductases. Directly upstream is a gene (BN592_00769) encoding a 250-amino-acid protein (CDD59474.1) annotated as a 7α-HSDH by homology in the SDR family, and upstream is a gene (BN592_00768) encoding a 280-amino-acid protein (CDD59473.1) predicted to encode an aldo/keto reductase. Downstream of BN592_00768 is a gene (BN592_00767) predicted to encode a gamma-glutamylcysteine synthetase, and upstream on the opposite strand of the gene cluster is an ORF (BN592_00771) encoding a hypothetical protein in the phasin superfamily involved in intracellular storage of polyhydroxyalkanoates (Fig. 1).
FIG 1.
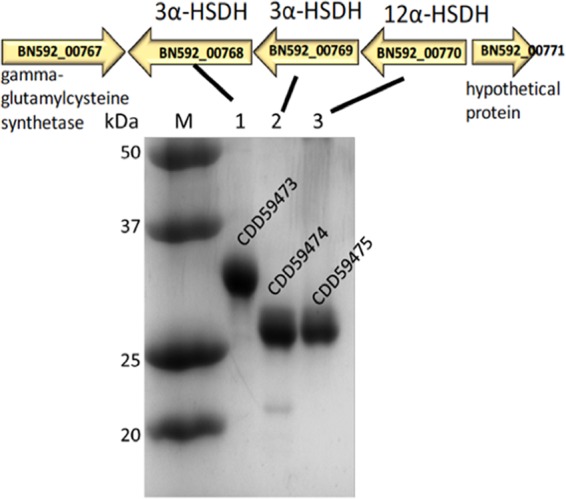
SDS-PAGE of purified recombinant 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from a gene cluster in Eggerthella sp. CAG:298.
Targeted gene synthesis, cloning, and overexpression of CDD59475 in E. coli.
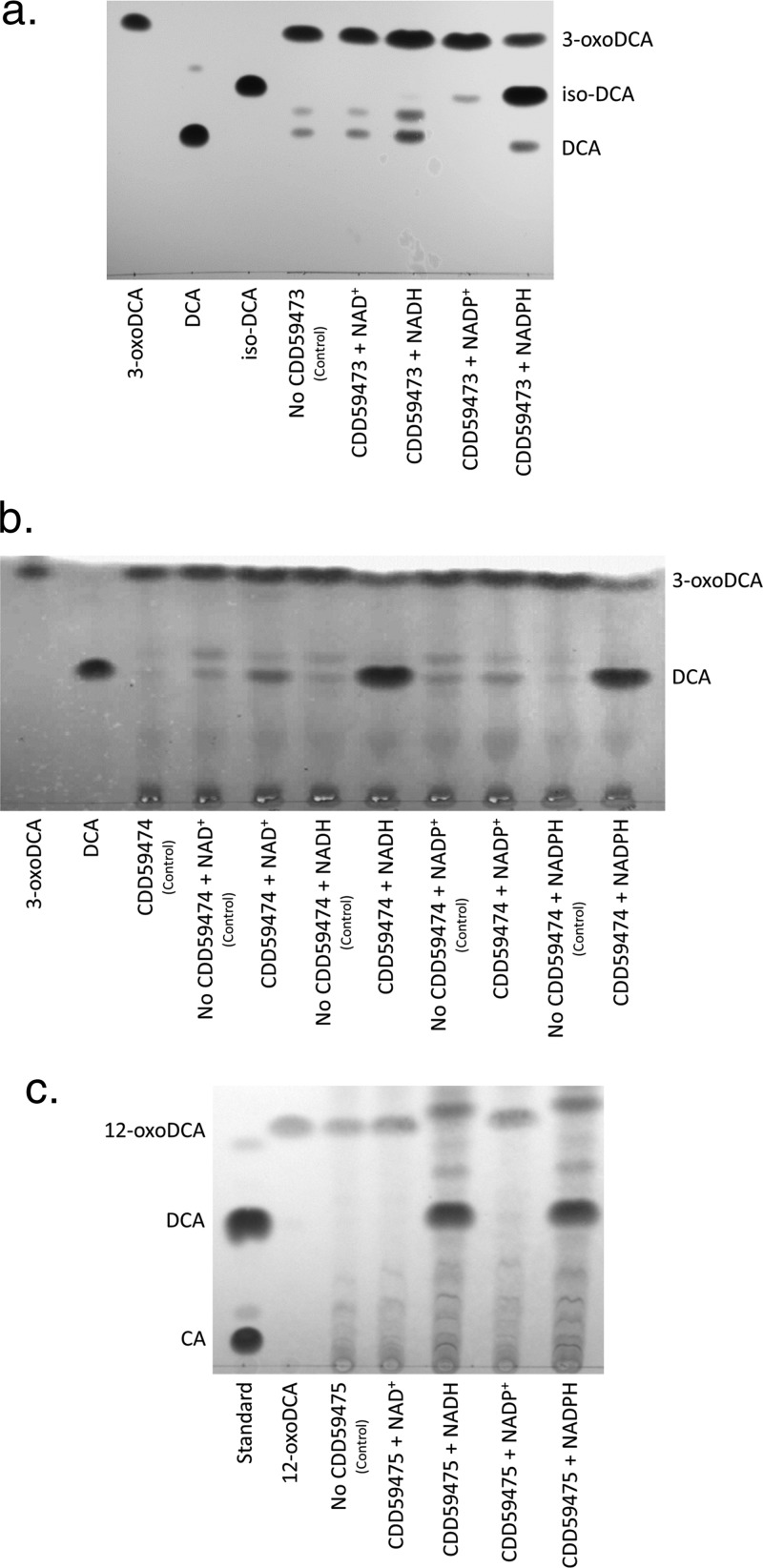
Since Eggerthella sp. CAG:298 is a metagenomic sequence and a microbial isolate is not available, the genes were synthesized (GenBlock IDT, Inc.) and PCR amplified for ligation-independent cloning (LIC) into pET46(+) for overexpression of the recombinant N-terminal His-tagged protein in E. coli (Table 1). Purified recombinant CDD59475 separated as a single band on SDS-PAGE with a subunit molecular mass of 28.0 ± 0.6 kDa (theoretical molecular mass, 28.30 kDa) (Fig. 1). We initially screened CDD59475 dehydrogenase activity spectrophotometrically at 340 nm in the presence of oxo-bile acids at a concentration of 50 μM, including 3-oxo-deoxycholic acid (3-oxo-DCA), 7-oxo-DCA, and 12-oxo-DCA, with 150 μM NADH or NADPH as a cofactor. We did not detect enzyme activity when 3-oxo-DCA or 7-oxo-DCA was the substrate; however, CDD59475 clearly displayed NADPH-dependent activity in the presence of 12-oxo-DCA (Table 2). To initially confirm activity, we ran several substrates in the oxidative direction and separated the reaction products by thin-layer chromatography (TLC), the products of which were subjected to mass spectrometry (MS) (see Fig. S1 in the supplemental material). In the presence of NADP+, CDD59475 was capable of oxidizing cholic acid (CA) (3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and deoxycholic acid (DCA) (3α,12α-dihydroxy-5β-cholan-24-oic acid) but not CDCA (3α,7α-dihydroxy-5β-cholan-24-oic acid). TLC of overnight reactions confirmed the formation of a product, comigrating with an authentic DCA standard, from 12-oxo-DCA in the presence of NAD(P)H (Fig. 2). The reaction product was scraped from the TLC plate, and MS identified a major mass ion in negative mode at m/z 391.2831 (the molecular mass of DCA is 392.57 amu) (see Fig. S2 in the supplemental material).
TABLE 1.
Oligonucleotides used for amplification and directional cloning
| Gene | Primer sequence (5′→3′) |
Protein mol mass (kDa) | Ext. coefficient (M−1 cm−1)a | |
|---|---|---|---|---|
| Forward | Reverse | |||
| CDD59473 | GACGACGACAAGATGCAAGATGTTTTTACGTTGAAGAACG | GAGGAGAAGCCCGGTTTAGAAGTCGATCTCATCCGCATCC | 31.59 | 46,785 |
| CDD59474 | GACGACGACAAGATGGGAAAGCTCGAAGGGAAAGT | GAGGAGAAGCCCGGTTTACGAGCGGGAAAGGCCG | 26.355 | 6,085 |
| CDD59475 | GACGACGACAAGATGGGATTTCTCGAAGGTAAGACCG | GAGGAGAAGCCCGGTCTAGGGACGCAGACCCATGC | 28.33 | 27,640 |
Ext. coefficient, extinction coefficient.
TABLE 2.
Kinetic parameters for Eggerthella CAG:298 HSDHsa
| Gene | Protein function | Substrate | Coenzyme | Vmax (μmol min−1 mg−1) | Km (μM) | Kcat (min−1) | Kcat/Km |
|---|---|---|---|---|---|---|---|
| CDD59473 | 3β-HSDH | Iso-DCA | NADP+ | 42.26 ± 6.34 | 282.70 ± 25.68 | 1.33 ± 0.20 | 4.7 × 10−3 ± 9.6 × 10−4 |
| 3-Oxo-DCA | NADPH | 13.39 ± 1.56 | 470.42 ± 34.75 | 0.42 ± 0.049 | 9 × 10−4 ± 1.44 × 10−4 | ||
| CDD59474 | 3α-HSDH | DCA | NADP+ | 126.94 ± 5.57 | 155.12 ± 3.53 | 3,351 ± 146.73 | 21.6 ± 1.23 |
| 3-Oxo-DCA | NADPH | 123.92 ± 2.88 | 179.1 ± 8.73 | 3,271.52 ± 75.92 | 18.27 ± 1.14 | ||
| CDD59475 | 12α-HSDH | DCA | NADP+ | 21.29 ± 17.12 | 139.71 ± 14.64 | 603.15 ± 48.52 | 4.32 ± 0.667 |
| 12-Oxo-DCA | NADPH | 14.04 ± 2.79 | 11.41 ± 0.68 | 397.59 ± 79.25 | 34.84 ± 8.4 |
Values represent mean ± standard error of the mean from five technical replicates.
FIG 2.
Representative TLC and ESI-IT-TOF-MS of bile acid reaction products from recombinant CDD59473 (a), CDD59474 (b), and CDD59475 (c). Standard reaction mixtures contained 50 μM bile acid substrates and 150 μM pyridine nucleotide, and experiments included a no-enzyme control. Reaction mixtures were incubated at 37°C for 12 h. Reaction products that comigrated with authentic standards were scraped from the TLC plate, extracted with ethyl acetate, dried, and subjected to MS analysis after resuspension in mobile phase. Experiments were repeated three times.
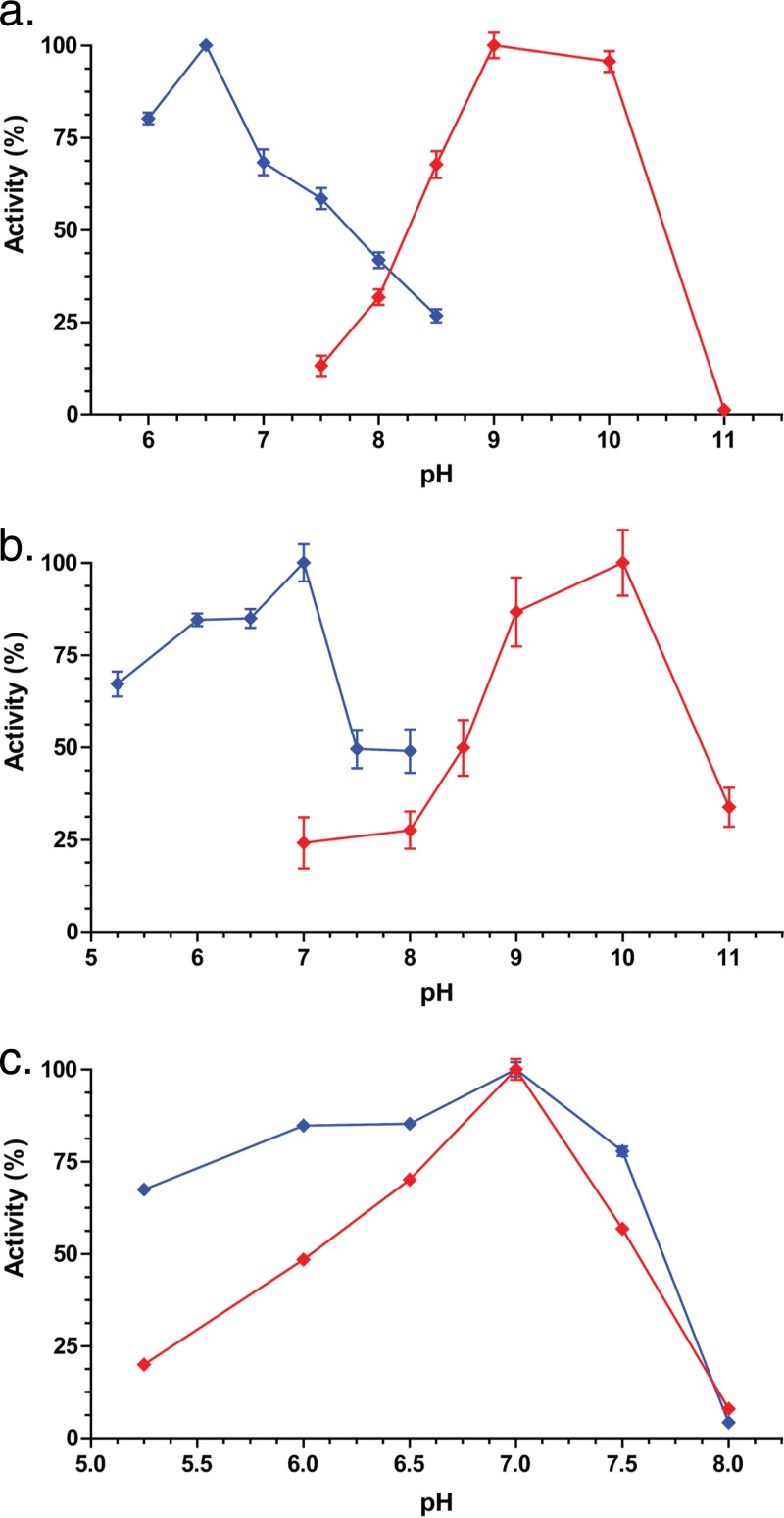
Next, we optimized pH in both the oxidative and reductive directions. In the oxidative direction, with DCA as a substrate and NADP+ as a cofactor, the optimum enzyme activity was observed at pH 7.0. Enzyme activity declined sharply from pH 7.0 to pH 8.0 (<10% relative activity), and ∼50% residual activity remained at pH 6.0 (Fig. 3). In the reductive direction, with 12-oxo-DCA as a substrate and NADPH as a cofactor, the optimum was also determined to be pH 7.0, with a similar sharp decline to <5% residual activity at pH 8.0 but ∼80% maximal activity between pH 6.5 and 6.0. We therefore chose pH 7.0 to examine steady-state kinetics in both directions.
FIG 3.
pH optima for purified recombinant CDD59473 (a), CDD59474 (b), and CDD59475 (c) in the oxidative (red) and reductive (blue) directions. Substrates for CDD59473 were 50 μM 3-oxo-DCA (reductive) and iso-DCA (oxidative). CDD59474 was tested with 50 μM 3-oxo-DCA (reductive) and DCA (oxidative). CDD59475 was tested with 50 μM 12-oxo-DCA (reductive) and DCA (oxidative). See Materials and Methods for buffer compositions. Experiments were repeated three times, and results are represented as mean ± standard error of the mean (SEM).
Kinetic parameters are listed in Table 2; substrate-saturation curves and Lineweaver-Burk plots are displayed in Fig. S2. Recombinant CDD59475 had Kms that were an order of magnitude higher for DCA (139.71 ± 14.64 μM) than for 12-oxo-DCA (11.41 ± 0.68 μM). The native 12α-HSDH characterized from Clostridium sp. strain ATCC 29733/VPI C48-50 had a comparable Km value for DCA in the presence of NADP+ (12 μM). The catalytic efficiency (Kcat/Km) for CDD59475 in the reductive direction was nearly an order of magnitude greater than that in the oxidative direction, but the turnover number (Kcat) was on the same order in both directions.
In the oxidative direction, CDD59475 recognized substrates regardless of side chain conjugation to glycine or taurine or presence/absence of a 7α-hydroxyl group, which was reported for the native 12α-HSDH from Clostridium sp. strain ATCC 29733/VPI C48-50. Interestingly, in the reductive direction, the enzyme displayed only 4.06% activity for 12-oxo-CA relative to 12-oxo-DCA, suggesting steric hindrance from the 7α-hydroxyl group (Table 3).
TABLE 3.
Substrate specificities of Eggerthella CAG:298 HSDHs
| Steroid | Coenzyme | Relative activity (%)a |
||
|---|---|---|---|---|
| CDD59473 | CDD59474 | CDD59475 | ||
| Iso-DCA | NAD(P)+ | 100 | ND | ND |
| DCA | NAD(P)+ | 22.37 ± 0.68 | 71.65 ± 0.78 | 51.93 ± 1.56 |
| GDCA | NAD(P)+ | 20.56 ± 0.59 | 57.84 ± 0.79 | 55.87 ± 1.17 |
| TDCA | NAD(P)+ | 21.83 ± 0.48 | 59.18 ± 0.06 | 58.90 ± 1.40 |
| CA | NAD(P)+ | 12.65 ± 0.17 | 65.53 ± 1.30 | 55.74 ± 1.23 |
| GCA | NAD(P)+ | 12.43 ± 0.30 | 61.18 ± 1.50 | 60.32 ± 1.37 |
| TCA | NAD(P)+ | 11.54 ± 0.31 | 63.90 ± 0.53 | 53.78 ± 0.35 |
| CDCA | NAD(P)+ | 11.65 ± 1.59 | 19.83 ± 0.82 | NA |
| GCDCA | NAD(P)+ | 13.15 ± 1.08 | 50.74 ± 0.36 | NA |
| TCDCA | NAD(P)+ | 11.51 ± 0.33 | 53.34 ± 1.52 | NA |
| 12-Oxo-DCA | NAD(P)H | NA | NA | 100 |
| 12-Oxo-CA | NAD(P)H | NA | NA | 4.06 ± 1.22 |
| 3-Oxo-DCA | NAD(P)H | 55.24 ± 2.08 | 100 | NA |
| 3-Oxo-CA | NAD(P)H | 10.27 ± 5.05 | 84.46 ± 1.07 | NA |
NA, no activity; ND, not determined. Values represent mean ± standard error of the mean from four technical replicates.
Characterization of recombinant CDD59473 and CDD59474 purified from E. coli.
We then overexpressed and purified recombinant CDD59473 (34.2 ± 0.4 kDa; theoretical molecular mass, 31.59 kDa) and CDD59474 (28.0 ± 0.6 kDa; theoretical molecular mass, 26.40 kDa) in E. coli (Fig. 1) and tested these enzymes against a panel of bile acids and their oxo-derivatives. Separation of CDD59473 and CDD59474 reaction products by TLC revealed formation of iso-DCA and DCA, respectively, when 3-oxo-DCA was the substrate (Fig. 2). The CDD59473 reaction product comigrated with authentic iso-DCA (392.57 amu) with a major mass ion of m/z 391.2849 in negative ion mode, consistent with iso-DCA formation. The reaction product from CDD59474 comigrated with DCA (Fig. 2), and the product in the oxidative direction comigrated with 3-oxo-DCA, with a major mass ion of m/z 389.2688 in negative ion mode (Fig. S2), consistent with formation of 3-oxo-DCA. A pH optimum for CDD59474 in the oxidative direction between pH 9 and 10 was observed, and the optimum in the reductive direction was pH 7.0. With NADP(H) as a cosubstrate, CDD59474 favored the reductive direction, but mass action should favor the oxidative direction in the anaerobic environment of the gut (Table 2; Fig. S2).
E. lenta DSM 2243T was shown to express two 3β-HSDHs (Elen_0198 and Elen_1325) in the SDR family, which share 38.1% amino acid sequence identity and are involved in iso-DCA formation (13). In contrast, the aldo/keto reductase family member, CDD59473, shares only 20% amino acid sequence identity with the 3β-HSDHs Elen_0198 and Elen_1325. The pH optimum of CDD59473 in the oxidative direction was between pH 9 and 10, as observed in CDD59474, with an optimum at pH 6.5 in the reductive direction (Fig. 3). The 3β-HSDH activity of CDD59473 displayed a Km ∼2.5-fold higher, a Vmax an order of magnitude lower, and a Kcat four orders of magnitude lower than those of CDD59474 (3α-HSDH) when 3-oxo-DCA was the substrate. CDD59473 shares with Elen_0198 and Elen_1325 the ability to form small quantities of DCA (Fig. 2) and thus low epimerase activity. Previous work (24) and unpublished data from our laboratory with cultures of E. lenta DSM 2243T demonstrate that iso-CDCA and iso-DCA are minor metabolites and that the mono-keto and di-keto intermediates predominate.
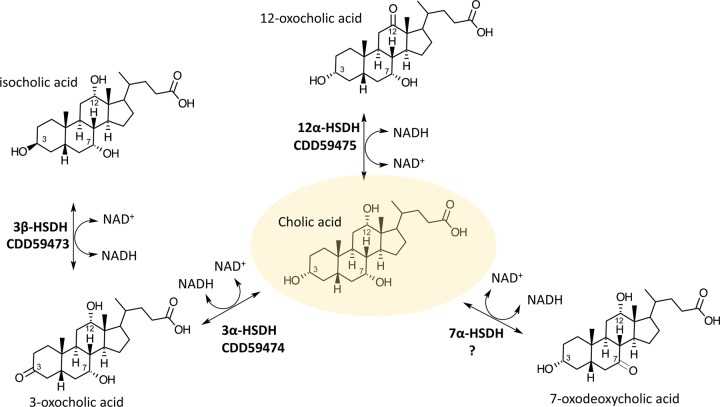
The high Km and low turnover number and catalytic efficiency of CDD59473 in the reductive direction (Table 2), which partially agree with values previously reported for E. lenta DSM 2243T 3β-HSDH, may further explain why iso-CDCA does not predominate in culture (13). Taken together, this three-gene cluster accounts for three of the four HSDH activities reported in E. lenta strains (13, 23, 24) (Fig. 4).
FIG 4.
Schematic representation of bile acid metabolism by Eggerthella lenta. Accession numbers of proteins that we identified in Eggerthella CAG:298 and the reactions they catalyze are shown.
Phylogeny of Eggerthella HSDH enzymes.
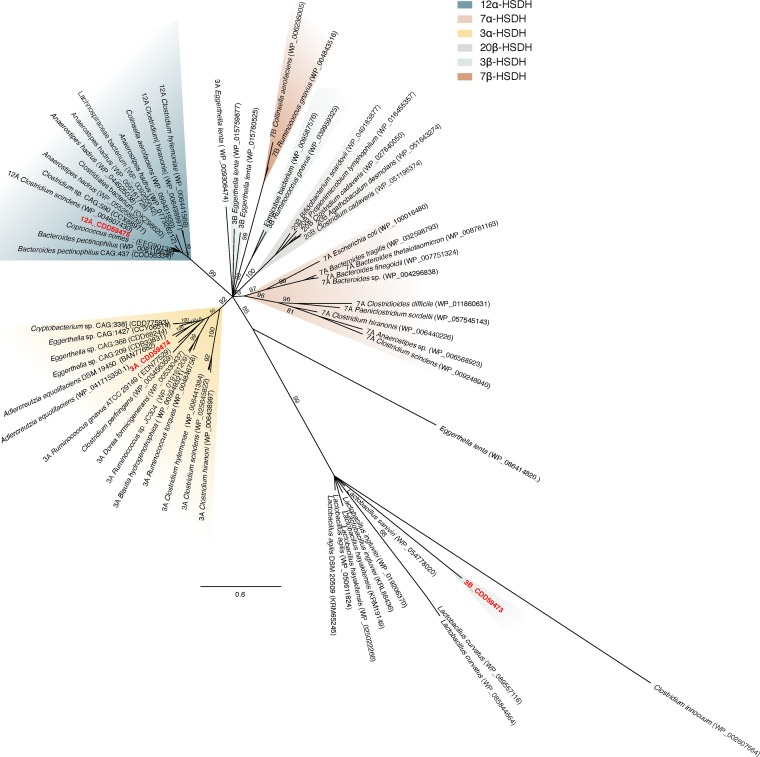
Next, the phylogenetic relationship of these novel Eggerthella HSDH enzymes to other E. lenta SDR family proteins and bacterial HSDH proteins was determined (Fig. 5; see Table S1 in the supplemental material). The 3β-HSDH, CDD59473, had by far the longest branch length of any cluster in the tree, distinct from other 3β-HSDHs characterized in E. lenta DSM 2243T and Ruminococcus gnavus ATCC 29149T (13). Proteins clustering with CDD59473 include predicted aldo/keto reductases and SDR family members. A BLAST search of CDD59473 against the nonredundant database revealed amino acid sequences (61 to 59% identity) that cluster with CDD59473 encoded by Lactobacillus spp. isolated from the gut of pigeon, such as L. agilis (KRM65245.1 and WP_050611824.1) and L. ingluviei (WP_019206370.1 and KRL88436.1) (31, 32), L. hayakitensis (WP_025022266.1 and KRM19149.1) from healthy equine (33), L. saniviri isolated form human feces (34), and L. curvatus isolated from kimchi (WP_089557116.1 and WP_085844664.1) (35) (Fig. 5).
FIG 5.
iTOl representation of FastTree maximum-likelihood phylogeny from MUSCLE alignment of bacterial hydroxysteroid dehydrogenases and the placement of CDD59473, CDD59474, and CDD59475. The phylogeny is the result of 100 bootstraps compared via Phylo.io. Clusters are shaded according to function, as indicated. See Table S1 in the supplemental material for additional sequence information.
CDD59474 is located within a cluster of confirmed and predicted 3α-HSDHs and shares a node and 65.46% amino acid sequence identity with a reported 3α-HSDH (Rumgna_02133) (13). The enzyme EDT24590.1 from C. perfringens clusters closely with Rumgna_02133, suggesting that this may be a 3α-HSDH whose activity was previously reported (27). Clustering closely with CDD59474 are uncharacterized potential 3α-HSDH candidates from species including the equol-utilizing bacterium Adlercreutzia equolifaciens (BAN77692.1), which shares many of its protein-coding genes with E. lenta strains (36), and the “Eubacterium-like” arginine utilizer Cryptobacterium (CDD77593.1), which is involved in periodontal disease (37). Also represented in this cluster are the BaiA enzymes from Clostridium scindens, Clostridium hylemonae, and Clostridium hiranonis, which are involved in the multienzyme bile acid 7α-dehydroxylation pathway responsible for conversion of CA to DCA and CDCA to lithocholic acid (LCA). BaiA enzymes are NAD(H)-dependent SDR family enzymes with strict specificity for bile acid coenzyme A thioesters, which is unique among bacterial HSDHs (38, 39). Our phylogeny also revealed distinct clustering of recently reported 20β-HSDHs from gut and urinary tract isolates (40).
Predicted 12α-HSDHs (30) also formed a separate branch which includes the 12α-HSDH CDD59475 and the characterized 12α-HSDH from Clostridium sp. strain ATCC 29733 (15, 29) (Fig. 5). We have recently confirmed 12α-HSDH activity in whole cells and pure CLOHYLEM_04236 and CLOSCI_02455 from C. hylemonae DSM 15053T and C. scindens ATCC 35704T, respectively (H. Doden, S. A. Sallam, S. Devendran, L. Ly, G. Doden, S. M. Mythen, S. L. Daniel, J. M. P. Alves, and J. M. Ridlon, unpublished data). Collinsella aerofaciens is reported to encode 7β-HSDH (COLAER_RS09325) (41) but is also predicted to encode a 12α-HSDH (COLAER_RS06615) based on homology with CDD59475. We also identified a putative 12α-HSDH in a strain of the butyrate-producing species Anaerostipes hadrus, an organism reported to exacerbate colitis in dextran sulfate sodium (DSS)-treated mice (42). Human gut metagenomic sequences from Clostridiales bacterium Nov_37_41 (OKZ96620.1), Lachnospiraceae bacterium 5_1_63FAA (WP_009265642.1), and Clostridium sp. strain CAG:590 (CCX88977.1) are also predicted to encode 12α-HSDH. Interestingly, while the majority of sequences were represented by Firmicutes and Actinobacteria, a human gut metagenomic sequence from a pectin-degrading bacterium, Bacteroides pectinophilus CAG:437 (CDD56334.1), was also represented.
Previously characterized 7α-HSDHs formed two distinct clusters in our phylogenetic analysis, one containing sequences from Firmicutes (Gram positive) and the other containing sequences from Bacteroidetes and E. coli (Gram negative); both clusters share a common node. While 7α-HSDH activity has been reported in numerous E. lenta strains, a gene encoding 7α-HSDH has yet to be reported in E. lenta (23, 24).
DISCUSSION
In the current study, we utilized targeted gene synthesis to clone and heterologously express genes from a gene cluster (BN592_00768 to BN592_00770) in Eggerthella CAG:298 that is hypothesized to encode multiple HSDHs (CDD59473, CDD59474, and CDD59475) (Fig. 1; Table 2). Characterization of the purified recombinant enzymes revealed that each enzyme catalyzed a distinct oxidation/reduction of bile acid hydroxyl groups. CDD59473 catalyzed NADPH-dependent reduction of 3-oxo-DCA yielding iso-DCA (3β-hydroxyl) as the main reaction product, comigrating on TLC and with mass spectrum consistent with iso-DCA (Fig. 2). CDD59473 is a member of the aldo-keto reductase family and is distantly related to previously characterized 3β-HSDHs from E. lenta DSM 2243T (Elen_0198 and Elen_1325) and R. gnavus (Rumgna_00694), all of which are SDR family enzymes (Fig. 5). Interestingly, despite the divergence in sequence, the epimerase activity of CDD59473 (Fig. 2; Table 2) is consistent with gut microbial 3β-HSDHs characterized previously (13).
The host generates α-hydroxyl bile acids at C-3, C-7, and, in half of the bile acid pool, C-12 (cholic acid). Each α-hydroxyl group and the carboxyl group (C-24) is on the same face of the ABCD ring system, resulting in a polar, hydrophilic side (Fig. 4). Below the ABCD ring system is the hydrophobic face, which interacts with lipids and cholesterol packaged into mixed micelles. The isomerization of the 3α-hydroxyl to 3β-hydroxyl decreases the amphipathic nature of bile acids, making them less effective detergents and also less toxic to bacteria as well as host cells (13, 42, 43). The high Km value (Table 1) suggests that this enzyme functions at high concentrations of toxic secondary bile acids. The low turnover number (Kcat) and catalytic efficiency (Kcat/Km) of CDD59473 versus SDR family enzymes (Elen_0198, Elen_1325, and Rumgna_00694) provide a contrast that structural biology might illuminate in an attempt to optimize the reductase direction of 3β-HSDH and also to establish the basis for the stereospecificity of the reaction that yields low 3α-HSDH (epimerase) activity. Currently, there are no structures reported for gut bacterial 3β-HSDH. Our phylogenetic analysis identified multiple Lactobacillus spp. isolated from diverse avian and mammalian gastrointestinal tracts encoding putative 3β-HSDHs with sequence identities at or below 71% (Fig. 5). A similar targeted gene synthesis approach would be useful in determining the functions and catalytic potentials of these putative 3β-HSDHs. Isomerization at C-3 of primary bile acids may be important in precluding bile acid 7α-dehydroxylation, a microbial biochemical pathway responsible for formation of DCA and LCA, bile acids that are causally associated with cancers of the colon and liver (4).
In contrast to CDD59473, CDD59474 converted 3-oxo-DCA to DCA, and thus BN592_00769 encodes 3α-HSDH (Fig. 4). DCA is generated by the multistep bile acid 7α-dehydroxylation of CA by a few species of intestinal clostridia (4) and is found in fecal water at ∼50 μM (44) but can reach several hundred micromolar (45). With Km values for CDD59474 at ∼150 μM and specificity for DCA, these results indicate that at high DCA concentrations, CDD59473 and CDD59474 work in concert to epimerize DCA to iso-DCA, potentially as a means of detoxification in E. lenta. Interestingly, CDD59474 was clustered with 3α-HSDH from R. gnavus (Rumgna_02133) but was more distantly related to a 3α-HSDH characterized in E. lenta DSM 2243T (Elen_0690) (Fig. 5) (13). E. lenta has been reported to express 7α-HSDH (23, 24); however, genes encoding 7α-HSDH in E. lenta have yet to be identified. CDD59474 was annotated as “7α-HSDH” due to sequence homology, yet expression and characterization of this protein revealed specificity for 3α-hydroxyl groups, providing a cautionary tale and further reinforcing the need to functionally characterize genes predicted to encode particular functions by annotation.
To our knowledge, BN592_00770 is the only reported sequence so far demonstrated to encode 12α-HSDH (CDD59475) in E. lenta (Fig. 2 and 4; Tables 2 and 3), and sequence comparison with CDD59475 is expected to elucidate further gut bacterial 12α-HSDHs (Fig. 5). CDD59475 shares with 12α-HSDH from Clostridium sp. strain ATCC 29733/VPI C48-50 and the 3α-HSDH CDD59474 a conserved N-terminal pyridine nucleotide binding domain (GX3GXG) and active-site catalytic triad (SYK) (see Fig. S3 in the supplemental material). Particular bile acid structures trigger germination of Clostridium difficile spores. Recent work on a germinant receptor (CspC) in C. difficile determined that bile acid specificity was dependent on 12α-hydroxylation (46). Binding to this receptor led to the release of Ca2+ dipicolinic acid from the inside of the spore and subsequent influx of water, ultimately leading to growth into a vegetative cell (46). Gut bacterial 12α-HSDH activity may therefore affect C. difficile germination.
Thus, our analysis demonstrates that E. lenta strains harbor distinct forms of 3α-HSDH, 3β-HSDH, and 12α-HSDH enzymes (Fig. 1 and 4). Critical at the intersection between next-generation sequencing data (metagenomics and metatranscriptomics) and metabolomics is biochemistry, providing the ability to experimentally demonstrate that a particular amino acid sequence catalyzes a particular biochemical reaction. A major task in gut microbiology research today is linking nucleic and amino acid sequences in databases to function and determining how functions affect microbiome structure and host physiology. A major question in the field of bile acid metabolism (47) that we are currently working to address is why bacteria, such as E. lenta, encode numerous HSDHs and oxidize bile acids in an anaerobic environment.
MATERIALS AND METHODS
Bacterial strains and chemicals.
Escherichia coli DH5α was obtained from New England Biolabs (NEB; Ipswich, MA), and E. coli BL21-CodonPlus(DE3) RIPL competent cells were obtained from Agilent, Santa Clara, CA. The pET-46 Ek/LIC vector kit was obtained from Novagen (San Diego, CA). The QIAprep Spin Miniprep kit was obtained from Qiagen (Valencia, CA). Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology (St. Louis, MO). Bile acid substrates were purchased from Steraloids, Inc (Newport, RI, USA).
Targeted gene synthesis, cloning, expression, and purification of recombinant proteins.
Gene fragments (GenBlock) encoding 3α-HSDH, 3β-HSDH, and 12α-HSDH from E. lenta CAG:298 (Bio Project PRJEB816) were synthesized by Integrated DNA Technologies (IDT) (Coralville, IA, USA). Gene fragments were amplified with primers synthesized by IDT (Table 1) using the Phusion high-fidelity polymerase (Stratagene, La Jolla, CA) and were cloned into the pET-46b vector (Novagen) as described by the manufacturer's protocol. Cultivation, plasmid isolation, and sequence confirmation were as previously described (40). HSDH enzymes were expressed as previously described (40). The recombinant proteins were then purified using Talon metal affinity resin (Clontech Laboratories, Mountain View, CA) as per the manufacturer's protocol. The recombinant protein was eluted using an elution buffer composed of 20 mM Tris-HCl, 150 mM NaCl, 20% glycerol, 10 mM 2-mercaptoethanol (pH 7.9), and 250 mM imidazole. The protein purity was assessed by SDS-PAGE, and protein bands were visualized by staining with Coomassie brilliant blue G-250. Subunit molecular mass was calculated using three independent SDS-polyacrylamide gels with Bio-Rad Precision Plus Protein Kaleidoscope prestained protein standards (Bio-Rad Laboratories, Inc., Hercules, CA) using the image processing software ImageJ (https://imagej.nih.gov/ij/index.html). Recombinant protein concentrations were calculated based on their molecular mass and extinction coefficients (see Table S1 in the supplemental material). Briefly, the deduced amino acid sequence was used as input for Expasy's ProtParam tool (https://web.expasy.org/protparam/), and the subunit mass and extinction coefficient (mM−1 cm−1) were utilized to determine enzyme concentration (mg ml−1) in a NanoDrop 2000c with a 10-mm-pathlength cuvette at 280 nm.
Enzyme assays.
The buffers used to study the pH profiling of each enzyme contained 150 mM NaCl, 20% glycerol, and 10 mM 2-mercaptoethanol and varied as follows: 50 mM sodium citrate, (pH 4.0 to 6.0), 50 mM sodium phosphate (pH 6.5 to 7.5), 50 mM Tris-Cl (pH 8 to 9), and 50 mM glycine-NaOH (pH 10 to 11). Linearity of enzyme activity with respect to time and enzyme concentration was determined aerobically by monitoring the oxidation/reduction of NAD(P)(H) at 340 nM (ε = 6,220 M−1 cm−1) in the presence of bile acid and steroid substrates. The standard reaction mixtures were 50 mM sodium phosphate buffer at pH 6.5 (CDD59473 in reductive direction) or pH 7.0 (CDD59474 in reductive direction and CDD59475 in both oxidative and reductive directions) and 50 mM Tris-HCl buffer at pH 9.5 (CDD59473 in oxidative direction) or pH 10 (CDD59474 in oxidative direction), each containing 150 μM pyridine nucleotide cofactor and with reactions initiated by addition of the enzyme (50 nM). Kinetic parameters were estimated by fitting the data to the Michaelis-Menten equation by the nonlinear regression method using the enzyme kinetics module in GraphPad Prism (GraphPad Software, La Jolla, CA). Substrate specificity studies were performed with 150 μM pyridine nucleotide and 50 μM bile acid substrate at the respective pH optimum for the enzyme and each reaction direction.
TLC and MS of bile acid metabolites.
The standard reaction mixtures with different pyridine nucleotide were set up as described above and incubated overnight at room temperature. The reactions were stopped by adding 100 μl of 1N HCl. The reaction mixtures were extracted by vortexing with 2× volumes of ethyl acetate for 1 to 2 min, and then the organic phase was recovered. The organic phase was evaporated under nitrogen gas. The residue was dissolved in 50 μl ethyl acetate and spotted along with standards on thin-layer chromatography (TLC) plate (Silica Gel IB2-F Flexible TLC sheet, 20 by 20 cm with 250-μm analytical layer; J.T. Baker, Avantor Performance Materials LLC, PA, USA). A mobile phase consisting of 70:20:2 (vol/vol/vol) toluene–1,4-dioxane–acetic acid was used. Bile acid metabolites separated on the TLC plate were visualized by spraying 10% phosphomolybdic acid in ethanol and heated at 100°C for 15 min. The spots corresponding to substrate and product were isolated and extracted and underwent mass spectrometry (MS) analysis. Liquid chromatography (LC)-MS analysis was run on a Shimadzu LC-MS-IT-TOF system (Shimadzu Corporation, Kyoto, Japan). The mass spectrometer was operated with an electrospray ionization (ESI) source in negative ionization mode. The nebulizer gas pressure was set at 150 kPa with a source temperature of 200°C and the gas flow at 1.5 liters/min. The detector voltage was 1.65 kV. High-purity nitrogen gas was used as the collision cell gas. The mass spectrogram data were processed with the LC solution Workstation software.
Phylogenetic reconstruction of bacterial SDR family and aldo-keto reductase family protein sequences.
Protein sequences were aligned with MUSCLE using the web service from the EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/muscle/). The alignment was manually explored to verify the absence of inaccuracies. Phylogeny reconstruction was performed using both FastTree v2.1 (48, 49) and IQ-TREE, a time-efficient tool to generate maximum-likelihood phylogenies (50). The two methods were used for comparative purposes. FastTree ran locally using default settings. IQ-TREE ran locally using the following options: -st AA -m TEST -bb 1000 -alrt 1000 (-st, sequence type; -m, automatic model selection; -bb, number of bootstrap replicates). The generated trees were exported in newick format and uploaded to iTOL v3 for visualization (51). The trees were compared using Phylo.io (52). Sequence information given in Table S1 includes a number of previously characterized gut bacterial HSDH enzymes (53–61).
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge the financial support provided to J.M.R. for new faculty startup through the Department of Animal Sciences at the University of Illinois at Urbana-Champaign (Hatch ILLU-538-916).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02475-17.
REFERENCES
- 1.Joice R, Yasuda K, Shafquat A, Morgan XC, Huttenhower C. 2014. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab 20:731–741. doi: 10.1016/j.cmet.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridlon JM, Bajaj JS. 2015. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B 5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. 2003. A G protein-coupled receptor responsive to bile acids. J Biol Chem 278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 7.Raufman JP, Cheng K, Saxena N, Chahdi A, Belo A, Khurana S, Xie G. 2011. Muscarinic receptor agonists stimulate matrix metalloproteinase 1-dependent invasion of human colon cancer cells. Biochem Biophys Res Commun 415:319–324. doi: 10.1016/j.bbrc.2011.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 9.Odermatt A, Da Cunha T, Penno CA, Chandsawangbhuwana C, Reichert C, Wolf A, Dong M, Baker ME. 2011. Hepatic reduction of the secondary bile acid 7-oxolithocholic acid is mediated by 11β-hydroxysteroid dehydrogenase 1. Biochem J 436:621–629. doi: 10.1042/BJ20110022. [DOI] [PubMed] [Google Scholar]
- 10.Ferrandi EE, Bertolesi GM, Polentini F, Negri A, Riva S, Monti D. 2012. In the search for sustainable chemical processes: cloning, recombinant expression, and functional characterization of the 7α- and 7β-hydroxysteroid dehydrogenases from Clostridium absonum. Appl Microbiol Biotechnol 95:1221–1233. doi: 10.1007/s00253-011-3798-x. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann AF, Sjövall J, Kurz G, Radominska A, Schteingart CD, Tint GS, Vlahcevic ZR, Setchell KDR. 1992. A proposed nomenclature for bile acids. J Lipid Res 33:599–604. [PubMed] [Google Scholar]
- 12.Heuman DM. 1989. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J Lipid Res 30:719–730. [PubMed] [Google Scholar]
- 13.Devlin AS, Fischbach MA. 2015. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paumgartner G, Beuers U. 2002. Ursodeoxycholic acid in cholestatic liver disease: mechanisms of action and therapeutic use revisited. Hepatology 36:525–531. doi: 10.1053/jhep.2002.36088. [DOI] [PubMed] [Google Scholar]
- 15.Macdonald IA, Jellet JF, Mahony DE. 1979. 12α-Hydroxysteroid dehydrogenase from Clostridium group P strain C48-50 ATCC #29733. J Lipid Res 20:234–239. [PubMed] [Google Scholar]
- 16.Edenharder R, Pfützner A. 1988. Characterization of NADP-dependent 12beta-hydroxysteroid dehydrogenase from Clostridium paraputrificum. Biochim Biophys Acta 962:362–370. doi: 10.1016/0005-2760(88)90266-4. [DOI] [PubMed] [Google Scholar]
- 17.Kageyama A, Benno Y, Nakase T. 1999. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49:1725–1732. doi: 10.1099/00207713-49-4-1725. [DOI] [PubMed] [Google Scholar]
- 18.Bokkenheuser VD, Winter J, Finegold SM, Sutter VL, Ritchie AE, Moore WE, Holdeman LV. 1979. New markers for Eubacterium lentum. Appl Environ Microbiol 37:1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha JR, Butler VP Jr, Neu HC, Lindenbaum J. 1983. Digoxin-inactivating bacteria: identification in human gut flora. Science 220:325–327. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 20.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. 2013. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feighner SD, Hylemon PB. 1980. Characterization of a corticosteroid 21-dehydroxylase from the intestinal anaerobic bacterium Eubacterium lentum. J Lipid Res 21:585–593. [PubMed] [Google Scholar]
- 22.Wegner K, Just S, Gau L, Mueller H, Gérard P, Lepage P, Clavel T, Rohn S. 2017. Rapid analysis of bile acids in different biological matrices using LC-ESI-MS/MS for the investigation of bile acid transformation by mammalian gut bacteria. Anal Bioanal Chem 409:1231–1245. doi: 10.1007/s00216-016-0048-1. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald IA, Jellett JF, Mahony DE, Holdeman LV. 1979. Bile salt 3α- and 12α-hydroxysteroid dehydrogenases from Eubacterium lentum and related organisms. Appl Environ Microbiol 37:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edenharder R, Mielek K. 1984. Epimerization, oxidation and reduction of bile acids by Eubacterium lentum. System Appl Microbiol 5:287–298. doi: 10.1016/S0723-2020(84)80031-4. [DOI] [Google Scholar]
- 25.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas ME, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. 2011. Colonization-induced host-gut microbial metabolic interaction. mBio 2:e00271-10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson B, Hedlund J, Jörnvall H. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65:3879–3894. doi: 10.1007/s00018-008-8587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macdonald IA, Meier EC, Mahony DE, Costain GA. 1976. 3alpha-, 7alpha-, 12alpha-Hydroxysteroid dehydrogenase activities in Clostridium perfringens. Biochim Biophys Acta 450:142–153. doi: 10.1016/0005-2760(76)90086-2. [DOI] [PubMed] [Google Scholar]
- 28.Harris JN, Hylemon PB. 1978. Partial purification and characterization of NADP-dependent 12alpha-hydroxysteroid dehydrogenase from Clostridium leptum. Biochim Biophys Acta 528:148–157. doi: 10.1016/0005-2760(78)90060-7. [DOI] [PubMed] [Google Scholar]
- 29.Aigner A, Gross R, Schmid R, Braun M, Mauer S. April 2011. Novel 12alpha-hydroxysteroid dehydrogenases, production and use thereof. US patent 20110091921A1.
- 30.Kisiela M, Skarka A, Ebert B, Maser E. 2012. Hydroxysteroid dehydrogenases (HSDs) in bacteria—a bioinformatic perspective. J Steroid Biochem Mol Biol 129:31–46. doi: 10.1016/j.jsbmb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Baele M, Devriese LA, Haesebrouck F. 2001. Lactobacillus agilis is an important component of the pigeon crop flora. J Appl Microbiol 91:488–491. doi: 10.1046/j.1365-2672.2001.01407.x. [DOI] [PubMed] [Google Scholar]
- 32.Baele M, Vancanneyt M, Devriese LA, Haesebrouck F. 2003. Lactobacillus ingluviei sp nov., isolated from the intestinal tract of pigeons. Int J Syst Evol Microbiol 53:133–136. doi: 10.1099/ijs.0.02206-0. [DOI] [PubMed] [Google Scholar]
- 33.Morita H, Nakano A, Shimazu M, Toh H, Nakajima F, Nagayama M, Hisamatsu S, Kato Y, Takagi M, Takami H, Akita H, Matsumoto M, Masaoka T, Murakami M. 2009. Lactobacillus hayakitensis, L. equigenerosi and L equi, predominant lactobacilli in the intestinal flora of healthy thoroughbreds. Anim Sci J 80:339–346. doi: 10.1111/j.1740-0929.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 34.Oki K, Kudo Y, Watanabe K. 2012. Lactobacillus saniviri sp. nov. and Lactobacillus senioris sp. nov., isolated from human feces. Int J Syst Evol Microbiol 62:601–607. doi: 10.1099/ijs.0.031658-0. [DOI] [PubMed] [Google Scholar]
- 35.Jo S, Noh E, Lee J, Kim G, Choi J, Lee M, Song J, Chang J, Park J. 2016. Lactobacillus curvatus WiKim38 isolated from kimchi induces IL-10 production in dendritic cells and alleviates DSS-induced colitis in mice. J Microbiol 54:503–509. doi: 10.1007/s12275-016-6160-2. [DOI] [PubMed] [Google Scholar]
- 36.Toh H, Oshima K, Suzuki T, Hattori M, Morita H. 2013. Complete genome sequence of the equol-producing bacterium Adlercreutzia equolifaciens DSM 19450T. Genome Announc 1:e00742-13. doi: 10.1128/genomeA.00742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uematsu H, Sato N, Djais A, Hoshino E. 2006. Degradation of arginine by Slackia exigua ATCC 700122 and Cryptobacterium curtum ATCC 700683. Oral Microbiol Immunol 21:381–384. doi: 10.1111/j.1399-302X.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 38.Mallonee DH, Lijewski MA, Hylemon PB. 1995. Expression in Escherichia coli and characterization of a bile acid-inducible 3alpha-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol 30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 39.Bhowmik S, Jones DH, Chiu HP, Park IH, Chiu HJ, Axelrod HL, Farr CL, Tien HJ, Agarwalla S, Lesley SA. 2014. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins 82:216–229. doi: 10.1002/prot.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devendran S, Méndez-García C, Ridlon JM. 2017. Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J Lipid Res 58:916–925. doi: 10.1194/jlr.M074914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. 2013. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res 54:3062–3069. doi: 10.1194/jlr.M039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann AF, Roda A. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J Lipid Res 25:1477–1489. [PubMed] [Google Scholar]
- 43.Roda A, Hofmann AF, Mysels KJ. 1983. The influence of bile salt structure on self-association in aqueous solutions. J Biol Chem 258:6362–6370. [PubMed] [Google Scholar]
- 44.Ditscheid B, Keller S, Jahreis G. 2009. Faecal steroid excretion in humans is affected by calcium supplementation and shows gender-specific differences. Eur J Nutr 48:22–30. doi: 10.1007/s00394-008-0755-2. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton JP, Xie G, Raufman JP, Hogan S, Griffin TL, Packard CA, Chatfield DA, Hagey LR, Steinbach JH, Hofmann AF. 2007. Human cecal bile acids: concentration and spectrum. Am J Physiol Gastrointest Liver Physiol 293:G256–G263. doi: 10.1152/ajpgi.00027.2007. [DOI] [PubMed] [Google Scholar]
- 46.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bokkenheuser VD, Winter J. 1980. Biotransformation of steroid hormones by gut bacteria. Am J Clin Nutr 33:2502–2506. doi: 10.1093/ajcn/33.11.2502. [DOI] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson O, Dylus D, Dessimoz C. 2016. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol Biol Evol 33:2163–2166. doi: 10.1093/molbev/msw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. 1996. Crystal structures of the binary and ternary complexes of 7alpha-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 54.Ridlon JM, Kang DJ, Hylemon PB. 2010. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells JE, Hylemon PB. 2000. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66:1107–1113. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirano S, Masuda N, Oda H, Mukai H. 1981. Transformation of bile acids by Clostridium perfringens. Appl Environ Microbiol 42:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coleman JP, Hudson LL, Adams MJ. 1994. Characterization and regulation of the NADP-linked 7 alpha-hydroxysteroid dehydrogenase gene from Clostridium sordellii. J Bacteriol 176:4865–4874. doi: 10.1128/jb.176.16.4865-4874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baron SF, Franklund CV, Hylemon PB. 1991. Cloning, sequencing, and expression of the gene coding for bile acid 7 alpha-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. J Bacteriol 173:4558–4569. doi: 10.1128/jb.173.15.4558-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Aigner A, Schmid RD. 2011. Identification, cloning, heterologous expression, and characterization of a NADPH-dependent 7β-hydroxysteroid dehydrogenase from Collinsella aerofaciens. Appl Microbiol Biotechnol 90:127–135. doi: 10.1007/s00253-010-3052-y. [DOI] [PubMed] [Google Scholar]
- 60.Savino S, Ferrandi EE, Forneris F, Rovida S, Riva S, Monti D, Mattevi A. 2016. June Structural and biochemical insights into 7β-hydroxysteroid dehydrogenase stereoselectivity. Proteins 84:859–865. doi: 10.1002/prot.25036. [DOI] [PubMed] [Google Scholar]
- 61.Sherrod JA, Hylemon PB. 1977. Partial purification and characterization of NAD-dependent 7alpha-hydroxysteroid dehydrogenase from Bacteroides thetaiotaomicron. Biochim Biophys Acta 486:351–358. doi: 10.1016/0005-2760(77)90031-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.