ABSTRACT
Robust genetic systems for the hyperthermophilic Thermococcales have facilitated the overexpression of native genes, enabled the addition of sequences encoding secretion signals, epitope, and affinity tags to coding regions, and aided the introduction of sequences encoding new proteins in these fast-growing fermentative heterotrophs. However, tightly controlled and easily manipulated systems facilitating regulated gene expression are limited for these hosts. Here, we describe an alternative method for regulatory control reliant on a cis-encoded functional riboswitch in the model archaeon Thermococcus kodakarensis. Despite the hyperthermophilic growth temperatures, the proposed structure of the riboswitch conforms to a fluoride-responsive riboswitch encoded in many bacteria and similarly functions to regulate a component-conserved fluoride export pathway. Deleting components of the fluoride export pathway generates T. kodakarensis strains with increased fluoride sensitivity. The mechanism underlying regulated expression suggested that the riboswitch-encoding sequences could be utilized as a tunable expression cassette. When appended to a reporter gene, the riboswitch-mediated control system provides fluoride-dependent tunable regulatory potential, offering an alternative system for regulating gene expression. Riboswitch-regulated expression is thus ubiquitous in extant life and can be exploited to generate regulated expression systems for hyperthermophiles.
IMPORTANCE Gene expression is controlled by a myriad of interconnected mechanisms that interpret metabolic states and environmental cues to balance cell physiology. Transcription regulation in Archaea is known to employ both typical repressors-operators and transcription activators to regulate transcription initiation in addition to the regulation afforded by chromatin structure. It was perhaps surprising that the presumed ancient mechanism of riboswitch-mediated regulation is found in Bacteria and Eukarya, but seemingly absent in Archaea. We demonstrate here that a fluoride-responsive riboswitch functions to regulate a detoxification pathway in the hyperthermophilic archaeon Thermococcus kodakarensis. The results obtained define a universal role for riboswitch-mediated regulation, adumbrate the presence of several riboswitch-regulated genes in Thermococcus kodakarensis, demonstrate the utility of RNA-based regulation at high temperatures, and provide a novel riboswitch-regulated expression system to employ in hyperthermophiles.
KEYWORDS: Archaea, riboswitch, Thermococcus, fluoride, hyperthermophile, gene expression
INTRODUCTION
Regulated transcription is the dominant mechanism used to control gene expression and rapidly respond to stress or changing environmental conditions. Many mechanisms to regulate the activity of RNA polymerase (RNAP) have been described and some themes—using DNA binding proteins to promote or block RNAP binding to promoter sequences, for example—are employed in extant life (1–3). Although protein-based regulation appears to dominate, RNA-based regulatory systems are present. Well-supported arguments claim that RNA-based riboswitch-mediated regulatory strategies evolved in early cells (4–8).
Riboswitches are regulatory elements within transcripts that differentially fold to affect the expression of the encoding transcript. Riboswitches fold immediately upon emerging from RNAP and are typically responsive to a small regulatory molecule(s) which favors a mutually exclusive RNA structure from the apo structure of the transcript (4–6, 8–10). In Bacteria, differential folding of the RNA can form structures that enable continued transcription elongation or mediate intrinsic transcription termination (5, 6, 8). Riboswitches can also promote or limit the access to sequences necessary for initiation of translation, and the absence of translation can also lead to premature factor-dependent transcription termination (5, 8, 11, 12). Riboswitches, both native and synthetic sequences—sometimes with quite intricate regulatory strategies—have been utilized in the construction of expression cassettes which provide tight and dynamic control of in vivo protein levels (12–14, 16–18).
Many riboswitches have been described in Bacteria and Eukarya, but experimental evidence for bona fide riboswitch-mediated regulation in Archaea has not previously been reported (5, 8, 9, 19). Although archaeal riboswitches have been bioinformatically predicted, their potential utility to control transcription termination—a common mechanism used by bacterial riboswitches—was compromised by the absence of a requirement for RNA structures to mediate archaeal intrinsic transcription termination (20, 21). The optimal growth temperatures of some (hyper)thermophilic archaeal species was used to justify concerns that RNA-mediated regulation may be difficult to control at elevated temperatures. The lack of experimental evidence for riboswitch-mediated regulation in mesophilic species was still difficult to comprehend, but certainly, the artificial introduction of a bacterium-derived riboswitch into a methanogen confirmed that riboswitch-mediated regulation is possible in Archaea (13).
We characterize here an RNA sequence that functions as a fluoride-responsive riboswitch (FRR) in the model hyperthermophile Thermococcus kodakarensis. The sequence and predicted structural elements are conserved with similar riboswitches that control fluoride-regulated regulons in Bacteria (9). The deletion of genes under FRR control results in hypersensitivity to fluoride, and bioinformatic analyses predict these genes encode components of a fluoride export pump, conserved in all domains of life (19, 22, 23). Mutations that disrupt the predicted riboswitch structure and capacity to bind fluoride also disrupt fluoride-regulated expression in vivo (9, 24, 25). The alternative structures likely formed by the FRR suggest a control of translation initiation by blocking access to Shine-Dalgarno sequences. This simple mechanism suggested that a riboswitch-encoding sequence could be transposed to regulate any gene of interest. We validate this hypothesis by constructing a cassette containing the T. kodakarensis FRR (Tk-FRR) and experimentally demonstrate fluoride-responsive expression of a reporter gene in vivo.
RESULTS
Identification of a fluoride-responsive riboswitch in T. kodakarensis.
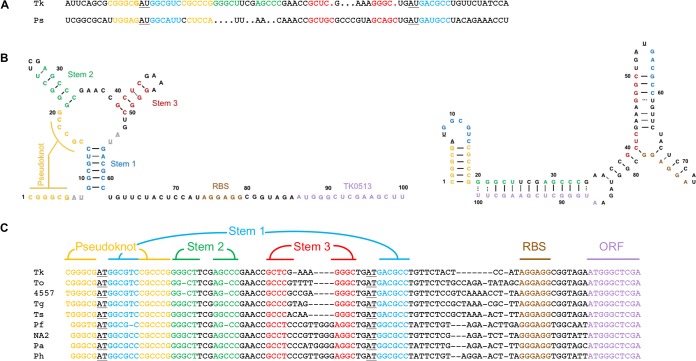
RNA sequences that can directly bind and differentially fold in response to fluoride have been characterized in Bacteria (9, 24–28). Conserved secondary structural elements and sequence homology suggest that the leader sequence of gene 0513 in T. kodakarensis (TK0513) contains a fluoride-responsive riboswitch (FRR) (Fig. 1A) (9, 26, 29, 30). This potential FRR not only shares the conserved structural and sequence elements, but is located adjacent to TK0514, a putative crcB homologue and predicted fluoride export pump (9, 22, 30). The T. kodakarensis FRR (Tk-FRR) retains sequences capable of pseudoknot formation as well as conserved sequences known to bind fluoride in the bacterial FRRs. In addition to conserved stem 1 and stem 3 sequences, the Tk-FRR contains complementary sequences that form stem 2. If stem 2 sequences are not paired, these sequences are entirely complementary to the translation initiation region of TK0513, and modeling with the mFOLD server (31) suggests that both the ribosome binding site (RBS) and first five codons of TK0513 would be paired in the folded structure (Fig. 1B). Comparisons of Thermococcales genome sequences show a near-universal conservation of the pseudoknot, stem 1, 2, and 3 sequences, and the initially translated regions of the adjacent downstream open reading frame (Fig. 1C) (32, 33). Furthermore, the Thermococcales FRR sequences are all upstream of but adjacent to genes encoding CrcB homologues.
FIG 1.
Thermococcales encode a fluoride-responsive riboswitch. (A) CLUSTAL W (1.83) sequence alignment of the Tk-FRR and Pseudomonas syringae FRR. Sequences are color-coded to match structural elements in adjacent panels, and underlined residues are known to be essential for coordinating fluoride in the folded RNA structure (9, 24, 25). (B) Predicted apo-bound (right) and fluoride-bound (left) structures of the Tk-FRR, highlighting the pairings of conserved structural elements. Predicted structures were determined with the mFold 2.0 in silico folding tool with default settings at 85°C (31). (C) FRR sequences and structural elements are highly conserved in Thermococcales. Tk, Thermococcus kodakarensis; To, Thermococcus onnurineus; 4557, Thermococcus sp. strain 4557; Ts, Thermococcus sibiricus; Pf, Pyrococcus furiosus; NA2, Pyrococcus sp. strain NA2; Pa, Pyrococcus abyssi; Ph, Pyrococcus horikoshii. Sequences were retrieved from the UCSC archaeal genome browser (32, 33).
Deletion of FRR-adjacent genes results in fluoride-sensitive T. kodakarensis strains.
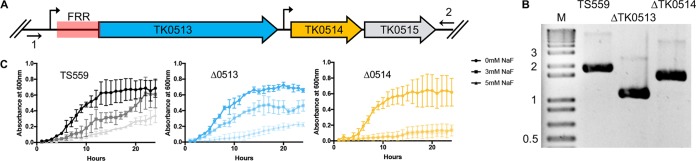
The evolutionary retention of putative FRR sequences suggested a simple model wherein exposure to fluoride would likely derepress or induce the expression of genes encoding fluoride detoxification machinery. The FRR is not immediately linked to the putative fluoride export pump-encoding gene in any Thermococcales genome (TK0514 in T. kodakarensis) but is separated by a conserved, yet hypothetical, long open reading frame (TK0513 in T. kodakarensis) (Fig. 2A). Deep sequencing of total RNAs and 5′ end mapping to identify transcription start sites (TSSs) do validate a TSS upstream of TK0513 that encodes the FRR and TK0513, but also another TSS between TK0513 and TK0514 (34). It is possible that the protein products of both TK0513 and TK0514 contribute to fluoride detoxification mechanisms, and to experimentally challenge this hypothesis, we determined the sensitivity of T. kodakarensis strains lacking or retaining TK0513 and TK0514 to fluoride addition. Using established procedures (35–37), we performed a markerless deletion of the sequences encoding either TK0513 (to the exclusion of sequences encoding the FRR) or TK0514 from the genome of strain TS559 (Fig. 2B). T. kodakarensis TS559 grows rapidly to an optical density of ∼1.0 in the absence of NaF, grows more slowly but to the same density when exposed to 3 mM NaF, and remains viable but grows linearly and slowly in the presence of 5 mM NaF (Fig. 2C). The deletion of either TK0513 or TK0514 did not affect growth rate or the final optical densities of cultures grown in the absence of NaF, but the deletion of either locus did limit growth in the presence of NaF. The growth of cells deleted for TK0514 was nearly and completely inhibited by the addition of 3 and 5 mM NaF, respectively. The deletion of TK0513 resulted in a reproducible but weaker sensitivity, with the addition of 3 mM NaF resulting in a modest decrease in final cell density and the addition of 5 mM NaF resulting in very slow and limited growth, even after 24 h (Fig. 2C).
FIG 2.
Deletion of genes regulated by the FRR confers fluoride sensitivity. (A) Schematic of the genetic organization of TK-FRR, TK0513, and TK0514; up- and downstream PCR primers and transcription start sites are indicated. (B) Diagnostic PCR-generated amplicons confirm markerless deletion of either TK0513 or TK0514. (C) Addition of NaF differentially slows and hinders growth of T. kodakarensis strains TS559, ΔTK0513, and ΔTK0514.
Tk-FRR is modular and can be exploited to provide F-dependent expression of a reporter gene.
Bacterial FRRs can be fused with reporter genes to provide fluoride-responsive regulation in vivo (6, 9, 27). We fused the Tk-FRR, in wild-type (WT) or mutationally altered forms, to TK1761, a reporter gene encoding a previously characterized β-glycosidase (Fig. 3A) (21, 36, 38, 39). The fusions were integrated into the T. kodakarensis TS559 genome at the normal TK1761 locus, and expression was driven by the constitutive heterologous hmtB promoter (39). β-Glycosidase activity assays were used to quantify reporter gene activity from parental (TS559) and WT FRR-TK1761 fusion-containing (TS800) strains in response to the addition of fluoride (Fig. 3B). T. kodakarensis encodes two β-glycosidases, both of which are nonessential and both of which have previously been characterized as reporter genes (36, 39). The collective activity of both β-glycosidases was not significantly influenced by the addition of fluoride in strain TS559. Replacing the native TK1761 promoter with the heterologous hmtB promoter was previously shown to increase β-glycosidase activity ∼5-fold (21, 39). Despite an identical promoter replacement in strain TS800, only a modest (∼10%) increase in β-glycosidase activity was observed in the absence of fluoride (Fig. 3B). The minor increase in β-glycosidase activity in TS800 suggests that in the absence of fluoride, the FRR effectively represses the known increased expression of TK1761 due to the promoter replacement. However, the addition of NaF to cultures of TS800 led to a dose-dependent increase in β-glycosidase activity (Fig. 3B). The ∼5-fold increase in β-glycosidase activity seen at 5 mM NaF matches the ∼5-fold change in β-glycosidase activity previously demonstrated by the promoter change alone, suggesting that the FRR no longer hinders expression at elevated fluoride levels.
FIG 3.
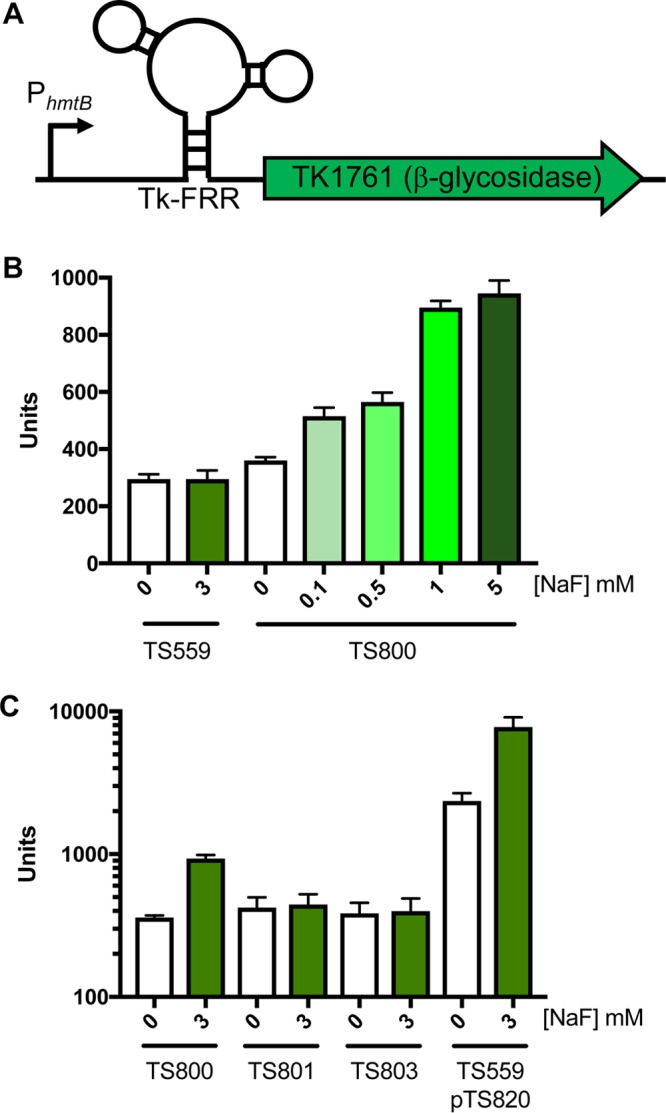
Tk-FRR cassette regulates expression of a reporter gene. (A) Diagram of the WT Tk-FRR expression cassette, consisting of a strong constitutive promoter PhmtB upstream of the Tk-FRR followed by an in-frame β-glycosidase (TK1761). (B) β-Glycosidase activity increases in TS800 cells exposed to increasing levels of NaF. TS800 contains the Tk-FRR cassette chromosomally integrated. (C) Tk-FRR-dependent β-glycosidase activity is reduced in TS801 (contains a mutation in the P2 stem) and TS803 (contains mutations in the fluoride binding pocket). TS559(pTS820) contains an autonomously replicating plasmid encoding the same expression cassette as TS800.
Site-directed mutagenesis of the Tk-FRR enabled a clarification of the roles different sequences and structures play in FRR-activity. Replacing a native C-G (nucleotide [nt] positions 11 and 12) sequence within the WT Tk-FRR stem 1 with a G-C sequence retains the overall GC content but was predicted to disrupt the ability of stem P1 to pair and thus the ability of the FRR to adopt a fluoride-bound structure (9, 24, 25). When this altered FRR-TK1761 fusion is integrated into the TS559 chromosome, resulting in T. kodakarensis strain TS801, the F responsiveness of the reported fusion is abolished and β-glycosidase levels return to baseline levels (Fig. 3C). A FRR-TK1761 fusion integrated into the TS559 genome with a substitution of a single U (nt position 54) between stem 3 and stem 1 with an A resulted in T. kodakarensis strain TS803. This residue had been previously established to be critical for fluoride binding in bacterial FRRs (9, 24, 25), and the substitution abolished fluoride-responsive β-glycosidase activity in the Tk-FRR as well.
T. kodakarensis can harbor autonomous expression vectors (37, 38, 40). Plasmids are maintained at ∼3-fold greater levels than chromosomes (38), and T. kodakarensis is oligoploid (∼7 to 19 genomes) (41). Gene dosage likely accounts for the increase in β-glycosidase activity observed when the Tk-FRR-TK1761 fusion reporter was introduced to strain TS559 with pTS820 (Fig. 3C). Fluoride-dependent increases in β-glycosidase activity were also noted when the fusion was episomally rather than chromosomally located.
The predicted apo- and fluoride-bound structures imply that regulation is imposed by limiting the access of the translation apparatus to the Shine-Dalgarno and initial translated sequences. The conservation of complementary sequences in stem 2 and the initial translated sequences of TK0513 suggested biological importance, but reporter fusions that encoded Tk-FRR sequences that contained or lacked the first five codons of TK0513 behaved nearly identically (data not shown), implying that sequestering the Shine-Dalgarno sequence alone was sufficient for FRR function in vivo.
DISCUSSION
The presence and regulatory potential of riboswitches in archaea had been predicted by in silico analyses (26, 42–46), and a bacterial riboswitch was introduced to offer regulatory potential to a methanogen (13), but no direct evidence for the retention and utility of archaeal riboswitches has been previously provided. We demonstrate here that T. kodakarensis employs a fluoride-responsive riboswitch (FRR) to regulate the expression of genes involved in fluoride detoxification. The deletion of TK0514 results in hypersensitivity to fluoride, supportive of the product of TK0514 (CrcB) functioning as a fluoride export pump. The deletion of TK0513 had a modest but reproducible fluoride-sensitive phenotype, implicating the protein product of TK0513 in fluoride detoxification. Our mutational analysis demonstrates that conserved RNA structures and sequences regulate expression in vivo, and the near-complete conservation of the FRR sequence in Thermococcales supports the use of riboswitch-mediated gene regulation in this clade (32, 33). Modeling the Tk-FRR structures suggested that the FRR would fold independently and that the FRR would be suitable to control the expression of any gene of interest. As a proof of concept, we fused the FRR-encoding sequences between a constitutive promoter and a reporter gene (TK1761) and monitored expression levels by measuring TK1761-encoded β-glycosidase activity. Fluoride-responsive increases in reporter gene activity were observed from both chromosomal and ectopic expressions.
Members of the order Thermococcales are commonly found in marine hydrothermal environments and are also regularly observed in cold oxygenated seawater, likely as a transient population searching out new hydrothermal environments to colonize (47–49). As such, the ∼50 μM [F−] in seawater (50) is likely tolerated by constitutive expression of a likely fluoride export pump from a promoter immediately upstream of TK0514 (34). The FRR is encoded in the upstream TK0513 regulatory unit, and our genetic results support a role for the protein product of TK0513 in supporting fluoride detoxification mechanisms. It is possible that the protein product of TK0513, a putative prenyltransferase, works with or modifies the activity of CrcB, or that translation of the TK0513-encoding transcript itself is sufficient to enable readthrough transcription from TK0513 into TK0514 to increase the expression of TK0514 in the presence of fluoride. Additional experimentation will be necessary to describe the full regulatory network associated with fluoride detoxification mechanisms in T. kodakarensis.
The addition of the Tk-FRR-encoding sequences immediately downstream of a known strong promoter limits the expression of the downstream genes in the absence of additional fluoride in the medium. A dose-dependent transition to an “on-state” is observed when fluoride is added, with maximal output matching that observed from the same promoter in the absence of an FRR (39) when fluoride is provided at high levels. The modular Tk-FRR thus provides a novel tunable regulatory module for use in hyperthermophiles. The inducer is inexpensive and abundant, and a significant level of expression can be stimulated. The dynamic expression range provided by FRR regulation will likely find utility in balancing and tuning synthetic platforms in Thermococcales. Given the absence of any required protein components, it is also likely that this FRR will function in any heterologous host and may be particularly well positioned to aid in the control of thermophilic hosts. This description of a functional archaeal riboswitch, coupled with the bioinformatics identification of many putative riboswitches in archaea (6, 26, 34), demonstrates that riboswitch-mediated regulation is ubiquitous in all extant life.
MATERIALS AND METHODS
Reagents and enzymes.
All chemicals were purchased from Sigma-Aldrich Corporation (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA). Restriction endonucleases and DNA polymerases were purchased from New England BioLabs (Ipswich, MA). DNA purification kits were purchased from Zymo Research (Orange, CA).
Strain construction and measurements of culture growth.
Strains and plasmids are detailed in Table 1. Deletion strains were constructed as described previously (35, 36). T. kodakarensis strains were maintained in rich medium (artificial seawater [ASW]-yeast extract-tryptone [YT]-elemental sulfur [S0]-pyruvate), supplemented with agmatine (51). Strains carrying the Tk-FRR cassette either chromosomally integrated (TS800, TS801, and TS803) or on autonomously replicating plasmids [TS559(pTS820)] were maintained in rich medium supplemented with agmatine and lovastatin (38). Using the structures of the Thermotoga petrophila (24) and Pseudomonas syringae (9) FRRs as guides, we identified critical positions within the Tk-FRR which, when mutated, abrogate FRR functionality; the respective mutations are outlined in Table 1 and in Fig. S2 in the supplemental material. Culture densities were monitored by changes in optical density at 600 nm (OD600) using a Genesys 20 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Where indicated, sodium fluoride was added to the medium at the time of inoculation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype(s) and/or relevant feature(s) | Reference or source |
|---|---|---|
| Strains | ||
| TS559 | ΔpyrF, ΔtrpE::pyrF, ΔTK0664, ΔTK0149 | 36 |
| ΔTK0513 | TS559, ΔTK0513 | This work (Fig. 2) |
| ΔTK0514 | TS559, ΔTK0514 | This work (Fig. 2) |
| TS800 | TS559, WT Tk-FRR cassette chromosomally integrated (PhmtB-Tk-FRR-TK1761) | This work (Fig. 3) |
| TS801 | TS559, mutant Tk-FRR cassette chromosomally integrated (nt 11, 12; CG→GC), disrupts P1 stem | This work (Fig. 3) |
| TS803 | TS559, mutant Tk-FRR cassette chromosomally integrated (nt 54; T→A), disrupts fluoride binding pocket | This work (Fig. 3) |
| Plasmids | ||
| pLC70 | Autonomously replicating Thermococcus plasmid | 38 |
| pTS820 | pLC70 containing the WT Tk-FRR cassette (PhmtB-Tk-FRR-TK1761) | This work (Fig. 3) |
β-Glycosidase expression reporter assays.
Exponentially growing cultures were used to inoculate medium supplemented with NaF, and cells from 10-ml aliquots were harvested by centrifugation at an OD600 of 0.2. Cells were lysed via successive freeze-thaw cycles in 0.2 ml of 10 mM Tris-HCl (pH 8). Cellular debris was removed by centrifugation (>16,000 × g for 15 min), and the protein concentrations of the resulting soluble fractions were determined by Bradford assays (52). The β-glycosidase activity present in each lysate was determined by monitoring the change in absorbance at 405 nm (A405) during incubation at 85°C after the addition of 2.8 mM ortho-nitrophenyl-β-galactoside (ONPG) dissolved in 50 mM sodium phosphate (pH 6.5). As described previously (21, 36, 38, 39), and to provide comparative values, one unit of activity was defined as catalyzing a ΔA405 of 1 in 1 min/pg of protein. The results reported are the averages, with standard errors, from minimally triplicate biological replicates with triplicate technical replicates.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the NIH (GM 100329) and DOE (004010-00002) to T.J.S. M.C.S. was supported in part by a merit scholarship from Colorado State University.
We thank members of the lab for constructive comments and help with strain construction.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02306-17.
REFERENCES
- 1.Gehring AM, Walker JE, Santangelo TJ. 2016. Transcription regulation in Archaea. J Bacteriol 198:1906–1917. doi: 10.1128/JB.00255-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peeters E, Charlier D. 2010. The Lrp family of transcription regulators in Archaea. Archaea 2010:750457. doi: 10.1155/2010/750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouhammouch M, Werner F, Weinzierl ROJ, Geiduschek EP. 2004. A fully recombinant system for activator-dependent archaeal transcription. J Biol Chem 279:51719–51721. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 4.Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherwood AV, Henkin TM. 2016. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70:361–374. doi: 10.1146/annurev-micro-091014-104306. [DOI] [PubMed] [Google Scholar]
- 6.Nelson JW, Breaker RR. 2017. The lost language of the RNA world. Sci Signal 10:eaam8812. doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallberg ZF, Su Y, Kitto RZ, Hammond MC. 2017. Engineering and in vivo applications of riboswitches. Annu Rev Biochem 86:515–539. doi: 10.1146/annurev-biochem-060815-014628. [DOI] [PubMed] [Google Scholar]
- 8.Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 9.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. 2012. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren GX, Guo XP, Sun YC. 2017. Regulatory 3′ untranslated regions of bacterial mRNAs. Front Microbiol 8:1276. doi: 10.3389/fmicb.2017.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. 2012. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A 109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etzel M, Mörl M. 2017. Synthetic riboswitches: from plug and pray toward plug and play. Biochemistry 56:1181–1198. doi: 10.1021/acs.biochem.6b01218. [DOI] [PubMed] [Google Scholar]
- 13.Demolli S, Geist MM, Weigand JE, Matschiavelli N, Suess B, Rother M. 2014. Development of β-lactamase as a tool for monitoring conditional gene expression by a tetracycline-riboswitch in methanosarcina acetivorans. Archaea 2014:725610. doi: 10.1155/2014/725610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D-S, Gusti V, Pillai SG, Gaur RK. 2005. An artificial riboswitch for controlling pre-mRNA splicing. RNA 11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.McKeague M, Wong RS, Smolke CD. 2016. Opportunities in the design and application of RNA for gene expression control. Nucleic Acids Res 44:2987–2999. doi: 10.1093/nar/gkw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berens C, Suess B. 2015. Riboswitch engineering—making the all-important second and third steps. Curr Opin Biotechnol 31:10–15. doi: 10.1016/j.copbio.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Berens C, Groher F, Suess B. 2015. RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression. Biotechnol J 10:246–257. doi: 10.1002/biot.201300498. [DOI] [PubMed] [Google Scholar]
- 19.Wachter A. 2010. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol 7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 20.Santangelo TJ, Reeve JN. 2006. Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. J Mol Biol 355:196–210. doi: 10.1016/j.jmb.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 21.Santangelo TJ, Cubonová L, Skinner KM, Reeve JN. 2009. Archaeal intrinsic transcription termination in vivo. J Bacteriol 191:7102–7108. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA. 2013. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc Natl Acad Sci U S A 110:19018–19023. doi: 10.1073/pnas.1310439110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockbridge RB, Lim H-H, Otten R, Williams C, Shane T, Weinberg Z, Miller C. 2012. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc Natl Acad Sci U S A 109:15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren A, Rajashankar KR, Patel DJ. 2012. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Guffy SL, Williams B, Zhang Q. 2017. An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol 13:968–974. doi: 10.1038/nchembio.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson JW, Plummer MS, Blount KF, Ames TD, Breaker RR. 2015. Small molecule fluoride toxicity agonists. Chem Biol 22:527–534. doi: 10.1016/j.chembiol.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wedekind JE, Dutta D, Belashov IA, Jenkins JL. 2017. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J Biol Chem 292:9441–9450. doi: 10.1074/jbc.R117.787713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieft JS, Chase E, Costantino DA, Golden BL. 2010. Identification and characterization of anion binding sites in RNA. RNA 16:1118–1123. doi: 10.1261/rna.2072710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan PP, Holmes AD, Smith AM, Tran D, Lowe TM. 2012. The UCSC archaeal genome browser: 2012 update. Nucleic Acids Res 40:D646–D652. doi: 10.1093/nar/gkr990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider KL. 2006. The UCSC archaeal genome browser. Nucleic Acids Res 34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäger D, Förstner KU, Sharma CM, Santangelo TJ, Reeve JN. 2014. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genomics 15:684. doi: 10.1186/1471-2164-15-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hileman TH, Santangelo TJ. 2012. Genetics techniques for Thermococcus kodakarensis. Front Microbiol 3:195. doi: 10.3389/fmicb.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santangelo TJ, Cubonová L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol 76:1044–1052. doi: 10.1128/AEM.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farkas JA, Picking JW, Santangelo TJ. 2013. Genetic techniques for the Archaea. Annu Rev Genet 47:539–561. doi: 10.1146/annurev-genet-111212-133225. [DOI] [PubMed] [Google Scholar]
- 38.Santangelo TJ, Cubonová L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl Environ Microbiol 74:3099–3104. doi: 10.1128/AEM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santangelo TJ, Cubonová L, Matsumi R, Atomi H, Imanaka T, Reeve JN. 2008. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J Bacteriol 190:2244–2248. doi: 10.1128/JB.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takemasa R, Yokooji Y, Yamatsu A, Atomi H, Imanaka T. 2011. Thermococcus kodakarensis as a host for gene expression and protein secretion. Appl Environ Microbiol 77:2392–2398. doi: 10.1128/AEM.01005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaans SK, van der Oost J, Kengen SWM. 2015. The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles 19:741–750. doi: 10.1007/s00792-015-0750-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bengert P, Dandekar T. 2003. A software tool-box for analysis of regulatory RNA elements. Nucleic Acids Res 31:3441–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bengert P, Dandekar T. 2004. Riboswitch finder–a tool for identification of riboswitch RNAs. Nucleic Acids Res 32:W154–W159. doi: 10.1093/nar/gkh352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang T-H, Huang H-D, Wu L-C, Yeh C-T, Liu B-J, Horng J-T. 2009. Computational identification of riboswitches based on RNA conserved functional sequences and conformations. RNA 15:1426–1430. doi: 10.1261/rna.1623809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havill JT, Bhatiya C, Johnson SM, Sheets JD, Thompson JS. 2014. A new approach for detecting riboswitches in DNA sequences. Bioinformatics 30:3012–3019. doi: 10.1093/bioinformatics/btu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Retwitzer MD, Kifer I, Sengupta S, Yakhini Z, Barash D. 2015. An efficient minimum free energy structure-based search method for riboswitch identification based on inverse RNA folding. PLoS One 10:e0134262. doi: 10.1371/journal.pone.0134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poli A, Finore I, Romano I, Gioiello A, Lama L, Nicolaus B. 2017. Microbial diversity in extreme marine habitats and their biomolecules. Microorganisms 5:E25. doi: 10.3390/microorganisms5020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dillon JG, Zhang Y, Wang H, Jian H, Leng H, Xiao X. 2017. Microbial community structure of deep-sea hydrothermal vents on the ultraslow spreading southwest Indian ridge. Front Microbiol 8:1012. doi: 10.3389/fmicb.2017.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huber JA, Butterfield DA, Baross JA. 2006. Diversity and distribution of subseafloor Thermococcales populations in diffuse hydrothermal vents at an active deep-sea volcano in the northeast Pacific Ocean. J Geophys Res Biogeosci 111:G04016. doi: 10.1029/2005JG000097. [DOI] [Google Scholar]
- 50.Seyfried WE Jr, Ding K. 1995. The hydrothermal chemistry of fluoride in seawater. Geochim Cosmochim Acta 59:1063–1071. doi: 10.1016/0016-7037(95)00023-S. [DOI] [Google Scholar]
- 51.Čuboňová L, Katano M, Kanai T, Atomi H, Reeve JN, Santangelo TJ. 2012. An archaeal histone is required for transformation of Thermococcus kodakarensis. J Bacteriol 194:6864–6874. doi: 10.1128/JB.01523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.