ABSTRACT
Few data have been published on the occurrence and functional role of acetic acid bacteria (AAB) in lambic beer production processes, mainly due to their difficult recovery and possibly unknown role. Therefore, a novel aseptic sampling method, spanning both the spatial and temporal distributions of the AAB and their substrates and metabolites, was combined with a highly selective medium and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as a high-throughput dereplication method followed by comparative gene sequencing for their isolation and identification, respectively. The AAB (Acetobacter species more than Gluconobacter species) proliferated during two phases of the lambic beer production process, represented by Acetobacter orientalis during a few days in the beginning of the fermentation and Acetobacter pasteurianus from 7 weeks until 24 months of maturation. Competitive exclusion tests combined with comparative genomic analysis of all genomes of strains of both species available disclosed possible reasons for this successive dominance. The spatial analysis revealed that significantly higher concentrations of acetic acid (from ethanol) and acetoin (from lactic acid) were produced at the tops of the casks, due to higher AAB counts and a higher metabolic activity of the AAB species at the air/liquid interface during the first 6 months of lambic beer production. In contrast, no differences in AAB species diversity occurred throughout the casks.
IMPORTANCE Lambic beer is an acidic beer that is the result of a spontaneous fermentation and maturation process. Acidic beers are currently attracting attention worldwide. Part of the acidity of these beers is caused by acetic acid bacteria (AAB). However, due to their difficult recovery, they were never investigated extensively regarding their occurrence, species diversity, and functional role in lambic beer production. In the present study, a framework was developed for their isolation and identification using a novel aseptic sampling method in combination with matrix-assisted laser desorption ionization–time of flight mass spectrometry as a high-throughput dereplication technique followed by accurate molecular identification. The sampling method applied enabled us to take spatial differences into account regarding both enumerations and metabolite production. In this way, it was shown that more AAB were present and more acetic acid was produced at the air/liquid interface during a major part of the lambic beer production process. Also, two different AAB species were encountered, namely, Acetobacter orientalis at the beginning and Acetobacter pasteurianus in a later stage of the production process. This developed framework could also be applied for other fermentation processes.
KEYWORDS: lambic beer fermentation, acetic acid bacteria, MALDI-TOF MS, metabolite target analysis
INTRODUCTION
Belgian beers are produced by four different types of fermentation: (i) bottom fermentation of water, barley malt, and hop with Saccharomyces bayanus or Saccharomyces pastorianus for lager beers; (ii) top fermentation of water and a variety of ingredients (barley malt, hops, cereals, herbs, and spices) with Saccharomyces cerevisiae for ales; (iii) nonspontaneous mixed fermentation, traditionally carried out with an in-house starter culture that consists of yeasts and lactic acid bacteria (LAB) followed by maturation in oak casks for red and red-brown acidic ales; and (iv) spontaneous mixed fermentation, traditionally obtained through air inoculation followed by fermentation and maturation in wooden casks for acidic ales (1–3). Among the latter, lambic beer production is probably the oldest surviving commercial brewing style, dating back to the Middle Ages. Lambic beers are obtained by spontaneous fermentation of water, barley malt, unmalted wheat, and aged dry hops for up to 3 years (3–6).
The microbiology of the lambic beer production process carried out by traditional breweries was studied several decades ago and has been characterized by a succession of Enterobacteriaceae, S. cerevisiae and/or S. pastorianus, Pediococcus damnosus and/or Lactobacillus brevis, and Dekkera bruxellensis (2, 3, 7–11). These studies made use of culture-dependent methods, often coupled to phenotypic characterizations, that are outdated now and that have a low throughput compared to that of the current state-of-the-art methodology for microbiological analyses (12). Recently, two lambic beer fermentation studies have been performed with up-to-date culture-dependent microbiological analysis techniques, in particular, regarding the yeast and LAB communities of traditional and industrial production processes (13, 14). New in these studies was the use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as a high-throughput technique for the dereplication of numerous microbial isolates obtained from a complex community. Dereplication was followed by identification through the comparative sequence analysis of 16S rRNA or housekeeping genes of genomic DNA from representative strains. MALDI-TOF MS has only recently been introduced into the field of food microbiology for the identification of microorganisms, leading to the initial construction of reference databases, in particular, for probiotic bacteria and LAB (15–23), yeasts (24, 25), and acetic acid bacteria (AAB) (13, 14, 26–28).
Until now, the occurrence and species diversity of AAB in lambic beer production has not been studied extensively (13, 14). Their role was considered limited, although the acidic taste of lambic beers is often linked to acetic acid besides the lactic acid produced by LAB. Yet, two new AAB species have been described recently that seem to be characteristic for lambic beers, namely, Acetobacter lambici (29) and Gluconobacter cerevisiae (30). The earlier sporadic isolation of AAB may be due to their difficult and inconsistent recovery, as has been shown during the study of many food fermentation processes in which they are involved (5, 31, 32). Such processes are also often subjected to a temporal metabolite target analysis, which was not the case during the most recent studies on lambic beer production (13, 14). Moreover, as AAB are obligate aerobic bacteria, oxidizing ethanol and glucose into acetic acid and gluconic acid, respectively, it is likely that they are concentrated at the air/liquid interface of the wooden casks and hence are missed by classical submerged sampling of the casks through the cork-plugged sampling hole positioned just above the bottom on the front panel of the casks. This sampling technique does not enable one to take into account potential spatial differences of microbial communities and their metabolites produced throughout the casks.
The aim of the present study was to examine the presence and role of AAB during the lambic beer production process at a traditional brewery, by applying a novel aseptic sampling method for investigating both the spatial and temporal distributions of the AAB and those of their substrates and metabolites, as well to improve the cultivation and high-throughput methodologies for their isolation and identification.
RESULTS
Enumerations of AAB.
Similar AAB counts, as determined after plating on modified deoxycholate-mannitol-sorbitol (mDMS) agar medium, were obtained for the two lambic beer production casks examined as biological duplicates (Table 1). AAB could not be detected in the wort used for the lambic beer production before its transfer into the casks, probably due to their very low counts. Once the wort was transferred to the casks, the AAB could be enumerated. Three phases could be distinguished when considering the AAB counts as a function of time (Table 1): a first phase, overlapping with the first week of (yeast) fermentation, containing less than log 4.0 CFU per ml; a second phase, lasting for a few weeks, during which no AAB counts could be enumerated; and a third phase, overlapping with the last part of the production process and lasting until the end of sampling (24 months), representing continuously decreasing counts of the AAB, starting from more than log 6.0 CFU/ml after 3 months of maturation to counts undetectable by selective plating at the end (Table 1). The latter was accompanied by visible pellicle formation by the yeasts. During the third phase, minor differences in enumerations between the three sampling depths were found. In general, the AAB counts were significantly higher (P < 0.05) at the air/liquid interface (top of the casks), except for the 13-month sample (Table 1). The latter anomaly could be ascribed to the brewery practices applied, such as volumetric adjustments of the liquor in the casks to account for evaporation losses. Those were performed by adding fermenting liquor of the same age and brew, thereby causing the mixing of the cask contents and influx of oxygen into the casks.
TABLE 1.
Counts of presumptive AAB as plated on mDMS agar medium present in samples taken at different heights of two wooden casks throughout a 24-month lambic beer production process
| Sampling time point | mDMS counts (log CFU/ml)a |
|||||
|---|---|---|---|---|---|---|
| Cask 1 |
Cask 2 |
|||||
| Top | Middle | Bottom | Top | Middle | Bottom | |
| Before filling | ULDb | ULD | ||||
| 1 h | 2.67 | 2.65 | ||||
| 24 h | 3.48 | 3.33 | 3.38 | 2.86 | 3.31 | 3.16 |
| 3 d | 3.54 | 3.62 | 3.52 | 3.36 | 3.45 | 3.41 |
| 1 wk | ULQc | ULQ | ULQ | ULQ | ULQ | ULQ |
| 2 wk | ULQ | ULQ | ULQ | ULQ | ULQ | ULQ |
| 3 wk | ULQ | ULQ | ULQ | ULQ | ULQ | ULQ |
| 7 wk | 6.08 ± 0.04 A | 6.10 ± 0.04 A | 5.16 ± 0.06 B | 6.01 ± 0.04 A | 5.78 ± 0.04 B | 5.40 ± 0.05 C |
| 3 mo | 6.30 ± 0.07 A | 6.08 ± 0.02 B | 6.12 ± 0.06 B | 6.71 ± 0.01 A | 6.67 0.01 ± B | 6.45 ± 0.02 C |
| 6 mo | 5.07 ± 0.05 | 4.98 ± 0.03 | 4.98 ± 0.03 | 6.10 ± 0.05 A | 5.75 ± 0.05 B | 5.68 ± 0.09 B |
| 9 mo | 5.09 ± 0.04 A | 4.56 ± 0.16 B | 4.67 ± 0.07 B | 4.63 ± 0.03 A | 3.90 ± 0.02 B | 3.83 ± 0.04 B |
| 13 mod | 4.53 ± 0.04 B | 4.82 ± 0.03 A | 3.81 ± 0.08 C | 3.34 ± 0.07 C | 4.12 ± 0.05 A | 3.54 ± 0.01 B |
| 18 mo | 3.19 ± 0.13 A | 2.48 ± 0.28 B | 2.43 ± 0.08 B | 3.62 ± 0.06 A | 2.80 ± 0.03 B | 2.43 ± 0.26 B |
| 24 mo | ULD | ULD | ULD | ULD | ULD | ULD |
mDMS, modified deoxycholate-mannitol-sorbitol. Statistically significant differences (P < 0.05) between samples taken at the tops, middles, and bottoms of the casks are indicated with uppercase letters.
ULD, under limit of detection.
ULQ, under limit of quantification (<30 CFU/ml).
Volumetric adjustment was applied.
Species diversity determination of AAB by MALDI-TOF MS.
A total of 371 bacterial isolates, picked randomly from the mDMS agar medium, were obtained from the two casks examined. These isolates were subjected to MALDI-TOF MS fingerprinting. The GenBank accession numbers for the sequences generated in this study are NR_028625.1 for Acetobacter orientalis, HG329531.1 for A. lambici, CP015168.1 for Acetobacter pasteurianus, HG329585.1 for G. cerevisiae, HG329584.1 for Gluconobacter wancherniae, KF537430.1 for Acetobacter cerevisiae, NR_113673.1 for Acetobacter aceti, NR_113551.1 for Acetobacter lovaniensis, and NR_028625.1 for Acetobacter indonesiensis.
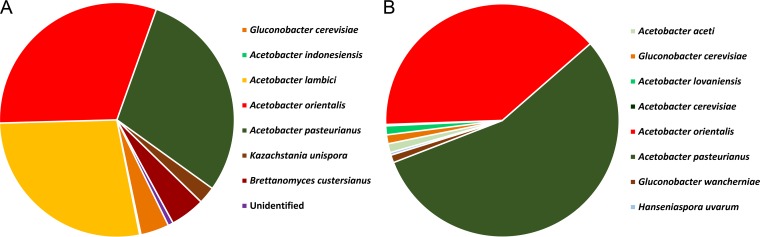
A total of 359 isolates were identified as AAB species, namely, A. orientalis (31%), A. pasteurianus (29%), A. lambici (28%), and G. cerevisiae (4%) as the most prevalent species in cask 1, and A. pasteurianus (54%) and A. orientalis (38%) as the most prevalent species in cask 2 (Fig. 1). One isolate could not be identified, and the remaining 11 isolates represented yeast species, although antifungal compounds were added to the mDMS agar medium.
FIG 1.
Normalized percental distributions of 371 microbial isolates, randomly picked from mDMS agar medium, from samples of cask 1 (A) and cask 2 (B), taken at the tops, middles, and bottoms of the casks during a 24-month lambic beer production process.
Community dynamics of AAB.
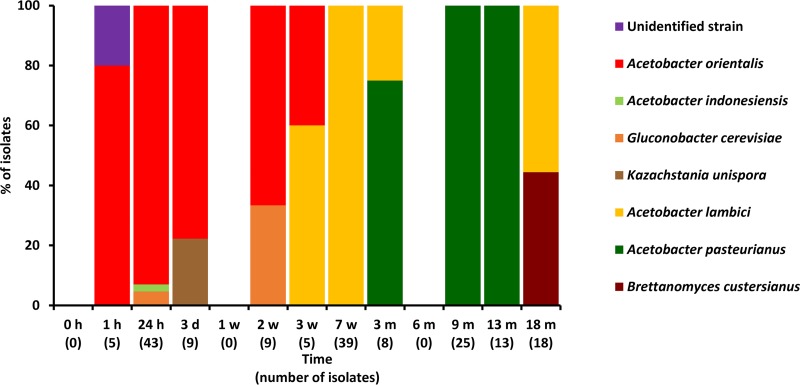
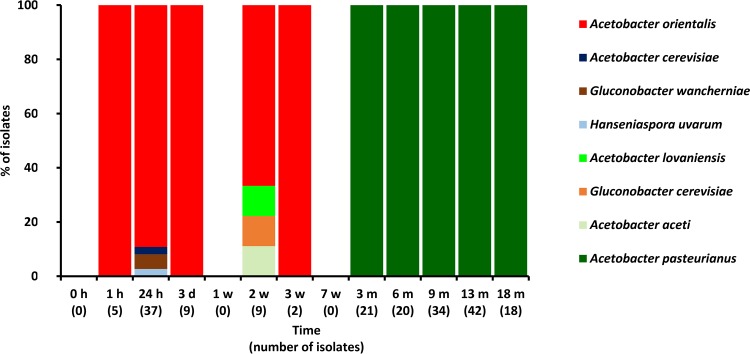
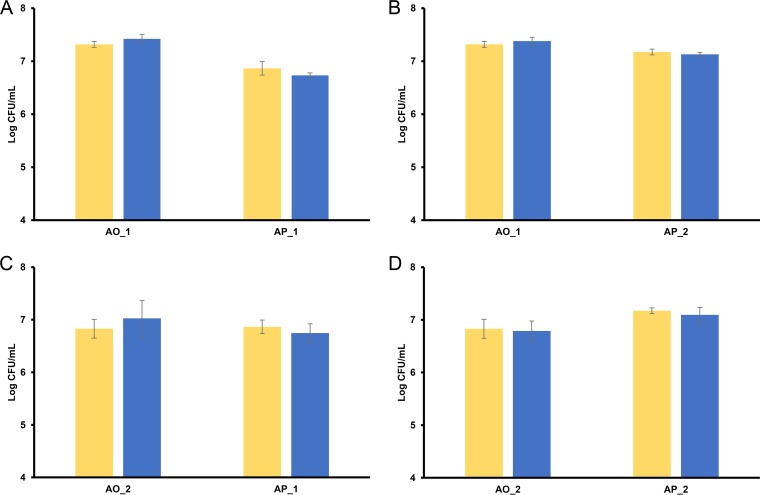
Different AAB species could be associated with the first and third phases described above. Acetobacter orientalis was prevalent during the first phase. However, several other AAB species belonging to genera of the family Acetobacteraceae were also isolated during this first phase (Fig. 2 and 3). During the third phase, A. pasteurianus and A. lambici were prevalent in cask 1 and solely A. pasteurianus in cask 2. The successive prevalence of A. orientalis and A. pasteurianus was not due to competitive exclusion, as strains of both species grown together on the same mDMS agar medium did not affect each other (Fig. 4).
FIG 2.
Identification of 174 microbial isolates, randomly picked from mDMS agar medium, from samples of cask 1 during a 24-month lambic beer production process. Since no differences were found regarding species diversity for the different sampling heights, isolates taken at the top, middle, and bottom of the cask were combined.
FIG 3.
Identification of 197 microbial isolates, randomly picked from mDMS agar medium, from samples of cask 2 during a 24-month lambic beer production process. Since no differences were found regarding species diversity for the different sampling heights, isolates taken at the top, middle, and bottom of the cask were combined.
FIG 4.
Competitive exclusion test for the simultaneous growth of strains of Acetobacter orientalis and Acetobacter pasteurianus. Yellow bars show the mDMS plate counts of the single strains. Blue bars show the mDMS plate counts of the mixed strains. (A) A. orientalis strain 1 (AO_1) and A. pasteurianus strain 1 (AP_1). (B) A. orientalis strain 1 (AO_1) and A. pasteurianus strain 2 (AP_2). (C) A. orientalis strain 2 (AO_2) and A. pasteurianus strain 1 (AP_1). (D) A. orientalis strain 2 (AO_2) and A. pasteurianus strain 2 (AP_2).
Comparative genomics.
The comparative genomic analysis revealed that 241 orthogroups were exclusively present in all four A. orientalis genomes available, and 139 orthogroups were exclusively present in all 22 A. pasteurianus genomes available. The orthogroups only present in A. orientalis contained extra gene families that were related to carbohydrate transport and metabolism, such as TonB receptors, major facilitator superfamily (MFS) transporters, carbohydrate porins, and a glucose-methanol-choline oxidoreductase. The orthogroups only present in A. pasteurianus also contained extra gene families that were related to acidic and ethanol stress tolerance mechanisms, such as squalene-hopene cyclase and cell wall biogenesis glycosyltransferases. They also contained extra alcohol and aldehyde dehydrogenase gene families as well as extra acetolactate synthase gene families.
Substrate consumption and metabolite production.
The concentrations and profiles of substrates and metabolites were comparable for the biological duplicates analyzed, unless stated otherwise. Furthermore, no significant differences within a certain time period were found for the concentrations of carbohydrates, ethanol, ethyl acetate, and gluconic acid when measured at the tops, middles, and bottoms of the casks.
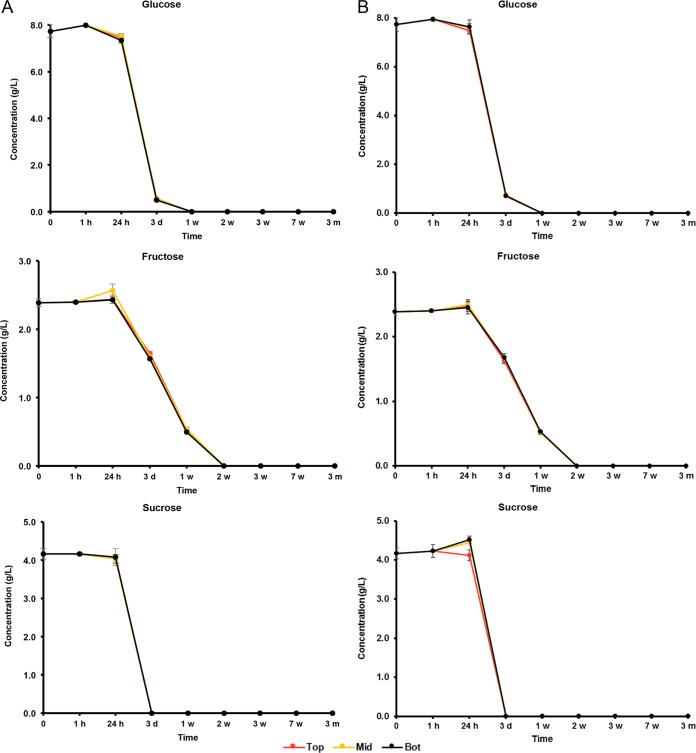
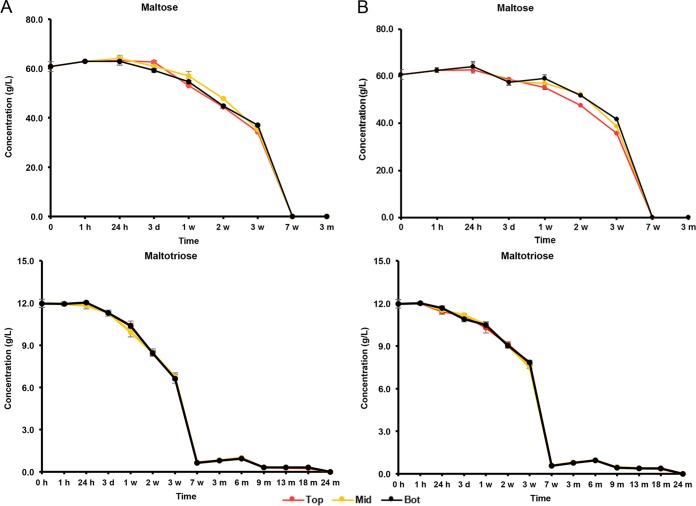
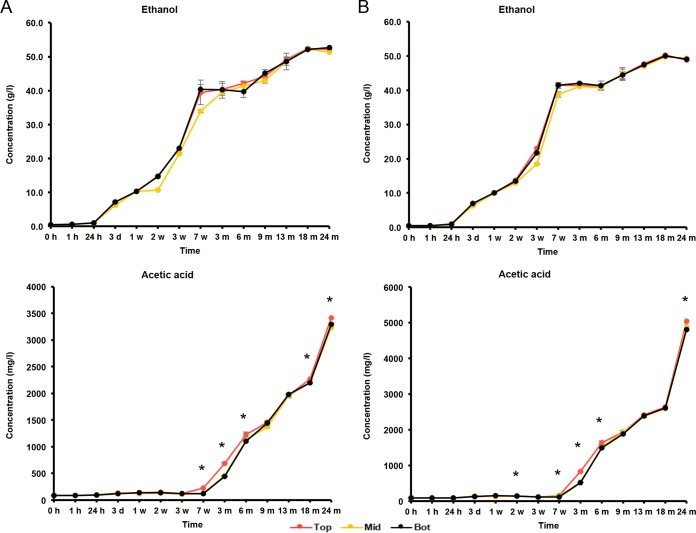
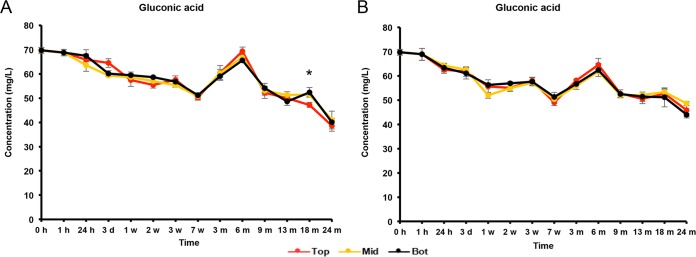
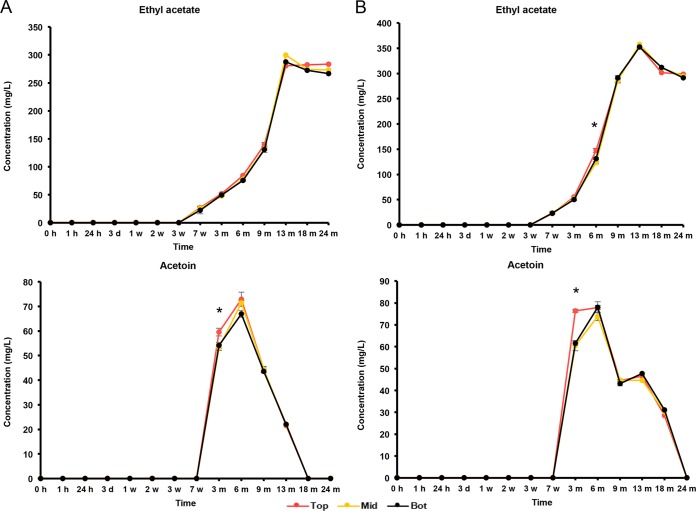
The initial wort was rich in carbohydrates, among which glucose (8.0 g/liter), fructose (2.5 g/liter), sucrose (4.0 g/liter), maltose (60.0 g/liter), and maltotriose (12.0 g/liter) were the most abundant (Fig. 5 and 6). During the first 7 weeks of fermentation, mono-, di-, and trisaccharides were nearly completely depleted and mostly converted into ethanol (main alcoholic fermentation phase carried out by yeasts). Due to this depletion, rapid ethanol and carbon dioxide production ceased, enabling the AAB to grow, since oxygen was no longer flushed out by carbon dioxide production. As soon as the AAB thrived, the ethanol concentrations thus stopped increasing, possibly reflecting a dynamic equilibrium between the low concentrations of ethanol still produced by yeasts and ethanol oxidation by the AAB (Fig. 7). From 6 months onwards, overlapping with the period that the AAB declined, the ethanol concentrations increased again, indicating that the production of ethanol from maltotriose and higher maltooligosaccharides by yeasts of the maturation phase was higher than the oxidation of ethanol into acetic acid by the AAB. Acetic acid was produced from 7 weeks onwards, exceeding concentrations of 2.0 g/liter in both casks (Fig. 7). Up to 6 months, acetic acid was produced in significantly higher concentrations (P < 0.05) at the tops than in the middles and at the bottoms of the casks. Gluconic acid was present from the start of the fermentations, and a slight increase was noticeable from 7 weeks until 6 months (Fig. 8). From 3 weeks onwards, ethyl acetate, the highly abundant ester present in lambic beers, was formed. Ethyl acetate reached maximal concentrations of 300 mg/liter on average for cask 1 and 350 mg/liter for cask 2 after 13 months of maturation (Fig. 9). Acetoin was produced from 7 weeks until 6 months, reaching concentrations between 70 and 80 mg/liter in both casks (Fig. 9). The largest increase in acetoin concentrations was found from 7 weeks until 3 months. During this period, acetoin was produced in significantly higher concentrations (P < 0.05) at the tops than in the middles and at the bottoms of the casks. Acetoin concentrations decreased after 6 months, coinciding with the decline of the AAB counts.
FIG 5.
Consumption of glucose, fructose, and sucrose in cask 1 (A) and cask 2 (B) at the tops (Top), middles (Mid), and bottoms (Bot) of the casks during a 24-month lambic beer production process.
FIG 6.
Consumption of maltose and maltotriose in cask 1 (A) and cask 2 (B) at the tops (Top), middles (Mid), and bottoms (Bot) of the casks during a 24-month lambic beer production process.
FIG 7.
Production of ethanol and acetic acid in cask 1 (A) and cask 2 (B) at the tops (Top), middles (Mid), and bottoms (Bot) of the casks during a 24-month lambic beer production process. Statistically significant differences (P < 0.05) between samples taken at the tops and those at the middles and bottoms of the casks are marked with asterisks.
FIG 8.
Production of gluconic acid in cask 1 (A) and cask 2 (B) at the tops (Top), middles (Mid), and bottoms (Bot) of the casks during a 24-month lambic beer production process. The statistically significant differences (P < 0.05) between samples taken at the top and those at the middle and bottom of the cask are marked with an asterisk.
FIG 9.
Production of ethyl acetate and acetoin in cask 1 (A) and cask 2 (B) at the tops (Top), middles (Mid), and bottoms (Bot) of the casks during a 24-month lambic beer production process. Statistically significant differences (P < 0.05) between samples taken at the tops and those at the middles and bottoms of the casks are marked with asterisks.
DISCUSSION
Recently performed studies revisited the microbiology of traditional and industrial lambic beer production processes (13, 14). However, few data were published on the occurrence and functional role of AAB, mainly due to their difficult recovery and identification, and possibly unknown contribution, during lambic beer production (5). The present study led to new insights into the dynamics and importance of AAB during the complex spontaneous fermentation and maturation stages of lambic beer production. Therefore, an appropriate aseptic sampling method, spanning both the spatial and temporal distributions of AAB, substrates, and metabolites, was performed. Also, the use of an optimal selective medium (mDMS [33, 34]) enabled a high recovery of AAB. Yet, yeast isolates were still obtained from mDMS agar containing cycloheximide during the main fermentation phase and maturation phase, when yeasts are indeed abundantly present in the fermenting liquor compared to AAB, given the cycloheximide-resistant nature of lambic beer yeasts such as Hanseniaspora and Brettanomyces (13, 14, 35). Further, high-throughput dereplication through MALDI-TOF MS, followed by comparative gene sequence analysis, enabled accurate identifications of the AAB isolates. By doing so, viable AAB could be retrieved throughout the different and major parts of the duplicate lambic beer production process studied. Acetobacter species seemed to be more prevalent than Gluconobacter species. These findings contrasted former studies stating that AAB are only found inconsistently throughout the lambic beer fermentation and maturation processes (13, 14). However, particular AAB species largely varied among the casks tested, reflecting not only their dependence on oxygen for growth (which, in turn, depends on the volumes and wood porosity of the casks used) but also their potential origin (environmental air, casks, etc.). Although the differences were small, the AAB generally occurred in higher numbers at the air/liquid interface of the casks, reflecting their aerobic nature. Moreover, they could be isolated at all sampling heights applied (tops, middles, and bottoms of the casks), confirming that AAB can survive under oxygen limitation (13, 14, 36–38). The presence of oxygen in the casks is mainly due to microoxygenation of the liquor through the wood of the casks (but depends on the porosity of the wood and the diameter of the casks), as has been shown for wine barrels (39).
During the initial stages of fermentation, a first peak of AAB occurred, with A. orientalis as the prevailing species. This AAB species was previously found during lambic beer fermentations (13, 14). Due to the combination of a monosaccharide-rich environment and low ethanol concentrations, some Gluconobacter species also occurred during this fermentation phase, which lasted only a couple of days. Among them was G. cerevisiae, which was indeed first isolated from a lambic beer fermentation (30). During the first AAB phase, oxygen was still widely present due to the filling of the casks, causing turbulence and hence explaining the absence of spatial distributions. An intermediate phase with no detectable AAB counts was characteristic of both casks examined, which suggested a decrease of the initially high oxygen levels due to the carbon dioxide production by yeasts that are more prevalent then (in particular, Saccharomyces species [13, 14]), causing oxygen to be flushed out. The increase in AAB numbers after 7 weeks of maturation marked the occurrence of a second AAB phase, which reached a peak of almost twice the initial numbers up to approximately 107 CFU/ml, lasted for several months, and harbored A. pasteurianus as the prevailing AAB species. Competitive exclusion of A. orientalis could be excluded, as strains of the two species could grow on mDMS agar medium simultaneously. The prevalence of A. pasteurianus during this phase of the production process could be due to a better adaption to the environment, which is characterized by the presence of high concentrations of ethanol and increasing concentrations of acetic acid. An indication for this better adaptation was the presence of extra genes coding for acidic and ethanol tolerance and oxidation mechanisms in the genomes of all strains of A. pasteurianus examined, as revealed through comparative genomics of the species A. orientalis and A. pasteurianus. Also, the lactic acid produced by LAB will contribute to the acid stress to which A. pasteurianus is exposed. In contrast, A. orientalis is less ethanol tolerant than A. pasteurianus (40), which explained its occurrence in the beginning of the production process. Moreover, its genome harbored more carbohydrate uptake and oxidation genes, possibly explaining its prevalence in that low-ethanol- and high-carbohydrate-level phase. Furthermore, A. orientalis can produce gluconic acid from glucose (40). The isolation of A. lambici (cask 1) confirmed its, until now, unique presence during lambic beer fermentation (29). However, given the spontaneous nature of the lambic beer production process, the AAB species present as well as their dynamics will be most likely brewery dependent and will be affected by the process conditions applied and the type and porosity of the wooden barrels used.
The occurrence of large numbers of AAB led to the oxidation of ethanol and hence a large increase in acetic acid concentrations and, concurrently, an increase in ethyl acetate concentrations. Acetic acid is the oxidation product of ethanol as the result of dehydrogenase activities of the AAB (41). Due to the limited presence of Gluconobacter species, almost no gluconic acid was produced, except in the second AAB phase (Acetobacter species) that was characterized by a limited gluconic acid production. The formation of ethyl acetate was probably an interplay of both chemical and enzymatic reactions that take place during lambic beer maturation. Indeed, it was postulated that nonenzymatic synthesis accounts for a portion of the esters formed (9). Alternatively, it has been shown that A. pasteurianus possesses esterases, among which esterase 1 is responsible for ethyl acetate biosynthesis (42, 43). The substantial production of acetoin during lambic beer maturation could be due to the oxidation of lactic acid via pyruvate by the AAB species present, as was shown for A. pasteurianus during cocoa bean fermentation (44, 45). The fact that acetoin production coincided with the second AAB peak and that more acetoin was produced at the air/liquid interface supports this biosynthesis potential of A. pasteurianus. Moreover, comparative genomics of A. orientalis and A. pasteurianus revealed the presence of extra genes coding for acetolactate synthase and acetolactate decarboxylase, which are responsible for acetoin production in all available genomes of A. pasteurianus strains. The decrease in the concentration of acetoin overlapped with the maturation phase and could be ascribed to the growth of Dekkera species, since it has been shown that these yeast species can use alternative external electron acceptors such as acetoin to reconcile their redox imbalance caused by the inability to reoxidize NADH plus H+ via the glycerol pathway (46, 47). Since the differences in bacterial enumerations were small, the significantly higher concentrations of acetic acid and acetoin found at the tops of the casks at certain time points of the lambic beer production process pointed toward a higher metabolic activity of the AAB species present, owing to the abundance of oxygen. The extent of these differences will likely increase with increasing cask volumes and decreasing wood porosity. No spatial differences in acetic acid concentrations occurred from 6 months of maturation onwards. As the AAB numbers decreased from this point onwards and acetic acid was still produced, this acetic acid production has to be primarily ascribed to the growth of Dekkera bruxellensis (2, 3, 8, 10, 13, 14). Dekkera bruxellensis is known to produce acetic acid under aerobic conditions (48). The AAB numbers decreased mainly due to pellicle formation by D. bruxellensis and other oxidative yeasts (2, 3). As pellicle formation only occurred after 6 months, when AAB started to decline, this pellicle has to be primarily ascribed to the yeast species present.
From a technological point of view, it is key to avoid excessive AAB growth during lambic beer production, as an influx of too much of oxygen will lead to high numbers of AAB and excessive concentrations of acetic acid and acetoin, which may create an undesirable flavor profile in the lambic beer. The rather high concentrations found during the present study are due to the lack of sufficient volumetric adjustments of the liquor in the casks to compensate for evaporation and sampling losses. Yet, acetic acid and ethyl acetate are desirable compounds for the complex lambic beer flavor profile, albeit below certain concentrations (9). The use of well-sealed wooden casks that enable microaerobic conditions as well as practices such as volumetric adjustments to compensate for evaporation helps to control AAB growth. Pellicle formation by yeasts is a natural phenomenon that helps prevent oxygen influx and thus AAB growth.
In conclusion, the novel aseptic isolation method combined with a highly selective agar medium and accurate dereplication enabled us to isolate and identify AAB from a complex spontaneous fermentation process. This study highlighted a two-phase evolution of AAB during the duplicate lambic beer production process examined. A comparative genomic analysis gave some insights into this two-phase evolution, whereby A. orientalis occurred during a few days in the beginning of the fermentation and A. pasteurianus occurred from 7 weeks until 24 months of maturation. Generally, the tops of the casks were characterized by higher AAB counts and higher concentrations of their metabolites, pointing toward a higher metabolic activity of the AAB species at the air/liquid interfaces of the casks. In contrast, no differences in AAB species diversity occurred throughout the casks. These data on AAB will enable us to monitor and hence modify the fermentation conditions to improve the quality of lambic beer.
MATERIALS AND METHODS
Sampling.
A lambic beer production process was started in a traditional lambic brewery southwest of Brussels in November 2014. Wort of 12.6°P was prepared according to the brewer's recipe and cooled overnight in a coolship open to the environmental air. Two identical 660-liter oak casks, previously used several times to produce lambic beer and located in the bottom row of tens of casks in a cellar at ambient temperature, ranging from 9°C to 20°C, were filled with wort of the same brew. The relative humidity of the cellar was controlled to prevent a drop below 70%. Both casks were cleaned with high pressurized water at 50°C and fumigated with sulfur sticks to inhibit mold growth after cleaning. Before filling, the casks were opened to the environmental air to remove the remaining sulfur vapors.
Both casks were sampled as a function of time, representing biological duplicates. Samples of 100 ml were taken from the cooled wort before and 1 h, 1 and 3 days, 1, 2, 3, and 7 weeks, and 3, 6, 9, 13 (time point with volumetric adjustment of the liquor in the cask), 18, and 24 months after its transfer to the wooden casks. Sampling was performed at different heights (top, middle, and bottom) in the casks by means of an aseptic isolation method, by applying flame sterilization and using γ-irradiated jumbo pipettes (900 mm, high-density polyethylene; VWR International, Darmstadt, Germany). The pipettes were inserted into the cork-plugged bungholes located at the tops of the casks and adjusted for sampling at the intended heights (10, 50, and 90 cm from the top) to take samples of 50 ml per insertion. Samples were put on ice before their transfer to the laboratory for analysis. Portions of the samples were analyzed immediately (plating); another portion (50 ml) was centrifuged (7,200 × g for 20 min at 4°C) to store cell-free culture supernatants at −20°C for metabolite target analysis.
Microbiological analyses. (i) Selective plating and culturing.
Selective plating of the chilled samples was performed immediately after sampling. Therefore, the samples were serially diluted in 0.85% (mass/vol) saline, and 100 μl of each dilution was plated on mDMS agar medium for the enumeration and isolation of presumptive AAB after aerobic incubation at 30°C (34). The agar medium was supplemented with 5 ppm of amphotericin B (Sigma-Aldrich, Bornem, Belgium) and 200 ppm of cycloheximide (Sigma-Aldrich) to inhibit fungal growth. To verify the AAB species diversity obtained through plating as a function of time, in particular, with respect to the successive dominance of A. orientalis and A. pasteurianus, competitive exclusion among these species was evaluated as follows. Two representative strains of both the A. pasteurianus (referred to as AP_1 and AP_2) and A. orientalis (referred to as AO_1 and AO_2) MALDI-TOF MS clusters were grown in mDMS medium. Third-generation cultures of these strains were mixed in equal concentrations based on their optical density at 600 nm (OD600) and plated in triplicates on mDMS agar medium. The single-strain cultures, adjusted to the same OD600 value, were plated in triplicates as a positive control. Samples of agar medium containing 30 to 300 CFU were counted after 7 days of incubation. The A. pasteurianus and A. orientalis strains could be easily distinguished on the basis of their colony morphology.
(ii) Enumeration and isolation of colonies.
Samples of agar medium containing 30 to 300 CFU were counted after 7 days of incubation to determine the presumptive AAB community dynamics. Subsequently, 10% of the total numbers of colonies were randomly picked from appropriate dilutions to determine the culture-dependent microbial species diversity.
(iii) Dereplication and identification of isolates by MALDI-TOF MS.
For dereplication and identification of the presumptive AAB isolates, each colony was subcultivated twice on fresh agar medium. The resulting third-generation colonies were used for MALDI-TOF MS fingerprinting (13). These cells were stored in cryovials at −80°C in 25% (vol/vol) glycerol after cultivation in mDMS medium. In short, an inoculation loop with a cell mass was suspended in 300 μl of ultrapure water, after which, 900 μl of ethanol was added. This cell suspension was centrifuged (21,000 × g for 3 min at 4°C) and stored at −20°C. Before analysis, the cell suspensions were centrifuged (21,000 × g for 3 min at 4°C), the cell-free supernatants were removed, and the cell pellets were resuspended in 50 μl of 70% (vol/vol) formic acid (Merck, Darmstadt, Germany). These suspensions were transferred to a 96-well plate and further handled by a spotting robot (Viaflo 96; Integra Biosciences, Zizers, Switzerland), which added 80 μl of acetonitrile and mixed the contents. After centrifugation (3,700 × g for 10 min at 4°C), 1-μl samples of these solutions were spotted in duplicates on an OPTI-TOF 384 stainless steel plate (AB SCIEX, Framingham, MA) and overlaid with 1 μl of matrix solution (5 mg/ml of α-cyano-4-hydroxycinnamic acid in water/acetonitrile/trifluoroacetic acid [48:50:2]). The mass spectra were measured by means of a 4800 Plus MALDI-TOF MS analyzer (AB SCIEX), as described previously (49).
Data Explorer 4.0 software (AB SCIEX) was used to convert the mass spectra into .txt files to import them into a database using BioNumerics 7 (Applied Maths, Sint-Martens-Latem, Belgium). The spectral profiles were compared by means of the Pearson product moment correlation coefficient, and a dendrogram was built using the unweighted pair group method with arithmetic mean (UPGMA) clustering algorithm. Homogeneous clusters consisting of isolates with visually identical and/or virtually identical mass spectra were delineated. Representative strains from each cluster were identified through comparative sequence analysis of the 16S rRNA gene and the housekeeping gene dnaK (13).
Genome mining and comparative genomics.
To be able to explain the switch between A. orientalis and A. pasteurianus and to underline the differences in fitness, the physiologies of strains of these two species were compared by genome mining. Therefore, a comparison was made between the four A. orientalis and the 22 A. pasteurianus genomes available in GenBank of the National Center for Biotechnology Information (NCBI, Bethesda, MD). OrthoFinder software was used as the analysis tool to identify orthologous protein sequence families or orthogroups, i.e., a group of genes descended from a single gene from the last common ancestor of a group of species, that were uniquely present in the genomes of strains of one of the two species compared (50). Diamond was used as a search tool for sequence alignment (51).
Metabolite target analysis.
Metabolite analysis was targeted toward potential substrates (carbohydrates and ethanol) consumed by and metabolites (acetic acid and gluconic acid) and flavor compounds (acetoin and ethyl acetate) produced by AAB. All samples were both prepared and analyzed in triplicates.
The concentrations of ethanol and acetic acid were measured by high-performance liquid chromatography with refractive index (HPLC-RI) detection, applying external calibration, as described previously (52). Briefly, a Waters chromatograph (Waters, Milford, MA) equipped with an ICSep ICE ORH-801 column (Transgenomic North America, Omaha, NE) and coupled to an RI detector (Waters) was used. The mobile phase consisted of ultrapure water containing 5 mM H2SO4 at 0.4 ml/min. Modifications included sample preparation, which involved a deproteinization step for both cell-free culture supernatants of the samples taken and the standards used. Then, 300 μl of Carrez A solution [36 g/liter of K4Fe(CN)6·3H2O] and 300 μl of Carrez B solution (72 g/liter of ZnSO4·7H2O) were added to 600 μl of analyte. All these solutions were vortexed, centrifuged (21,912 × g for 15 min at 4°C), and filtered (0.2-μm-pore-size Whatman filters; GE Healthcare Life Sciences, Bucks, UK) before they were injected (30 μl) into the column.
The concentrations of acetoin and ethyl acetate were measured by gas chromatography with flame ionization detection (GC-FID), applying internal standardization as described before (45). Briefly, a Focus gas chromatograph (Interscience, Breda, The Netherlands) equipped with a Stabilwax-DA column (Restek, Bellefonte, PA) coupled to an FID-80 detector (Interscience) was used. Hydrogen gas was used as the carrier gas and nitrogen gas was used as the make-up gas. The injector and detector temperatures were set at 240°C and 250°C, respectively. The following temperature gradient was used: 0.0 to 10.0 min, linear gradient at 10°C/min until 140°C; 10.0 to 11.8 min, linear gradient at 50°C/min until 230°C; and 11.8 to 21.8 min, 230°C. All samples and standards were vortexed, centrifuged, and filtered, as described above, before they were injected (1 μl; split 40) into the column.
The concentration of gluconic acid was measured with ultraperformance liquid chromatography with tandem mass spectrometry (UPLC-MS/MS) detection, applying external calibration as described before (53). Briefly, an Acquity system chromatograph (Waters) equipped with an HSS T3 column (Waters) and coupled to a TQ tandem mass spectrometer with a ZSpray electrospray ionization source (Waters) was used. The mobile phase consisted of 980 ml of ultrapure water, 20 ml of methanol, and 2 ml of formic acid (eluent A), and 50 ml of ultrapure water, 950 ml of methanol, and 2 ml of formic acid (eluent B), with the following gradient: 0.0 to 1.5 min, isocratic 10% B; 1.5 to 3.0 min, linear from 10 to 90% B; 3.0 to 4.0 min, isocratic 90% B; 4.0 to 4.1 min, linear from 90 to 10% B; and 4.1 to 6.0 min, isocratic 10% B. Modifications included a constant flow rate of 0.2 ml/min and sample preparation, which involved predilution of the samples in ultrapure water followed by a deproteinization step for both cell-free culture supernatants of the samples taken and the standards used. Then, 500 μl of acetonitrile was added to 500 μl of analyte. All samples and standards were vortexed, centrifuged, and filtered, as described above, before they were injected (10 μl) into the column.
The concentrations of glucose, fructose, sucrose, and maltose were measured with high-performance anion-exchange chromatography coupled to pulsed amperometric detection (HPAEC-PAD) with internal standardization, as described before (54). Briefly, an ICS3000 chromatograph (Dionex, Sunnyvale, CA) equipped with a Carbopac PA10 column (Dionex) and coupled to a pulsed amperometric detector (Dionex) was used. The mobile phase consisted of ultrapure water (eluent A), 167 mM NaOH (eluent B), and 500 mM NaOH (eluent C) at 1 ml/min. Modifications included the sample preparation, which involved the predilution of the samples in ultrapure water followed by deproteinization of both samples and the standards with Carrez A and Carrez B solutions. Then, 300 μl of Carrez A solution and 300 μl of Carrez B solution were added to 300 μl of prediluted analyte plus 300 μl of internal standard (rhamnose; Fluka Chemie, Buchs, Switzerland). All samples and standards were vortexed, centrifuged, and filtered, as described above, before they were injected (10 μl) into the column, with the following gradient of the mobile phase: 0.0 to 20.0 min, isocratic 13% B; 20.0 to 25.0 min, linear from 13% to 0% B and from 0% to 100% C; 25.0 to 30.0 min, isocratic 100% C; 30.0 to 31.0 min, linear from 0% to 13% B and from 100% to 0% C; and 31.0 to 35.0 min, isocratic 13% B.
The concentration of maltotriose was measured with HPAEC-PAD with internal standardization, applying a different methodology. The preparation of the samples and the standards involved their predilution in ultrapure water followed by deproteinization. A 50-μl volume of the prediluted analyte was added to 950 μl of the deproteinization solution consisting of 50% (vol/vol) acetonitrile, 49.5% (vol/vol) ultrapure water, and 0.5% (vol/vol) rhamnose solution (50 mg of rhamnose in 1 ml of ultrapure water). All samples and standards were vortexed, centrifuged, filtered, and analyzed, as described above. They were injected (10 μl) into a Carbopac PA100 column (Dionex) and eluted at 1 ml/min. The mobile phase consisted of ultrapure water (eluent A), 100 mM NaOH (eluent B), 1,000 mM NaOH (eluent C), and 100 mM NaOH with 380 mM Na acetate (eluent D) with the following gradient: 0.0 to 5.0 min, isocratic 96% B and 4% C; 5.0 to 15.0 min, linear from 96% to 60% B and from 4% to 40% C; 15.0 to 20.0 min, linear from 60% to 30% B and from 40% to 70% C; 20.0 to 20.1 min, linear from 30% to 0% B and from 70% to 100% C; 20.1 to 25.0 min, isocratic 100% C; 25.0 to 25.1 min, linear from 100% to 0% C and from 0% to 100% D; 25.1 to 33.0 min, isocratic 100% D; and 33.0 to 40.0 min, linear from 0% to 96% B, from 0% to 4% C, and from 100% to 0% D.
Statistical analysis.
One-way analyses of variance (ANOVAs) were conducted for the determination of differences in enumerations and metabolite concentrations between samples taken at the tops, middles, and bottoms of the casks, followed by a series of post hoc pairwise comparisons with Tukey's tests. A probability level of 0.05 was considered to be significant for all statistical procedures. All statistical analyses and tests performed were executed through the SPSS v.20 package (IBM, Chicago, IL).
ACKNOWLEDGMENTS
This work was financially supported by the Research Council of the Vrije Universiteit Brussel (SRP7 and IOF342 projects), the Hercules Foundation (projects UABR09004 and UAB13002), and the KMO Portefeuille (projects 2014KMO084991, 2015KMO091056, and 2016KMO149170). J.D.R. is the recipient of a PhD fellowship of the Vrije Universiteit Brussel. M.V. is the recipient of a PhD fellowship of the Research Foundation Flanders (FWO).
We thank Wim Borremans for providing technical assistance.
REFERENCES
- 1.Spitaels F, Wieme AD, Snauwaert I, De Vuyst L, Vandamme P. 2017. Microbial ecology of traditional beer fermentations, p 179–196. In Bokulich N, Bamforth C (ed), Brewing microbiology: current research, omics and microbial ecology. Caister Academic Press, Poole, UK. [Google Scholar]
- 2.Van Oevelen D, Spaepen M, Timmermans P, Verachtert H. 1977. Microbiological aspects of spontaneous wort fermentation in the production of lambic and gueuze. J Inst Brew 83:356–360. doi: 10.1002/j.2050-0416.1977.tb03825.x. [DOI] [Google Scholar]
- 3.Verachtert H, Iserentant D. 1995. Properties of Belgian acid beers and their microflora. Part I. The production of gueuze and related refreshing acid beers; Cerevisia: 20:37–41. [Google Scholar]
- 4.De Keersmaecker J. 1996. The mystery of lambic beer. Sci Am 275:74–80. doi: 10.1038/scientificamerican0896-74. [DOI] [Google Scholar]
- 5.Pothakos V, Illeghems K, Laureys D, Spitaels F, Vandamme P, De Vuyst L. 2016. Acetic acid bacteria in fermented food and beverage ecosystems, p 73–100. In Matsushita K, Toyama H, Tonouchi N, Okamoto-Kainuma A (ed), Acetic acid bacteria: ecology and physiology. Springer, Tokyo, Japan. [Google Scholar]
- 6.Verachtert H, Derdelinckx G. 2005. Acidic beers: enjoyable reminiscences of the past. Cerevisia 30:38–47. [Google Scholar]
- 7.Martens H, Dawoud E, Verachtert H. 1991. Wort enterobacteria and other microbial populations involved during the first month of lambic fermentation. J Inst Brew 97:435–439. doi: 10.1002/j.2050-0416.1991.tb01082.x. [DOI] [Google Scholar]
- 8.Shanta Kumara HMC, Verachtert H. 1991. Identification of lambic superattenuating microorganisms by the use of selective antibiotics. J Inst Brew 97:181–185. doi: 10.1002/j.2050-0416.1991.tb01064.x. [DOI] [Google Scholar]
- 9.Van Oevelen D, L'Escaille F, Verachtert H. 1976. Synthesis of aroma components during the spontaneous fermentation of lambic and gueuze. J Inst Brew 82:322–326. doi: 10.1002/j.2050-0416.1975.tb06953.x. [DOI] [Google Scholar]
- 10.Verachtert H, Dawoud E. 1984. Microbiology of lambic-type beers. J Appl Bacteriol 57:R11–R12. [Google Scholar]
- 11.Verachtert H, Dawoud E, Shanta Kumara HMC. 1989. Interactions between Enterobacteriaceae and Saccharomyces cerevisiae during wort fermentation. Yeast 5:67–72. [Google Scholar]
- 12.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl Environ Microbiol 79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spitaels F, Wieme AD, Janssens M, Aerts M, Daniel H-M, Van Landschoot A, De Vuyst L, Vandamme P. 2014. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS One 9:e95384. doi: 10.1371/journal.pone.0095384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitaels F, Wieme AD, Janssens M, Aerts M, Van Landschoot A, De Vuyst L, Vandamme P. 2015. The microbial diversity of an industrially produced lambic beer shares members of a traditionally produced one and reveals a core microbiota for lambic beer fermentation. Food Microbiol 49:23–32. doi: 10.1016/j.fm.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Angelakis E, Million M, Henry M, Raoult D. 2011. Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J Food Sci 76:M568–M572. doi: 10.1111/j.1750-3841.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 16.De Bruyne K, Slabbinck B, Waegeman W, Vauterin P, De Baets B, Vandamme P. 2011. Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst Appl Microbiol 34:20–29. doi: 10.1016/j.syapm.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Doan NT, Van Hoorde K, Cnockaert M, De Brandt E, Aerts M, Le Thanh B, Vandamme P. 2012. Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in northern Vietnam. Lett Appl Microbiol 55:265–273. doi: 10.1111/j.1472-765X.2012.03287.x. [DOI] [PubMed] [Google Scholar]
- 18.Dušková M, Šedo O, Kšicová K, Zdráhal Z, Karpíšková R. 2012. Identification of lactobacilli isolated from food by genotypic methods and MALDI-TOF MS. Int J Food Microbiol 159:107–114. doi: 10.1016/j.ijfoodmicro.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 19.Kuda T, Izawa Y, Yoshida S, Koyanagi T, Takahashi H, Kimura B. 2014. Rapid identification of Tetragenococcus halophilus and Tetragenococcus muriaticus, important species in the production of salted and fermented foods, by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Food Control 35:419–425. doi: 10.1016/j.foodcont.2013.07.039. [DOI] [Google Scholar]
- 20.Sedo O, Vavrova A, Vad'urova M, Tvrzova L, Zdrahal Z. 2013. The influence of growth conditions on strain differentiation within the Lactobacillus acidophilus group using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry profiling. Rapid Commun Mass Spectrom 27:2729–2736. doi: 10.1002/rcm.6741. [DOI] [PubMed] [Google Scholar]
- 21.Snauwaert I, Papalexandratou Z, De Vuyst L, Vandamme P. 2013. Characterization of strains of Weissella fabalis sp. nov. and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int J Syst Evol Microbiol 63:1709–1716. doi: 10.1099/ijs.0.040311-0. [DOI] [PubMed] [Google Scholar]
- 22.Zeller-Péronnet V, Brockmann E, Pavlovic M, Timke M, Busch U, Huber I. 2013. Potential and limitations of MALDI-TOF MS for discrimination within the species Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides. J Verbrauch Lebensm 8:205–214. [Google Scholar]
- 23.da Cruz Pedrozo Miguel MG, de Castro Reis LV, Efraim P, Santos C, Lima N, Freitas Schwan NR. 2017. Cocoa fermentation: microbial identification by MALDI-TOF MS, and sensory evaluation of produced chocolate. Lebenson Wiss Technol 77:362–369. doi: 10.1016/j.lwt.2016.11.076. [DOI] [Google Scholar]
- 24.Usbeck JC, Kern CC, Vogel RF, Behr J. 2013. Optimization of experimental and modelling parameters for the differentiation of beverage spoiling yeasts by matrix-assisted-laser-desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) in response to varying growth conditions. Food Microbiol 36:379–387. doi: 10.1016/j.fm.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo JA, Miranda P, Flores-Felix JD, Sanchez-Juanes F, Ageitos JM, Gonzalez-Buitrago JM, Velazquez E, Villa TG. 2013. Atypical yeasts identified as Saccharomyces cerevisiae by MALDI-TOF MS and gene sequencing are the main responsible of fermentation of chicha, a traditional beverage from Peru. Syst Appl Microbiol 36:560–564. doi: 10.1016/j.syapm.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Andres-Barrao C, Benagli C, Chappuis M, Ortega Perez R, Tonolla M, Barja F. 2013. Rapid identification of acetic acid bacteria using MALDI-TOF mass spectrometry fingerprinting. Syst Appl Microbiol 36:75–81. doi: 10.1016/j.syapm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Wieme AD, Spitaels F, Aerts M, De Bruyne K, Van Landschoot A, Vandamme P. 2014. Effects of growth medium on matrix-assisted laser desorption-ionization time of flight mass spectra: a case study of acetic acid bacteria. Appl Environ Microbiol 80:1528–1538. doi: 10.1128/AEM.03708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Cleenwerck I, De Vuyst L, Vandamme P. 2017. Identification of acetic acid bacteria through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and report of Gluconobacter nephelii Kommanee et al. 2011 and Gluconobacter uchimurae Tanasupawat et al. 2012 as later heterotypic synonyms of Gluconobacter japonicus Malimas et al. 2009 and Gluconobacter oxydans (Henneberg 1897) De Ley 1961 (approved lists 1980) emend. Gosselé et al. 1983, respectively. Syst Appl Microbiol 40:123–134. doi: 10.1016/j.syapm.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Spitaels F, Li L, Wieme A, Balzarini T, Cleenwerck I, Van Landschoot A, De Vuyst L, Vandamme P. 2014. Acetobacter lambici sp. nov., isolated from fermenting lambic beer. Int J Syst Evol Microbiol 64:1083–1089. doi: 10.1099/ijs.0.057315-0. [DOI] [PubMed] [Google Scholar]
- 30.Spitaels F, Wieme A, Balzarini T, Cleenwerck I, Van Landschoot A, De Vuyst L, Vandamme P. 2014. Gluconobacter cerevisiae sp. nov., isolated from the brewery environment. Int J Syst Evol Microbiol 64:1134–1141. doi: 10.1099/ijs.0.059311-0. [DOI] [PubMed] [Google Scholar]
- 31.De Vuyst L, Camu N, De Winter T, Vandemeulebroecke K, Van de Perre V, Vancanneyt M, De Vos P, Cleenwerck I. 2008. Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int J Food Microbiol 125:79–90. doi: 10.1016/j.ijfoodmicro.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 32.Papalexandratou Z, Cleenwerck I, De Vos P, De Vuyst L. 2009. (GTG)5-PCR reference framework for acetic acid bacteria. FEMS Microbiol Lett 301:44–49. doi: 10.1111/j.1574-6968.2009.01792.x. [DOI] [PubMed] [Google Scholar]
- 33.Camu N, Gonzalez A, De Winter T, Van Schoor A, De Bruyne K, Vandamme P, Takrama JS, Addo SK, De Vuyst L. 2008. Influence of turning and environmental contamination on the dynamics of populations of lactic acid and acetic acid bacteria involved in spontaneous cocoa bean heap fermentation in Ghana. Appl Environ Microbiol 74:86–98. doi: 10.1128/AEM.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papalexandratou Z, Lefeber T, Bahrim B, Lee OS, Daniel HM, De Vuyst L. 2013. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol 35:73–85. doi: 10.1016/j.fm.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Morneau AD, Zuehlke JM, Edwards CG. 2011. Comparison of media formulations used to selectively cultivate Dekkera/Brettanomyces. Lett Appl Microbiol 53:460–465. doi: 10.1111/j.1472-765X.2011.03133.x. [DOI] [PubMed] [Google Scholar]
- 36.Bartowsky EJ, Henschke PA. 2008. Acetic acid bacteria spoilage of bottled red wine: a review. Int J Food Microbiol 125:60–70. doi: 10.1016/j.ijfoodmicro.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Papalexandratou Z, Camu N, Falony G, De Vuyst L. 2011. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol 28:964–973. doi: 10.1016/j.fm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Papalexandratou Z, Vrancken G, De Bruyne K, Vandamme P, De Vuyst L. 2011. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol 28:1326–1338. doi: 10.1016/j.fm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 39.De Rosso M, Cancian D, Panighel A, Dalla Vedova A, Flamini R. 2009. Chemical compounds released from five different woods used to make barrels for aging wines and spirits: volatile compounds and polyphenols. Wood Sci Technol 43:375–385. doi: 10.1007/s00226-008-0211-8. [DOI] [Google Scholar]
- 40.Lisdiyanti P, Kawasaki H, Seki T, Yamada Y, Uchimura T, Komagata K. 2001. Identification of Acetobacter strains isolated from Indonesian sources, and proposals of Acetobacter syzygii sp. nov., Acetobacter cibinongensis sp. nov., and Acetobacter orientalis sp. nov. J Gen Appl Microbiol 47:119–131. [DOI] [PubMed] [Google Scholar]
- 41.Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat Biotechnol 23:195–200. doi: 10.1038/nbt1062. [DOI] [PubMed] [Google Scholar]
- 42.Kashima Y, Iijima M, Okamoto A, Koizumi Y, Udaka S, Yanagida F. 1998. Purification and characterization of intracellular esterases related to ethyl acetate formation in Acetobacter pasteurianus. J Ferment Bioeng 85:584–588. doi: 10.1016/S0922-338X(98)80009-3. [DOI] [Google Scholar]
- 43.Kashima Y, Iijima M, Okamoto A, Nakano T, Tayama K, Koizumi Y, Udaka S, Yanagida F. 2000. Role of intracellular esterases in the production of esters by Acetobacter pasteurianus. J Biosci Bioeng 89:81–83. doi: 10.1016/S1389-1723(00)88055-X. [DOI] [PubMed] [Google Scholar]
- 44.Adler P, Frey LJ, Berger A, Bolten CJ, Hansen CE, Wittmann C. 2014. The key to acetate: metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation simulating conditions. Appl Environ Microbiol 80:4702–4716. doi: 10.1128/AEM.01048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens F, Lefeber T, De Vuyst L. 2014. Oxidation of metabolites highlights the microbial interactions and role of Acetobacter pasteurianus during cocoa bean fermentation. Appl Environ Microbiol 80:1848–1857. doi: 10.1128/AEM.03344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheffers WA. 1961. On the inhibition of alcoholic fermentation of Brettanomyces yeasts under anaerobic conditions. Experientia 17:40–42. doi: 10.1007/BF02157944. [DOI] [PubMed] [Google Scholar]
- 47.Steensels J, Daenen L, Malcorps P, Derdelinckx G, Verachtert H, Verstrepen KJ. 2015. Brettanomyces yeasts–from spoilage organisms to valuable contributors to industrial fermentations. Int J Food Microbiol 206:24–38. doi: 10.1016/j.ijfoodmicro.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Blomqvist J, Eberhard T, Schnurer J, Passoth V. 2010. Fermentation characteristics of Dekkera bruxellensis strains. Appl Microbiol Biotechnol 87:1487–1497. doi: 10.1007/s00253-010-2619-y. [DOI] [PubMed] [Google Scholar]
- 49.Wieme A, Cleenwerck I, Van Landschoot A, Vandamme P. 2012. Pediococcus lolii DSM 19927T and JCM 15055T are strains of Pediococcus acidilactici. Int J Syst Evol Microbiol 62:3105–3108. doi: 10.1099/ijs.0.046201-0. [DOI] [PubMed] [Google Scholar]
- 50.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 52.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Bruyn F, Zhang S, Pothakos V, Torres J, Lambot C, Moroni AV, Callanan M, Sybesma W, Weckx S, De Vuyst L. 2017. Exploring the impact of post-harvest processing on the microbiota and metabolite profiles during a case of green coffee bean production. Appl Environ Microbiol 83:e02398-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Meulen R, Scheirlinck I, Van Schoor A, Huys G, Vancanneyt M, Vandamme P, De Vuyst L. 2007. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl Environ Microbiol 73:4741–4750. doi: 10.1128/AEM.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]