Abstract
Epigenetic remodeling is one of the major features of the aging process. We recently demonstrated that DNA methylation of ELOVL2 and FHL2 CpG islands is highly correlated with age in whole blood. Here we investigated several aspects of age-associated hypermethylation of ELOVL2 and FHL2. We showed that ELOVL2 methylation is significantly different in primary dermal fibroblast cultures from donors of different ages. Using epigenomic data from public resources, we demonstrated that most of the tissues show ELOVL2 and FHL2 hypermethylation with age. Interestingly, ELOVL2 hypermethylation was not found in tissues with very low replication rate. We demonstrated that ELOVL2 hypermethylation is associated with in vitro cell replication rather than with senescence. We confirmed intra-individual hypermethylation of ELOVL2 and FHL2 in longitudinally assessed participants from the Doetinchem Cohort Study. Finally we showed that, although the methylation of the two loci is not associated with longevity/mortality in the Leiden Longevity Study, ELOVL2 methylation is associated with cytomegalovirus status in nonagenarians, which could be informative of a higher number of replication events in a fraction of whole-blood cells. Collectively, these results indicate that ELOVL2 methylation is a marker of cell divisions occurring during human aging.
Keywords: Epigenetics, Methylation, Biomarker, FHL2
In our aging society, the availability of reliable biomarkers of aging is important for an efficient and well-prioritized management of health services and resources to patients. A biomarker of aging is defined as “a biological parameter of an organism that either alone or in some multivariate composite will, in the absence of disease, better predict functional capability at some late age than will chronological age” (1), and its ideal features have been extensively drawn up (2,3). A major requisite of a biomarker of aging is that it should have a good correlation with the chronological age of the individuals in a population, and in addition reflect the inter-individual heterogeneity in the aging rates of the individuals, that is, their biological age. Biological age is a difficult concept to define, as an individual could display a youthful phenotype for some health parameters but not for others. However, it could be agreed that the ultimate readout of successful biological aging is a long life span (longevity) or a long (disease-free) health span. This means that appropriate human study designs such as middle aged and elderly prospective populations as well as family studies are essential to demonstrate the effectiveness of a biomarker of aging (4). To date, although a number of biological parameters, such as leukocyte telomere length, deletion of mtDNA, protein modifications, and changes in the expression of specific proteins, have been proposed as potential biomarkers of aging (5,6), their effectiveness remains to be proven.
Cross-sectional and longitudinal studies have demonstrated the profound remodeling of DNA methylation patterns that occurs in the human genome as a function of chronological age in several tissues (reviewed in reference (7)). Based on these results, reliable age predictors were defined by combining CpG sites whose methylation status was age associated (8–11). Besides the strong correlation with age, two of these epigenetic age predictors have recently been associated with mortality (12,13) and longevity (14), posing them as potential markers of biological aging. The first age predictor (9) combines 71 CpG sites from the Infinium HumanMethylation450 BeadChip (Infinium 450k) microarray, whose methylation status in blood varies with aging in a cohort of 656 participants aged 19 to 101 years. The second age-associated epigenetic signature (10) was built starting from several DNA methylation profiles available in public databases and includes 353 CpG sites from the Infinium HumanMethylation27 BeadChip (Infinium 27k) microarray (which are also included in the Infinium 450k microarray). This epigenetic clock works in most human cell types and tissues and is correlated not only with mortality but also with physical and cognitive fitness (15).
Currently there is a lack of cost-effective methods to analyze several dozens of CpG sites, such as those included in Hannum’s and Horvath’s epigenetic clocks, and the most affordable approach is to use the Infinium 450k microarray. The definition of epigenetic biomarkers including a smaller number of CpG probes, assessable by high-throughput methods such as EpiTYPER MassARRAY or pyrosequencing, would allow for translation of these findings toward actual clinical applications. Along this line, Wagner and co-authors tested blood predictors of age including few or single Infinium CpG probes (11). Compared with Horvath’s and Hannum’s clocks, these simple biomarkers were weakly correlated with chronological age, probably also because they are more affected by technical variations between independent microarray data sets (16). In addition, their association with mortality, although statistically significant in some data sets, was rarely reproducible in independent cohorts (12,16).
In one of the first studies that used the Infinium 450k platform to analyze age-associated changes in blood DNA methylation (17), we identified two genomic regions, localized within the CpG islands of the promoters of ELOVL2 and FHL2 genes, whose methylation status was highly correlated with age. These findings were later confirmed by similar studies (18–22). Both ELOVL2 and FHL2 showed continuous increase in methylation levels with age, with methylation values ranging from 7% to 91% for ELOVL2 and from 12% to 53% for FHL2. The CpGs identified by our group within ELOVL2 and FHL2 are not part of Horvath’s epigenetic clock, which was built using the CpG probes common to the Infinium HumanMethylation27 BeadChip and the Infinium450k, while are part of Hannum’s age predictor.
In this article, we investigated the potential of ELOVL2 and FHL2 as simple epigenetic biomarkers of chronological and biological aging. To this aim, we first confirmed the correlation of ELOVL2 and FHL2 methylation with age in different tissues and cell types, using also in vitro models of cell replication and senescence. Then, we measured ELOVL2 and FHL2 methylation in whole blood from a longitudinal cohort that is part of the Doetinchem Cohort Study. Finally, we evaluated for the first time the association of whole-blood ELOVL2 and FHL2 methylation with familial longevity, mortality, and other biological parameters in the Leiden Longevity Study (LLS). This cohort was established to identify biomarkers related to familial longevity (23,24) and includes long-lived siblings, their offspring, and the spouses of the offspring. Collectively these analyses represent a first step to disentangle the mechanisms that underlie ELOVL2 and FHL2 age-dependent hypermethylation and to evaluate their validity as biomarkers of aging.
Methods
Study Population
For the comparison of ELOVL2 and FHL2 DNA methylation between blood and granulocytes, we used samples from 92 participants aged 22 to 98 years (mean age = 55 years). The cohort included healthy volunteers recruited between 2004 and 2005 in Bologna and neighboring districts. Informed consent, approved by the Medical Ethics Committee of the S. Orsola-Malpighi Polyclinic (Bologna, Italy), was obtained from all participants.
Longitudinal assessment of ELOVL2 and FHL2 DNA methylation in whole blood was performed on 19 participants from the Doetinchem Cohort Study, a population-based prospective study on the impact of (changes in) lifestyle factors and biological risk factors on various aspects of health and well-being of Dutch adults. In 1987–1991 (t1), self-completed questionnaires were collected and a physical examination was performed on a random sample of 12,405 men and women aged 20–59 years (response rate 62%) from the town of Doetinchem. Of those, a two-third random sample of 7,768 participants was reinvited to be examined in 1993–1997 (t2, n = 6,113), 1998–2002 (t3, n = 4,916), 2003–2007 (t4, n = 4,520), and 2008–2012 (t5, n = 4,017). The response rate for the second, third, fourth, and fifth round was 75% and more. Informed consent was obtained from all participants, and ethical clearance was obtained from the Medical Ethics Committees of the Netherlands Organization of Applied Scientific Research. Further details of the study design have been described elsewhere (25). The 19 participants assessed for ELOVL2 and FHL2 methylation (9 women and 10 men) were a selection of healthy volunteers (baseline age range: 22–57 years) whose body mass index remained below 25kg/m2 during the entire follow-up (t1–t5). Details of this group are reported in Supplementary Table 1.
For the LLS, long-lived siblings of European descent were recruited together with their offspring and the spouses of the offspring. Families were included if at least two long-lived siblings were alive and fulfilled the age criterion of 89 years or older for men and 91 years or older for women, representing less than 0.5% of the Dutch population in 2001 (26). In total, 944 long-lived siblings with a mean age of 94 years (range 89–104), 1,671 offspring (61 years, 39–81), and 744 spouses thereof (60 years, 36–79) were included. For this study, we removed the individuals for whom DNA was not available for measurement (n = 96), the identity was not confirmed after genotyping (n = 81), or ELOVL2 and FHL2 were not successfully measured (n = 8), leaving 872 nonagenarians, 1,574 offspring, and 728 spouses for analysis. White blood cell (WBC) counts were measured using the Sysmex XE-2100 (TOA Medical Electronics) (27). Cytomegalovirus (CMV) serostatus was determined on blind samples using the CMV-IgG-ELISA PKS assay (Medac GmbH) (28).
Publicly Available DNA Methylation Profiles of Healthy Tissues
We considered all DNA methylation data sets available on October 2015 in GEO DataSets repository (http://www.ncbi.nlm.nih.gov/gds) that fulfilled the following requirements: (i) generated with Infinium 450k platform (GPL13534); (ii) analyzing healthy human tissues; and (iii) including information regarding the age of the involved participants (attribute name “age” or “age (yrs)” or “age (years)” or “age (y)”). The list of selected data sets is reported in Supplementary Table 2. Beta values were extracted for the CpG probes reported in Supplementary File 1.
Cell Cultures
Primary dermal fibroblasts (DFs) were established from biopsies of sun-protected forearm skin from healthy donors. All the donors gave written informed consent to the biopsy. In total, 10 participants were studied: 3 young donors (3 women, age range: 30–31 years), 3 middle-aged donors (2 men and 1 women, age range: 58–68 years), and 4 centenarian donors (4 women, age range: 100–105 years). DFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) containing 25mM of glucose supplemented with 10% fetal bovine serum (FBS, Life Technologies) at 37 °C in 5% CO2 humidified atmosphere. In addition, the medium contained 100U/mL of penicillin, 100 µg/mL of streptomycin (Life Technologies), and 4mM of glutamine.
Human embryonic diploid lung fibroblast cells (LF1) were grown at 37 °C in 5% CO2 and 2.5% O2 atmosphere in F-10 nutrient medium (Thermo Scientific) supplemented with 15% FBS, penicillin, streptomycin, and l-glutamine (29). Cells were serially passaged at 1:4 dilutions when reaching 80%–90% confluence. In early passage cultures, the time between passages was constant at approximately 3 days; as culture approached senescence, it increased up to 2–3 weeks. At this point, cells were passaged upon reaching 80% confluence at 1:2 dilution. Under these experimental conditions, cultures were monitored by microscopic observation every 2–3 days to assess cell division. Cell cultures that could not reach 80% confluence were considered senescent. Subsequently, cultures were incubated further up to 4 months with replating at 4, 8, 12, and 16 weeks in senescence. Experiments were performed with cells harvested at early passages, after 8 weeks in senescence (senescent) and 16 weeks in senescence (deep senescent).
Measurement of ELOVL2 and FHL2 Methylation
The EpiTYPER assay (Sequenom) was used to quantitatively measure the methylation level of individual CpG sites in ELOVL2 and FHL2 CpG islands, as previously described (17). The amplified regions were chr6:11,044,680-11,045,053 (GRCh37/hg19) within ELOVL2 locus and chr2:106,015,678-106,016,008 (GRCh37/hg19) within FHL2 locus. ELOVL2 and FHL2 amplicons included respectively probes: cg16867657, cg24724428, cg21572722, cg16323298 and cg06639320, cg22454769, cg24079702, cg26344233, cg06907053 from the Infinium 450k design (Supplementary Figure 1). PCR products were processed following manufacturer’s instructions to measure the percentage of methylation of single CpG sites or of group of adjacent CpG sites (indicated as CpG units), depending on the genomic sequence. For each locus, CpG units with missing values in more than 20% of the samples were removed, as well as samples with missing values in more than 20% of the CpG units.
Statistical Analyses
For each CpG unit, correlations between methylation level (measured as β value in data from Infinium450k data sets and as percentage of methylation in data from EpiTYPER assay) and chronological age or replicative passage were calculated using Spearman’s correlation.
To study longitudinal changes in ELOVL2 CpG_11.12.13.14 and FHL2 CpG_9.10 methylation in the Doetinchem Cohort Study, we used the R package lmerTest to construct mixed models with subject-specific random effects for the intercept and the slope. Importantly, this approach allows to account for missing data in our data set (Supplementary Table 1). For both the CpG units, we first constructed a model testing the association between methylation and time, expressed as years, which included an interaction term (Time × Age at the recruitment) to estimate the effect of the age at the recruitment on the rate of change of DNA methylation (slope) over time; subject-specific random effects were estimated on the intercept and on the slope. In ELOVL2 CpG_11.12.13.14 starting model, we observed no variability for the random effects on the intercept and on the slope of the interaction between participant and age at the recruitment. These two terms were therefore removed from the overspecified starting model, resulting in the subsequent model: ELOVL2_CpG_11.12.13.14~time*Age_at_recruitement+(0+time|Person.ID). Analysis of variance confirmed that the two models did not significantly differ. The complete model for FHL2 was equivalent, but the only nonsignificant effect was in the random effect of the slope between participant and age of recruitment, leading to the final model: FHL2_CpG_19.20~time+Age_at_recruitement + (1|Person.ID)+(0+time|Person.ID).
To study the association between familial longevity and ELOVL2 and FHL2 methylation in LLS offspring and their spouses, we performed linear regression, adjusted for age, gender, bisulphite date, and WBC counts, in STATA/SE 11.2 (StataCorp LP). Robust standard errors were used to account for sibship relations. Prospective analysis of ELOVL2 and FHL2 methylation was performed separately in the LLS nonagenarians and the LLS offspring and their spouses using a left-truncated Cox proportional hazards model, adjusted for age, gender, bisulphite date, and WBC counts, in STATA/SE 11.2 (StataCorp LP). Robust standard errors were used to account for sibship relations. After a mean follow-up time of 9.5 years, 92.5% (n = 807, LLS nonagenarians) and 6.4% (n = 147, LLS offspring and spouses) of the individuals had died.
To determine the association of ELOVL2 and FHL2 methylation with CMV infection in the LLS, we performed linear regression, separately for the LLS nonagenarians and the LLS offspring and their spouses, adjusted for age, gender, bisulphite date, and WBC counts, in STATA/SE 11.2 (StataCorp LP). Robust standard errors were used to account for sibship relations.
Results
Association of ELOVL2 and FHL2 DNA Methylation With Chronological Age Across Different Cell Types and Tissues
As the epigenetic clock described by Horvath is a multitissue age predictor, we considered ELOVL2 and FHL2 methylation in different cell types from healthy individuals of different ages.
First, considering that blood is composed of different cell types whose relative proportion can greatly vary among individuals, we studied age-associated changes in ELOVL2 and FHL2 DNA methylation in blood and purified granulocytes from the same participant. EpiTYPER analysis on 92 participants aged 22 to 98 years showed similar methylation levels and age-dependent changes in blood and granulocytes (Supplementary Figure 2). Within ELOVL2, a group of adjacent CpG units (from CpG_8 to CpG_15.16.17) showed highly significant hypermethylation associated with age. Both in blood and granulocytes, the highest correlation was observed for CpG_11.12.13.14 (Supplementary Figure 2A and C; Spearman’s ρ = .91 in blood and .93 in granulocytes). A similar behavior was observed for FHL2, where the region of age-associated differential methylation included CpG sites from CpG_9.10 to CpG_19.20 (Supplementary Figure 2B and D), with the highest values for CpG_9.10 in blood (Spearman’s ρ = .73) and in granulocytes (Spearman’s ρ = .71). Paired t test was used to compare DNA methylation profiles between blood and granulocyte samples from the same individual. Several CpG sites resulted in significantly different methylation profiles (p < .05), but the differences between the two tissues were very small (Supplementary Figure 3).
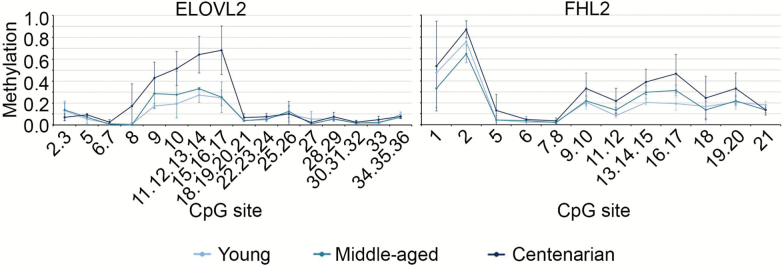
To evaluate whether ELOVL2 and FHL2 methylation correlated with chronological age in cell types other than blood, we measured their DNA methylation levels in primary DF cultures established from three young, three middle-aged, and four centenarian donors. Within ELOVL2, the group of CpG units from CpG_8 to CpG_15.16.17 showed significant hypermethylation in DFs from centenarians compared with DFs from young and middle-aged donors (Figure 1). The difference reached the maximum for CpG_11.12.13.14 (Student’s t test, p = .017 for centenarians vs young donors comparison and p = .026 for centenarians vs middle-aged donor comparison), which was also the CpG unit that better correlated with donors’ age (Supplementary Figure 4; Spearman’s ρ = .79). A similar but nonsignificant trend was observed when comparing DFs from middle-aged and young donors. DFs from centenarian and middle-aged donors showed high standard deviations for the CpG sites whose DNA methylation status is age dependent, whereas DFs from young donors were more homogeneous. Although the differences in standard deviations could be ascribed to the small number of observations and/or to different age ranges in the three groups (30–31, 58–68, and 100–105 years old), it is also possible to suppose that biopsies from older participants include cells which are more heterogeneous for their DNA methylation patterns. Within the FHL2 target region, no significant differences were observed between the three groups of DFs. However, a trend toward hypermethylation from young to centenarian donors was evident for the region spanning from CpG_9.10 to CpG_19.20 (Figure 1). Also in this case, DFs from centenarians displayed higher inter-individual variability than DFs from younger donors. CpG_16.17 was the most correlated with donors’ age (Supplementary Figure 4; Spearman’s ρ = .91).
Figure 1.
ELOVL2 and FHL2 DNA methylation profiles in dermal fibroblasts from young, middle-aged, and centenarian donors. Mean methylation values and standard deviations of the CpG sites assessed by the EpiTyper assay are reported.
To complete these results, we searched GEO DataSet repository for Infinium 450k experiments in which DNA methylation was assessed in healthy human tissues (Supplementary Table 2). For comparison, we also considered ELOVL2 and FHL2 DNA methylation values in a whole-blood data set. We extracted β values for the probes mapping within the ELOVL2 and FHL2 genes (15 and 34 probes, respectively) and calculated Spearman’s correlation with age. The methylation of several CpG sites correlated with age in different tissues (Supplementary File 2). In particular, the methylation of cg16867657, cg21572722, and cg24724428 within ELOVL2 and of cg06639320, cg22454769, and cg24079702 within FHL2, which was previously shown to be age associated in whole blood, correlated significantly with age in most tissues (Supplementary File 2). We focused our attention on cg16867657 (ELOVL2; Figure 2 and Supplementary Figure 5) and cg06639320 (FHL2; Supplementary Figure 6), which showed the best correlation with age in blood. For cg16867657, a trend toward hypermethylation was evident in most of the analyzed tissues and reached statistical significance in peripheral blood mononuclear cells, CD4 and CD8 T cells, monocytes, abdominal and gluteal subcutaneous adipose tissue, bone, temporal, frontal motor, occipital, parietal, sensory and visual cortex, frontal lobe white matter, cingulate gyrus hippocampus, midbrain, skeletal muscle, liver, dermis, epidermis, and mesenchymal stem cells. The only tissues in which we did not observe a trend toward hypermethylation were cerebellum and aortic tissue. A similar scenario was evident for cg06639320, with the exception of neutrophilis, abdominal subcutaneous adipose tissue, caudate nucleus, and epidermis that did not show a trend toward hypermethylation, whereas cerebellum and hip cartilage did. In this case, aortic tissue DNA methylation was also stable over time.
Figure 2.
Age-dependent hypermethylation of ELOVL2 cg16867657 in 30 normal tissues analyzed using the Infinium 450k platform (1: whole blood; 2: peripheral blood mononuclear cells; 3: neutrophils; 4: CD4 T cells; 5: CD8 T cells: 6: monocytes; 7: subcutaneous adipose tissue abdominal; 8: subcutaneous adipose tissue gluteal; 9: hip cartilage; 10: bone; 11: prefrontal cortex; 12: temporal cortex; 13: cerebellum; 14: frontal lobe white matter; 15: caudate nucleus; 16: cingulate gyrus; 17: frontal cortex; 18: hippocampus; 19: midbrain; 20: motor cortex; 21: occipital cortex; 22: parietal cortex; 23: sensory cortex; 24: visual cortex; 25: skeletal muscle; 26: liver; 27: aortic tissue; 28: dermis; 29: epidermis; 30: mesenchymal stem cells). For each tissue, linear regression between age and DNA methylation is plotted.
DNA Methylation of ELOVL2 and FHL2 in Cell Cultures at Different Replicative Passages
The observation that ELOVL2 and FHL2 are linearly hypermethylated with age in almost all tissues prompted us to think that the underlying mechanism could be linked to common cellular activities. A possibility could be that methylation of the two loci is associated with cellular replication. To test this hypothesis, we measured ELOVL2 and FHL2 methylation by EpiTYPER in DFs during in vitro cell divisions. DFs from the two middle-aged donors (indicated as DF1 and DF2) were expanded until medium (Passage 13, P13) and late (Passage 22, P22) passages. For all the ELOVL2 CpGs tested, P3 and P13 methylation profiles were largely similar (Figure 3A and Supplementary Figure 7A). On the contrary, in both DF cultures, the region spanning from CpG_9 to CpG_15.16.17 resulted in 10%–30% more methylation in P22 compared with previous passages. The correlation between DNA methylation and cell passage was high in both DF cultures (Supplementary Table 3), although it did not reach statistical significance, which may be due to the limited number of points taken into consideration. For the FHL2 target region, the results of the analysis were less straightforward, as passage-dependent hypermethylation was evident only in DF2 (Figure 3A and Supplementary Figure 7A).
Figure 3.
(A) ELOVL2 and FHL2 DNA methylation profiles in dermal fibroblasts (DFs) from middle-aged donor 1 (DF1) at replicative Passages 3, 13, and 22. (B) ELOVL2 and FHL2 DNA methylation profiles in F1 fetal lung fibroblasts at early passages, after 8 weeks in senescence (senescent LF1) and after 16 weeks in senescence (deeply senescent LF1).
Similar results were observed for human umbilical vein endothelial cells (HUVECs), which were cultured until growth arrest (Passage 12) as previously described (30). The methylation of region enclosed between CpG_9 and CpG_15.16.17 of ELOVL2 progressively increased with cell passages (Supplementary Figure 7B). Spearman’s correlation was maximum for CpG_10 (Spearman’s ρ = .86, p = 3×10–4, methylation range from 2% to 32%), whereas CpG_11.12.13.14 showed similar hypermethylation range but lower correlation (Spearman’s ρ = .52). Also in this case, the passage-dependent increase in FHL2 methylation retained statistical significance for CpG_9.10 (Spearman’s ρ = .63; p = .03), despite reduced magnitude (methylation range 14% to 22%, Supplementary Figure 7B).
HUVECs entered senescence approximately at Passage 9 (30), while methylation of ELOVL2 and, to a lesser extent, of FHL2 already increased at lower passages. This observation suggests that hypermethylation of the two loci is associated with cell replication rather than with cellular senescence. To validate this observation, we measured DNA methylation of ELOVL2 and FHL2 in LF1 fetal lung fibroblasts at early passages, after 8 weeks in senescence (senescent LF1) and after 16 weeks in senescence (deeply senescent LF1). EpiTYPER assay showed a striking increase in methylation level of ELOVL2 region enclosed from CpG_8 to CpG_15.16.17 and in FHL2 region enclosed from CpG_9.10 to CpG_19.20 (Figure 3B) in senescent cells compared with young cells. On the contrary, methylation profiles of senescent cells and deeply senescent cells were totally comparable, indicating that senescence progression does not affect per se the epigenetic status of the two loci.
Whole-Blood DNA Methylation of ELOVL2 and FHL2 in a Longitudinal Cohort
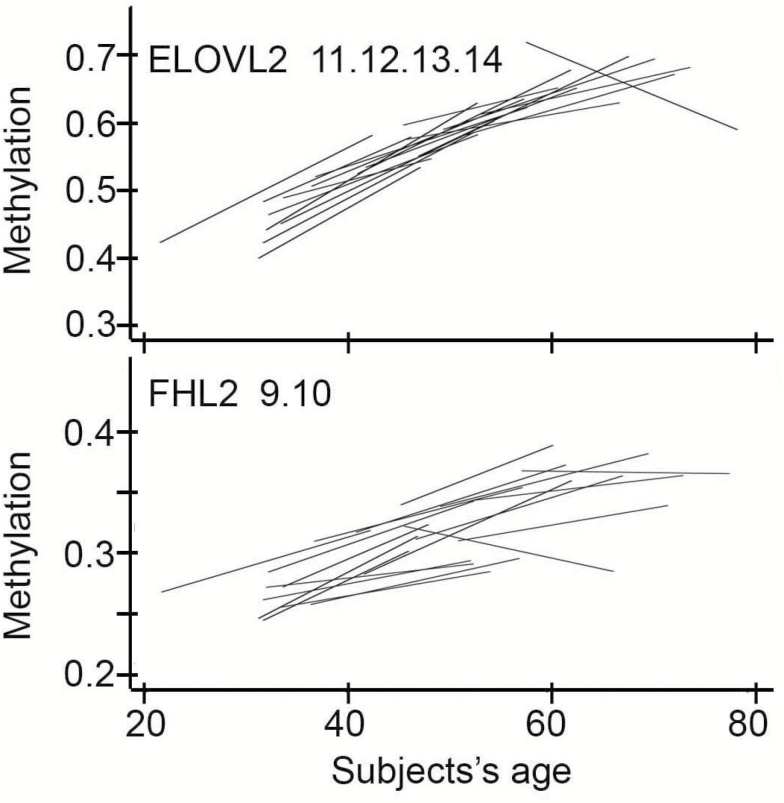
We measured whole-blood DNA methylation levels of ELOVL2 and FHL2 in a cohort of 19 participants tested longitudinally at intervals of 5 years, for five measurements (t1, t2, t3, t4, and t5; age at recruitment ranging from 22 to 57 years). We used a mixed-effects model including time and age at recruitment to evaluate longitudinal changes in methylation of ELOVL2 CpG_11.12.13.14 and FHL2 CpG_9.10, the two CpG sites with the highest correlation values with age in whole blood. DNA methylation profiles confirmed time-dependent hypermethylation of the two loci, with a clearer trend for ELOVL2 (Figure 4 and Supplementary Figure 8).
Figure 4.
Regression lines between DNA methylation of ELOVL2 CpG_11.12.13.14 or FHL2 CpG_9.10 and age of 19 participants longitudinally assessed at five intervals of 5 years.
For ELOVL2 CpG_11.12.13.14, we found a significant association between methylation and time (estimate = 0.016, SE = 0.0027; p = 8.62×10–8) and age at recruitment (estimate = 0.008, SE = 0.00077; p = 3.55×10–15). There was a significant methylation-by-time interaction with age at recruitment (estimate = −0.00027, SE = 0.000065; p = 1.03×10–4), indicating that the slope of association between ELOVL2 methylation and time decreases with age at recruitment.
Similarly, hypermethylation of FHL2 CpG_9.10 was associated with time (estimate = 0.0019, SE = 0.00038; p = 2.82×10–5) and age at recruitment (estimate = 0.003, SE = 0.00047; p = 5.19×10–6), but no interaction between the two terms was observed.
Notably, random effects on the intercept were statistically significant in FHL2 but not in ELOVL2 model, suggesting that in the latter subject-specific effects were negligible.
Association Between Age, Familial Longevity, Mortality, and CMV Infection in the LLS and ELOVL2 and FHL2 DNA Methylation
Whole-blood ELOVL2 and FHL2 methylation was measured in the LLS using EpiTYPER assay, and Spearman’s correlation with age was calculated for each CpG unit. Figure 5 shows the increase in the methylation of CpG units CpG_11.12.13.14 (ELOVL2) and CpG_9.10 (FHL2) with age in the LLS. As depicted in Supplementary Figure 9, the highest correlations in the combined group of LLS nonagenarians, offspring, and spouses were 0.749 (ELOVL2 CpG_11.12.13.14) and 0.634 (FHL2 CpG_9.10 and CpG_19.20).
Figure 5.
Scatterplot of age and methylation level for CpG unit CpG_11.12.13.14 in ELOVL2 and CpG_9.10 in FHL2 in the Leiden Longevity Study. Black circles (blue circles online) indicate male nonagenarians, gray circles (orange circles online) indicate female nonagenarians, gray diamonds (green diamonds online) indicate male offspring, black diamonds (purple diamonds online) indicate female offspring, gray squares (blue squares online) indicate male spouses, and black squares (red squares online) indicate female spouses.
To determine whether the methylation levels of ELOVL2 and FHL2 were also associated with familial longevity, we compared the methylation of CpG units CpG_11.12.13.14 (ELOVL2) and CpG_9.10 (FHL2) between the LLS offspring and their spouses. Although these groups have shown a wide range of metabolism-related differences (23) indicating a difference in metabolic health and biological age, the methylation levels of these CpG units did not differ (β = 0.27 (95% confidence interval [CI] −0.20 to 0.74), p = .257 and β = −0.03 (95% CI −0.33 to 0.27), p = .849, respectively), indicating that the methylation levels of ELOVL2 and FHL2 are not associated with biological age represented by familial longevity.
Next, we determined whether the methylation levels of ELOVL2 and FHL2 associate with prospective survival at high ages (LLS nonagenarians (n = 862 [798 deaths; ELOVL2] or n = 864 [801 deaths; FHL2]) and middle age (LLS offspring and spouses (n = 2,258 [147 deaths; ELOVL2] or n = 2,283 [147 deaths; FHL2]). We found no association with survival, neither for the methylation level of CpG units CpG_11.12.13.14 (ELOVL2) (hazard ratio [HR] = 1.01, 95% CI 0.99 to 1.02, p = .445 [LLS nonagenarians]; HR = 1.01, 95% CI 0.98 to 1.05, p = .382 [LLS offspring + spouses]) nor for CpG_9.10 (FHL2) (HR = 1.01, 95% CI 0.99 to 1.02, p = .486 [LLS nonagenarians]; HR = 1.01, 95% CI 0.95 to 1.07), p = .724 [LLS offspring + spouses]).
Methylation levels could be influenced by infections (31–33). Therefore, we determined the association of CMV serostatus with the methylation level of CpG units CpG_11.12.13.14 (ELOVL2) and CpG_9.10 (FHL2) in the LLS participants. LLS nonagenarians infected with CMV exhibit increased methylation levels of ELOVL2, comparable with an increase of ~3.5 years (β = 1.11, 95% CI 0.22 to 1.99, p = .015) (Table 1). The association of age with the methylation level of CpG unit CpG_11.12.13.14 is not affected by adding CMV serostatus to the model (data not shown).
Table 1.
Association of CpG_11.12.13.14 (ELOVL2) and CpG_9.10 (FHL2) Methylation With CMV Infection
| LLS Offspring + Spouses | LLS Nonagenarians | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | n | β | SE | 95% CI | p | n | β | SE | 95% CI | p | |
| CpG_11.12.13.14 | CMV infection | 2,131 | 0.35 | 0.25 | −0.13 to 0.84 | .15 | 790 | 1.11 | 0.45 | 0.22 to 1.99 | .01 |
| CpG_9.10 | CMV infection | 2,155 | −0.05 | 0.14 | −0.33 to 0.23 | .74 | 792 | 0.15 | 0.35 | −0.54 to 0.85 | .66 |
Note: 95% CI = 95% confidence interval; CMV = cytomegalovirus; LLS = Leiden Longevity Study; SE = standard error.
Discussion
Epigenomic studies on whole blood from individuals of different age were concordant in demonstrating that ELOVL2 and FHL2 CpG islands get hypermethylated during aging. We used different models (publicly available DNA methylation repositories, in vitro cellular models and human cohorts) to explore several aspects of age-dependent epigenetic shift of ELOVL2 and FHL2.
First, we confirmed age-dependent hypermethylation of ELOLV2 and FHL2 in blood and granulocytes. Then, we investigated whether this methylation pattern could be reproduced in other human tissues. A careful analysis of currently available Infinium 450k data demonstrated that this is the case, that is, ELOVL2 and FHL2 DNA methylation increases with age in most of the analyzed tissues (24/30 for cg16867657 within ELOVL2 and 22/30 for cg06639320 within FHL2). Differently from Horvath’s epigenetic clock, ELOVL2 cg16867657 did not show any trend toward hypermethylation in cerebellum. In this tissue, Horvath’s epigenetic clock predicts age with reasonable accuracy, although showing a slower tissue aging (34). Even if this point deserves deeper investigation through more representative cohorts, these results suggest that, differently from Horvath’s epigenetic clock, ELOVL2 cg16867657 methylation is related to cell replication. Indeed, it is well established that cell turnover greatly varies between different tissues: beside tissues with high proliferative rate (such as the hematopoietic system and the epidermis) there are tissues that, although not continuously replicating, have a substantial replicative potential in response to injury (such as the skeletal muscle and the liver). Estimations of replacement rates in different tissues through the (14)C bomb-pulse method have shown that cell turnover occurs in tissues like skeletal muscle and cerebral cortex (35,36), but that is definitely lower in cerebellum (36), consistent with the higher ration neurons/glial cells compared with the other brain regions (37). These observations perfectly fit with the trend of age-associated hypermethylation of ELOVL2 cg16867657, which was evident in all the tissues but cerebellum and aortic tissue, another tissue with low replication rate (38). As far as it concerns FHL2, the relationship between methylation and tissue cell turnover was less straightforward. For example, cg06639320 methylation was age correlated in cerebellum but not in epidermis, a tissue with higher replication rate. This could indicate that FHL2 methylation is not strictly associated to cell replication and that it can be affected by other age-associated factors.
Using two different in vitro models (DFs and HUVECs), we showed that the target regions within ELOVL2 and, to a lesser extent, FHL2 get hypermethylated with cell divisions. The increase in cellular methylation is not a consequence of senescence, as it is already evident at low-middle passages and it does not increase in ultra-senescent cells compared with senescent cells. Also the DNAm age estimated on the basis of Horvath’s epigenetic clock correlates with in vitro cell passages (10), confirming that cell division is tracked by methylation of multiple CpG sites within the genome (39–41). However, as aforementioned, Horvath’s epigenetic clock is not a measure of mitotic age, as it tracks age in nonproliferative tissues such as cerebellum (10).
We used a cohort of 19 participants, assessed every 5 years for 20 years, to evaluate intra-individual changes of ELOVL2 and FHL2 methylation over time. Consistently with previous results, participants displayed a progressive hypermethylation of ELOVL2 with time, while they were more heterogeneous regarding FHL2 changes in methylation.
Several studies, carefully gathered in a recent review (42), suggest that the number of cell divisions within a tissue affects aging, with a decrease in division rate contributing to extend life span. Therefore, we evaluated if DNA methylation of ELOVL2 and FHL2 CpG islands was associated with longevity and/or mortality in the LLS cohort. We previously demonstrated that LLS offspring are metabolically healthier and biologically younger than their spouses in terms of morbidity prevalence (23,24). However, ELOVL2 and FHL2 methylation levels did not show significant differences between nonagenarian siblings and their partners. Then we tested ELOVL2 and FHL2 methylation status at baseline for association with prospective mortality across two generations in LLS families, but also in this case we did not find a significant association. Collectively, these results indicate that ELOVL2 and FHL2 methylation is not associated with longevity/mortality in the LLS. In previous studies, Horvath’s and Hannum’s age predictors have been successfully associated with physical/cognitive fitness and mortality and have therefore been proposed as reliable markers of biological age (12–15). However, it must be considered that the mean age of the cohorts analyzed by Marioni and colleagues and Christensen and colleagues ranges from 66 to 79 and 33 to 76 years, respectively, and it is therefore definitely lower than that of nonagenarians (94 years). It is conceivable that the performance of a biomarker in predicting mortality can vary according to the baseline age of individuals, and therefore our results are not directly comparable with those of Marioni and colleagues. Recently telomere length, which is a marker of cell replication, has been investigated in the LLS (27). Leukocyte telomere length did not differ between middle-aged LLS offspring and their spouses, but it was significantly associated with mortality. However, it must be considered that age-associated telomere shortening could be a consequence not only of DNA replication during cell divisions, but also of accumulation of oxidative stress (43), which on the contrary could not affect ELOVL2 methylation. This consideration could explain why different markers of cell replication could provide different results when correlated to mortality.
Importantly, the hypothesis that the increase in ELOVL2 methylation occurs with cell replication fits well with the observation that in LLS nonagenarians ELOVL2 is hypermethylated in individuals infected with CMV. Chronic CMV infection leads to oligoclonal expansion of CD8+ T cells which tend to accumulate in megaclones, filling the immunological space at the expense of the TCR repertoire for other antigens (44,45). Therefore, ELOVL2 hypermethylation in CMV-positive nonagenarians could be informative of a higher number of replication events in a fraction of WBC.
In a previous study, Kim and colleagues demonstrated that hypermethylation of Nkx2-5 in endometrium mirrored somatic cell divisions and assumed that the observed changes were the result of stochastic error (46). At present, we are not able to say if the progressive increase of cells methylated in ELOVL2 and FHL2 CpG island with age is part of a developmental program (47) or a stochastic event. It is reasonable to suppose that, at least for ELOVL2, DNA methylation occurs in concomitance with DNA replication. Dnmt1 has been shown to interact with the replication machinery (48–50) and, although it has been classically considered a maintenance methyltransferase, it has been shown to have also de novo activity both in vitro and in vivo (51). While Dnmt1 activity decreases during in vitro replication of WI-38 fibroblasts, Dnmt3b shows the opposite trend (52,53) and could therefore have a role in hypermethylation of ELOVL2 with cellular passages. The fact that ELOVL2 hypermethylation occurs in several tissues suggests that this genomic region has a chromatin structure with a given probability of being target of methyltransferase activity throughout cell replications (54,55).
In conclusion, we suggest that methylation of ELOVL2 could be a calculator (monitor) of cell divisions. Further investigations should clarify the biological relevance of this clock, in particular in the context of aging cells and organisms.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was supported by the European Union’s Seventh Framework Programme (grant agreements 259679 “IDEAL,” 602757 “HUMAN,” 305280 “MIMOMICS,” and 613979 “MyNewGut), by the European Union’s H2020 Programme (grant agreement 634821 “PROPAG-AGEING”), and in part by the National Institute for Public Health and the Environment and the Ministry of Health, Welfare and Sport of The Netherlands (S/340005). J.M.S. was supported in part by the National Institutes of Health grant R37 AG016694. M.D.C. was supported in part by Glenn/AFAR Postdoctoral Fellowship for Translational Research on Aging.
Conflict of Interest
The authors declare that there are no conflicts of interest.
Supplementary Material
Acknowledgments
We thank B. Nagarajah and A. de Klerk for DNA isolations of the Doetinchem cohort.
References
- 1. Baker GT, 3rd, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–239. doi:10.1016/0531-5565(88)90025-3 [DOI] [PubMed] [Google Scholar]
- 2. Johnson TE. Recent results: biomarkers of aging. Exp Gerontol. 2006;41:1243–1246. doi:10.1016/j.exger.2006.09.006 [DOI] [PubMed] [Google Scholar]
- 3. Sprott RL. Biomarkers of aging and disease: introduction and definitions. Exp Gerontol. 2010;45:2–4. doi:10.1016/j.exger.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 4. Deelen J, Beekman M, Capri M, Franceschi C, Slagboom PE. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. BioEssays News Rev Mol Cell Dev Biol. 2013;35:386–396. doi:10.1002/bies.201200148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engelfriet PM, Jansen EHJM, Picavet HSJ, Dollé MET. Biochemical markers of aging for longitudinal studies in humans. Epidemiol Rev. 2013;35:132–151. doi:10.1093/epirev/mxs011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meissner C, Ritz-Timme S. Molecular pathology and age estimation. Forensic Sci Int. 2010;203:34–43. doi:10.1016/j.forsciint.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 7. Bacalini MG, Friso S, Olivieri F, et al. Present and future of anti-ageing epigenetic diets. Mech Ageing Dev. 2014;136–137:101–115. doi:10.1016/j.mad.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 8. Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi:10.1371/journal.pone.0014821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi:10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi:10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weidner CI, Lin Q, Koch CM, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24. doi:10.1186/gb-2014-15-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi:10.1186/s13059-015-0584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christiansen L, Lenart A, Tan Q, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. doi:10.1111/acel.12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horvath S, Pirazzini C, Bacalini MG, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging. 2015;7:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marioni RE, Shah S, McRae AF, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44:1388–1396. doi:10.1093/ije/dyu277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Q, Weidner CI, Costa IG, et al. DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy. Aging. 2016;8:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garagnani P, Bacalini MG, Pirazzini C, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi:10.1111/acel.12005 [DOI] [PubMed] [Google Scholar]
- 18. Bacalini MG, Boattini A, Gentilini D, et al. A meta-analysis on age-associated changes in blood DNA methylation: results from an original analysis pipeline for Infinium 450k data. Aging. 2015;7:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi:10.1093/hmg/ddt531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marttila S, Kananen L, Häyrynen S, et al. Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics. 2015;16:179. doi:10.1186/s12864-015-1381-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rönn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–3813. doi:10.1093/hmg/ddv124 [DOI] [PubMed] [Google Scholar]
- 22. Steegenga WT, Boekschoten MV, Lute C, et al. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age. 2014;36:9648. doi:10.1007/s11357-014-9648-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slagboom PE, Beekman M, Passtoors WM, et al. Genomics of human longevity. Philos Trans R Soc Lond B Biol Sci. 2011;366:35–42. doi:10.1098/rstb.2010.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westendorp RGJ, van Heemst D, Rozing MP, et al. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: The Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. doi:10.1111/j.1532-5415.2009.02381.x [DOI] [PubMed] [Google Scholar]
- 25. Verschuren WM, Blokstra A, Picavet HS, Smit HA. Cohort profile: the Doetinchem Cohort Study. Int J Epidemiol. 2008;37:1236–1241. doi:10.1093/ije/dym292 [DOI] [PubMed] [Google Scholar]
- 26. Schoenmaker M, de Craen AJ, de Meijer PH, et al. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet. 2006;14:79–84. doi:10.1038/sj.ejhg.5201508 [DOI] [PubMed] [Google Scholar]
- 27. Deelen J, Beekman M, Codd V, et al. Leukocyte telomere length associates with prospective mortality independent of immune-related parameters and known genetic markers. Int J Epidemiol. 2014;43:878–886. doi:10.1093/ije/dyt267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Derhovanessian E, Maier AB, Beck R, et al. Hallmark features of immunosenescence are absent in familial longevity. J Immunol. 2010;185:4618–4624. doi:10.4049/jimmunol.1001629 [DOI] [PubMed] [Google Scholar]
- 29. Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. [DOI] [PubMed] [Google Scholar]
- 30. Olivieri F, Lazzarini R, Recchioni R, et al. MiR-146a as marker of senescence-associated pro-inflammatory status in cells involved in vascular remodelling. Age. 2013;35:1157–1172. doi:10.1007/s11357-012-9440-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212:1563–1573. doi:10.1093/infdis/jiv277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kananen L, Nevalainen T, Jylhävä J, et al. Cytomegalovirus infection accelerates epigenetic aging. Exp Gerontol. 2015;72:227–229. doi:10.1016/j.exger.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 33. Pacis A, Tailleux L, Morin AM, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015. doi:10.1101/gr.192005.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horvath S, Mah V, Lu AT, et al. The cerebellum ages slowly according to the epigenetic clock. Aging. 2015;7:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinemeier KM, Schjerling P, Heinemeier J, Magnusson SP, Kjaer M. Lack of tissue renewal in human adult Achilles tendon is revealed by nuclear bomb (14)C. FASEB J. 2013;27:2074–2079. doi:10.1096/fj.12-225599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi:10.1016/j.cell.2005.04.028 [DOI] [PubMed] [Google Scholar]
- 37. Azevedo FAC, Carvalho LRB, Grinberg LT, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–541. doi:10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- 38. Neese RA, Misell LM, Turner S, et al. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci USA. 2002;99:15345–15350. doi:10.1073/pnas.232551499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koch CM, Wagner W. Epigenetic biomarker to determine replicative senescence of cultured cells. Methods Mol Biol Clifton NJ. 2013;1048:309–321. doi:10.1007/978-1-62703-556-9_20 [DOI] [PubMed] [Google Scholar]
- 40. Sidler C, Woycicki R, Kovalchuk I, Kovalchuk O. WI-38 senescence is associated with global and site-specific hypomethylation. Aging. 2014;6:564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi:10.1186/gb-2014-15-2-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Magalhães JP, Faragher RGA. Cell divisions and mammalian aging: integrative biology insights from genes that regulate longevity. Bioessays. 2008;30:567–578. doi:10.1002/bies.20760 [DOI] [PubMed] [Google Scholar]
- 43. Kawanishi S, Oikawa S. Mechanism of telomere shortening by oxidative stress. Ann NY Acad Sci. 2004;1019:278–284. doi:10.1196/annals.1297.047 [DOI] [PubMed] [Google Scholar]
- 44. Capri M, Monti D, Salvioli S, et al. Complexity of anti-immunosenescence strategies in humans. Artif Organs. 2006;30:730–742. doi:10.1111/j.1525-1594.2006.00295.x [DOI] [PubMed] [Google Scholar]
- 45. Franceschi C, Bonafè M, Valensin S. Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine. 2000;18:1717–1720. [DOI] [PubMed] [Google Scholar]
- 46. Kim JY, Tavaré S, Shibata D. Counting human somatic cell replications: methylation mirrors endometrial stem cell divisions. Proc Natl Acad Sci USA. 2005;102:17739–17744. doi:10.1073/pnas.0503976102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Magalhães JP. Programmatic features of aging originating in development: aging mechanisms beyond molecular damage? FASEB J. 2012;26:4821–4826. doi:10.1096/fj.12-210872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. [DOI] [PubMed] [Google Scholar]
- 49. Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. [DOI] [PubMed] [Google Scholar]
- 50. Schermelleh L, Haemmer A, Spada F, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi:10.1093/nar/gkm432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39:310–318. doi:10.1016/j.tibs.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 52. Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. [DOI] [PubMed] [Google Scholar]
- 53. Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. [DOI] [PubMed] [Google Scholar]
- 54. Landan G, Cohen NM, Mukamel Z, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–1214. doi:10.1038/ng.2442 [DOI] [PubMed] [Google Scholar]
- 55. Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi:10.1038/ng.946 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.