Abstract
Animals, particularly poikilotherms, exhibit distinct physiologies at different environmental temperatures. Here, we hypothesized that temperature-based differences in physiology could affect the amount of variation in complex quantitative traits. Specifically, we examined, in Caenorhabditis elegans, how different temperatures (15°C, 20°C, and 25°C) affected the amount of interindividual variation in life span and also expression of three reporter genes—transcriptional reporters for vit-2, gpd-2, and hsp-16.2 (a life-span biomarker). We found the expected inverse relationship between temperature and average life span. Surprisingly, we found that at the highest temperature, there were fewer differences between individuals in life span and less interindividual variation in expression of all three reporters. We suggest that growth at 25°C might canalize (reduce interindividual differences in) life span and expression of some genes by eliciting a small constitutive heat shock response. Growth at 25°C requires wild-type hsf-1, which encodes the main heat shock response transcriptional activator. We speculate that increased chaperone activity at 25°C may reduce interindividual variation in gene expression by increasing protein folding efficiency. We hypothesize that reduced variation in gene expression may ultimately cause reduced variation in life span.
Keywords: Variation, Biomarker, Aging, Nongenetic, Proteostasis
Genetically identical organisms of the same age grown in the same environment are physiologically distinct. One particularly striking illustration of this is that life span in isogenic populations of Caenorhabditis elegans in the same environment varies by an order of magnitude (1). Interindividual differences in gene expression can be even greater (2) and these differences in early adult life can predict differences in later life span (3–6). These differences in gene expression and attendant differences in life span operationally define persistent physiological states. Here, we sought to understand how the environment, in this case environmental temperature, contributes to the manifestation of variation in complex quantitative traits.
For C elegans and other poikilotherms, temperature modulates physiology, and development and aging occur more rapidly at higher temperatures (7–10). These differences in rates of development and aging have sometimes been attributed to an increase in chemical reaction rates (8–10). However, molecular analysis reveals that the types and numbers of molecular species inside cells change with temperature. Such differences in molecular physiology at different temperatures have been observed in organisms ranging from bacteria (different membrane lipid compositions) (11) to snakes (different isoforms of enzymes) (12). Thus, while an increase in reaction rates might (via some series of steps) contribute to shortened life span at higher temperatures, it is also possible that differences in life span might be due to the existence of different compositions of molecular species at different temperatures. In any case, the molecular physiology of a cell, defined by the complement of biochemicals and enzymes present at a given temperature is not identical to the molecular physiology of that cell at a different temperature. This observation is reflected by the broad general observation that many organisms have temperature-specific genetic requirements.
In C elegans, life-span determinations are routinely conducted at temperatures between 15°C and 25°C, a fact which would seemingly make consideration of temperature-induced differences in physiology relevant to interpreting life-span results. In fact, distinct genes affect life span at different temperatures. For example, trpa-1, which encodes a TRP channel and sod-1, which encodes a superoxide dismutase, are required for wild-type life span at 15°C (7) or 16°C (13), respectively, are not required at higher temperatures. Similarly, loss of function mutations in hif-1, encoding hypoxia-induced transcription factor 1, extend life span but only at 25°C (14). Finally, animals bearing the hsf-1 (ok600) (a complete loss of function mutation in the master transcription factor controlling the heat shock response) do not live past larval stages at any growth temperature (15), whereas animals bearing hsf-1(sy441) (a partial loss of function mutation) raised at lower temperatures (below 25°C) grow to adulthood, produce progeny and age (16).
In this work, to study how temperature dependent differences in physiology affect quantitative traits, we measured the amount of interindividual variation in life span and gene expression in genetically identical C elegans. We grew animals at different temperatures. We determined individual animals’ life spans and variation in life span. We determined levels of expression for reporter genes relevant to reproduction (Pvit-2::gfp; vit-2 encodes a yolk protein, vitellogenin), metabolism (Pgpd-2::gfp; gpd-2 encodes a glycolytic enzyme, glyceraldehyde 3-phosphate dehydrogenase), and stress response/life-span prediction (Phsp-16.2::gfp; hsp-16.2 encodes a chaperone, a small heatshock protein of 16.2 kDa), and interindividual variation in these quantities. We found that variation in life span and variation in expression of genes (including the life-span biomarker [A life-span biomarker is a trait one can measure in an individual that predicts subsequent life span better than chronological age (17). In C elegans, there are a number of reporter genes, including the stress response inducible reporter Phsp-16.2::gfp, whose expression defines such life-span biomarkers (3,4,5,6).]) was reduced (canalized) (18) at elevated temperature (25°C). This interindividual variation was associated with increased chaperone pathway activity, which we speculate may cause the reduction in variation.
Results
To determine how temperature affects the amount of physiological variation among genetically identical C elegans in homogeneous environments, we measured expression of reporter genes and life spans in individual animals. We measured reporter gene expression in animals grown at 16°C, 20°C, and 25°C using methods we developed that allow high technical reproducibility (2). In these experiments, we measured more than 500 animals with each reporter at each temperature in five separate trials, analyzing over 25,000 individuals (see Table 1). We measured life span for animals grown and aged at 15°C, 20°C, and 25°C; to do so, we conducted 50 individual life-span trials surveying more than 4,500 animals (see Table 2). We detail experimental methods in Supplementary Materials.
Table 1.
Average Reporter Gene Expression Level and Average CV for Expression at Different Temperatures
| Reportera | 16°C | 20°C | 25°C |
|---|---|---|---|
| Constitutive Phsp-16.2::gfp | 0 | 0 | 8.9 (0.218) |
| Induced Phsp-16.2::gfp | 23.0 (0.210) | 22.0 (0.205) | 21.7 (0.185) |
| Constitutive Pvit-2::gfp | 97 (0.268) | 133 (0.199) | 127 (0.179) |
| Constitutive Pgpd-2::gfp | 309 (0.224) | 513 (0.235) | 530 (0.194) |
Note: CV = coefficient of variation.
aAverage expression level (1 Average from approximately 500 animals per reporter per experiment; 5 total experiments) shown in Photomultiplier Tube (PMT) counts, with the average coefficient of variation (1 CV value per experiment; 5 total experiments) shown in parenthesis.
Table 2.
Average Life Span and Coefficient of Variation for Life Span at Different Temperatures
| 15°C (11, 1,278)a | 20°C (26, 2,634)a | 25°C (13, 779)a | |
|---|---|---|---|
| Mean life span (days) | 31.1 | 22.8 | 17.9 |
| Coefficient of variation | .255 | .235 | .184 |
Note: aNumber of experiments and total number of animals in parentheses (experiments, animals).
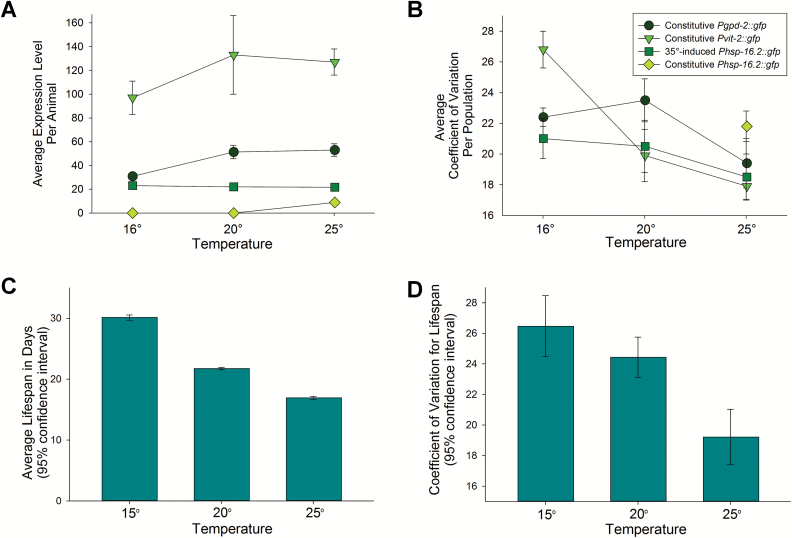
Table 1 and Figure 1A show mean gene expression at different temperatures. Expression of Green Fluorescent Protein (GFP) from the vit-2 and gpd-2 promoters increased with temperature (between 16°C and 20°C), whereas the induced expression of GFP from the hsp-16.2 promoter was not (Figure 1A, Table 1; see Supplementary Materials section for induction conditions). Animals raised at 25°C that were not previously heat shocked showed basal, constitutive expression from the hsp-16.2 promoter but animals raised at 16°C and 20°C did not (Figure 1A, Table 1). Figure 1C and Table 2 show life-span data. As was expected from previous work (9,19), animals raised and aged at 15°C had the longest life spans followed by animals at 20°C and 25°C (Figure 1C, Table 1).
Figure 1.
Interindividual variation in gene expression and life span as a function of temperature. This figure shows that the amount of interindividual variation in three kinds of reporter gene expression, including a life-span biomarker, decreases at 25°C (B); interestingly, the amount of interindividual variation in life span also decreased at 25°C (D). Top graphs (A, B) depict mean ± SEM; bottom graphs (C, D) depict 95% confidence intervals (CI). To determine statistical significance, two-way analysis of variance (ANOVA) followed by the Holm–Sidak test for multiple comparisons was used for comparing the effects of temperature and strain on mean or CV, unless otherwise stated. CV is the coefficient of variation (SD/mean); CV is displayed as a whole-number percentage. Five independent trials consisting of approximately 500 individuals of each strain (one per reporter) at each temperature were used to generate the graphs of reporter gene expression. Fluorescence levels for animals expressing Pgpd-2::gfp are divided by 10 for plotting purposes; actual values are shown in Supplementary Table S1. (A) All reporters were significantly different in expression level from one another within each temperature. All p values from two-way ANOVA followed by Holm–Sidak testing for effects of strain and temperature on mean expression are shown in Supplementary File F1. Measured Pgpd-2::gfp expression was significantly different between 16°C and 20°C (p = .001) and 16°C and 25°C (p = .0003). Pvit-2::gfp was significantly different between 16°C and 20°C (p = .007). No significant differences were detected in induced Phsp-16.2::gfp expression between any temperatures; constitutive Phsp-16.2::gfp expression at 25°C was significantly different from all other gene expression levels at 25°C (p = .02, one-way repeated measures ANOVA, Student–Newman–Keuls test). Temperature had a significant effect on mean expression between 16°C and 20°C and between 16°C and 25°C, when the entire profile of reporters is considered (p = .00009 for 16°C vs 25°C and 16°C vs 20°C). Also, see Table 1. (B) Pvit-2::gfp CV was significantly greater than Phsp-16.2::gfp CV at 16°C (p = .011). Pgpd-2::gfp CV significantly changed between 20°C and 25°C (p = .0003). Pvit-2::gfp CV significantly changed between 16°C and 25°C (p = .008). CV measured for 25°C Phsp-16.2::gfp expression was significantly higher than for the 35°C -induced expression of Phsp-16.2::gfp (p = .02, paired t test). Temperature has a significant effect on CV of the analyzed reporters between 16°C and 25°C (p = .00028) and between 20°C and 25°C (p = .019); increasing temperature decreased worm-to-worm variation in gene expression. Explicit p values for every comparison from the two-way ANOVA followed by Holm–Sidak testing for effects of temperature and strain on CV are detailed in Supplementary File F2. Also, see Table 1. (C) Mean life span significantly decreased with increasing temperature; 95% CI shown. Fifty independent life-span experiments at various temperatures comprised the Life span × Temperature data set. Also, see Table 2. (D) Interindividual variation in life span significantly decreased at 25°C; 95% CI shown. Also, see Table 2. Raw data for life spans and individual life-span trial CVs is available in Supplementary File F3.
Interestingly, compared with animals grown at lower temperatures, animals grown at 25°C also had lower interindividual variation in gene expression (Figure 1B, Table 1). Variation was lower for all three genes, including the life-span biomarker Phsp-16.2::gfp (Figure 1B). After correcting for bias in mean and coefficient of variation introduced by the discreet sampling of the continuous variable of life span, we found that animals grown at 25°C also showed decreased interindividual variation in life span (for details about bin corrections, see Supplementary Material and Supplementary Figures S1–4). Thus, for C elegans, developing and aging at 25°C canalizes variation in gene expression and life span.
Discussion
Despite considerable study (2,20–26), the reasons that isogenic organisms in homogeneous environments manifest phenotypic variation and physiological differences are not well understood. Here, we studied how temperature affects the manifestation of the phenotypic variation in gene expression and life span. Differences in these traits can define distinct physiological states (3,6,25). To quantify gene expression, we cultured animals at different temperatures. We then used methods that we developed (2) to precisely quantify average whole-animal gene expression level of a number of reporter genes, including a Phsp-16.2::gfp life-span biomarker and animal-to-animal variation in these quantities.
We also correlated these measurements with parallel measurements of life span and variation in life span at different temperatures. Performing such measurements required large numbers of trials. Here, due to the level of trial-to-trial variation in measured variation in life span at different temperatures, we needed to analyze more than 50 independent life-span experiments (involving 94 hermaphrodites per experiment on average) to resolve statistically significant differences. These experiments also required careful attention to experimental detail. Specifically, we had to correct for the bias in mean and coefficient of variation that is introduced by sampling life span at discreet points in time. Sampling life-span noncontinuously results in an intrinsic positive skewing of both mean and standard deviation. We discuss this in more detail and show mathematical simulations demonstrating these effects in the Supplementary Material and Supplementary Figures S1–4.
How might different environmental temperatures bring about these different effects? Unlike mammals (but like other poikilotherms), C elegans maintains a body temperature close to that of its environment and aspects of its physiology differ with temperature. For example, insulin signaling decreases with increasing temperature (27), whereas gene expression directed by a chaperone promoter is only induced at higher temperature (this report; Figure 1A). Our results here showed that growth at 25°C, near the top of C elegans fitness range (12°C–26°C) (28), shortened life span and lowered variation in life span and in gene expression.
In poikilotherms, physiological effects arising from differences in environmental temperature are well described (12). In animals ranging from Daphnia (29) to flies (10) to alligators (30), increased environmental temperatures increase the pace of development and aging. C elegans also develops (31) and ages (9,19) more rapidly at higher temperatures. The reason for the increased tempo is not clear but it has often been explained with the idea that development and aging depend on chemical reactions and these work more quickly at higher temperatures (8,9). In fact, the picture is more complex: in poikilotherms, most metabolic reactions depend on enzymes with particular temperature optima and their expression is differently affected by temperature (12). In addition to changes in enzyme expression level to compensate for temperature-dependent differences in activity, many organisms express temperature specific enzyme isoforms (12). For these reasons, the complement of enzymes in a particular cell in a C elegans raised at 15°C is different than in the same cell in an otherwise-identical animal raised at 25°C. For example, in this work (Figure 1A), GAPDH promoter activity increases with temperature; thus, there is presumably a higher concentration of GAPDH in cells at higher temperatures then at lower.
Consistent with the previous findings, we note that at 25°C, the sy441 loss of function allele of hsf-1, which encodes the protein that activates expression of heat shock response genes, arrests during larval development (16), suggesting that expression of heatshock response genes might be needed for adult survival at this temperature. In this work, we have shown that expression directed by one particular heat shock responsive promoter, hsp16.2, is induced at 25°C (Figure 1A). We therefore suggest that the diminution in phenotypic variation for both gene expression and life span at 25°C may be a consequence of low level hsf-1-dependent expression of HSP-16.2 and other chaperones at the higher temperature. Restated, the protein folding aspect of proteostasis may be contributing to reduced cell-to-cell and animal-to-animal variation in gene expression, which in turn may cause reduced animal-to-animal variation in life span.
Finally, we note that this work establishes an intriguing correlation between variation in life-span biomarker expression and variation in life span itself. One simple explanation for this correlation between decreased variation in these traits is that heightened expression of chaperones at elevated temperatures might decrease phenotypic variation in both processes. Increased chaperone expression could reduce variation in gene expression and life span by increasing both absolute number and proportion of functional molecules of particular protein species expressed in individual animals.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Author Contributions
A.M., T.E.J., R.B., and M.K. designed experiments. S.L. and G.S. performed the life-span experiments. A.M. made the DNA and worms for TJ3003. P.M.T. sequenced and outcrossed TJ3003. A.M. performed gene expression measurements. M.M.C. and A.M. analyzed the data. A.M., M.M.C., and R.B. wrote the final version of the paper. A.M. and R.B. wrote initial and final versions of the article and guarantee the integrity of the results.
Funding
This work was supported by the National Institute on Aging and the National Institute of General Medicine at the National Institutes of Health by grants 4R00AG045341 to A.M. R01AG038518 to M.K., R00AG045200 to S.L., and R01GM97479 to R.B.
Supplementary Material
Acknowledgments
The authors thank Daniel Promislow and George Martin for thoughtful discussions and comments on the manuscript. The authors also thank anonymous reviewers whose comments resulted in us correcting the life-span data for bias introduced by bin size, which increased the accuracy of our measurements of life span and variation in life span.
References
- 1. Kirkwood TB, Finch CE. Ageing: the old worm turns more slowly. Nature. 2002;419:794–795. doi:10.1038/419794a [DOI] [PubMed] [Google Scholar]
- 2. Mendenhall AR, Tedesco PM, Sands B, Johnson TE, Brent R. Single cell quantification of reporter gene expression in live adult Caenorhabditis elegans reveals reproducible cell-specific expression patterns and underlying biological variation. PLoS One. 2015;10:e0124289. doi:10.1371/journal.pone.0124289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mendenhall AR, Tedesco PM, Taylor LD, Lowe A, Cypser JR, Johnson TE. Expression of a single-copy hsp-16.2 reporter predicts life span. J Gerontol A Biol Sci Med Sci. 2012;67:726–733. doi:10.1093/gerona/glr225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sánchez-Blanco A, Kim SK. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002047. doi:10.1371/journal.pgen.1002047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002306. doi:10.1371/journal.pgen.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi:10.1038/ng1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao R, Zhang B, Dong Y, et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. 2013;152:806–817. doi:10.1016/j.cell.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suda H, Sato K, Yanase S. Timing mechanism and effective activation energy concerned with aging and lifespan in the long-lived and thermosensory mutants of Caenorhabditis elegans. Mech Ageing Dev. 2012;133:600–610. doi:10.1016/j.mad.2012.07.007 [DOI] [PubMed] [Google Scholar]
- 9. Shoyama T, Ozaki T, Ishii N, Yokota S, Suda H. Basic principle of the lifespan in the nematode C. elegans. Mech Ageing Dev. 2007;128:529–537. doi:10.1016/j.mad.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 10. Loeb J, Northrop JH. Is there a temperature coefficient for the duration of life? Proc Natl Acad Sci USA. 1916;2:456–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinensky M. Temperature control of phospholipid biosynthesis in Escherichia coli. J Bacteriol. 1971;106:449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hazel JR, Prosser CL. Molecular mechanisms of temperature compensation in poikilotherms. Physiol Rev. 1974;54:620–677. [DOI] [PubMed] [Google Scholar]
- 13. Yen K, Patel HB, Lublin AL, Mobbs CV. SOD isoforms play no role in lifespan in ad lib or dietary restricted conditions, but mutational inactivation of SOD-1 reduces life extension by cold. Mech Ageing Dev. 2009;130:173–178. doi:10.1016/j.mad.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 14. Leiser SF, Begun A, Kaeberlein M. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell. 2011;10:318–326. doi:10.1111/j.1474-9726.2011.00672.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morton EA, Lamitina T. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell. 2013;12:112–120. doi:10.1111/acel.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi:10.1534/genetics.104.028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baker GT, 3rd, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–239. [DOI] [PubMed] [Google Scholar]
- 18. Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [DOI] [PubMed] [Google Scholar]
- 19. Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. [DOI] [PubMed] [Google Scholar]
- 20. Gärtner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim. 1990;24:71–77. [DOI] [PubMed] [Google Scholar]
- 21. Delbruck M. The burst size distribution in the growth of bacterial viruses (bacteriophages). J Bacteriol. 1945;50:131–135. [DOI] [PubMed] [Google Scholar]
- 22. Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi:10.1073/pnas.162041399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi:10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- 24. Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi:10.1038/nature10665 [DOI] [PubMed] [Google Scholar]
- 25. Colman-Lerner A, Gordon A, Serra E, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi:10.1038/nature03998 [DOI] [PubMed] [Google Scholar]
- 26. Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi:10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. [DOI] [PubMed] [Google Scholar]
- 28. Riddle DL Blumenthal T Meyer BJ Priess JR, eds. C. elegans II. Cold Spring Harbor Monograph Series. Vol. 33 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 29. Macarthur JW, Baillie WH. Metabolic activity and duration of life. I. Influence of temperature on longevity in Daphnia magna. J Exp Zool. 1929;53:221–242. [Google Scholar]
- 30. Lance VA. Alligator physiology and life history: the importance of temperature. Exp Gerontol. 2003;38:801–805. [DOI] [PubMed] [Google Scholar]
- 31. Byerly L, Cassada RC, Russell RL. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev Biol. 1976;51:23–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.