Abstract
The human gastrointestinal microbiota plays a key homeostatic role in normal functioning of physiologic processes commonly undermined by aging. We used a previously validated 34-item frailty index (FI34) to identify changes in gut microbiota community structure associated with biological age of community-dwelling adults. Stool 16S rRNA cDNA libraries from 85 subjects ranging in age (43–79) and FI34 score (0–0.365) were deep sequenced, denoised, and clustered using DADA2. Subject biological age but not chronological age correlated with a decrease in stool microbial diversity. Specific microbial genera were differentially abundant in the lower, middle, and upper 33rd percentiles of biological age. Using Sparse Inverse Covariance Estimation for Ecological Association and Statistical Inference (SPIEC-EASI) and Weighted Gene Co-Expression Network Analysis (WGCNA), we identified modules of coabundant microbial genera that distinguished biological from chronological aging. A biological age-associated module composed of Eggerthella, Ruminococcus, and Coprobacillus genera was robust to correction for subject age, sex, body mass index, antibiotic usage, and other confounders. Subject FI34 score positively correlated with the abundance of this module, which exhibited a distinct inferred metagenome as predicted by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). We conclude that increasing biological age in community-dwelling adults is associated with gastrointestinal dysbiosis.
Keywords: Frailty, Dysbiosis, Diversity, Coabundance
Since 1950, the growth rate of the global population of persons aged 60 and older has exceeded that of the world population as a whole. This global trend, due in part to worldwide decreases in fertility and mortality rates, toward a predominantly aged population is considered irreversible and unlikely to change (1). Widespread healthy human aging may support the transition to an era of expanded elder health care.
Human aging is characterized by a progressive decline in organ function and associated physical function. The term frailty is regarded as one’s risk for age-related adverse health outcomes including but not limited to falls, disability, institutionalization, and mortality (2,3). Clinical characteristics of frailty are broadly defined and include anorexia, sarcopenia, cognitive decline, relative immobility, and subsequent dependence in performing activities of daily living (4,5). Frailty is not necessarily a function of one’s chronological age. Persons of a given age are not at equal risk for adverse outcomes or shortened life expectancy (6). Frailty, related to one’s “biological age,” is thus considered a distinct clinical syndrome that may be specially monitored and addressed to promote healthy aging in at-risk individuals (7).
The human gastrointestinal (GI) microbiota, comprising more than 1 trillion organisms, has been shown to play a key homeostatic role in normal functioning of physiologic processes commonly undermined by aging. Germ-free and germ-attenuated animal models have implicated characteristic age-associated derangements such as chronic inflammation, decreased bowel motility, altered locomotion, and impaired cognition with changes in the gut microbiota (8–11). Dysbiosis in older persons has been likewise attributed to age-related changes in physiology and lifestyle such as diminished bowel enzymatic activity, barrier function, as well as changes in community residence, diet, and pharmacotherapy consistent with geriatric conditions (12–14).
Recent studies conducted in the United Kingdom suggest that frailty as measured by the accumulated health deficit (Rockwood) model is associated with decreased GI microbiota alpha-diversity in community-dwelling adults (15). In this same population, changes in GI microbiota have been suggested to more closely correlate with biological age than chronological age (16). Specific determinants of these observations and their potential significance in the frailty process remain to be fully explored. Moreover, geographic location has been shown to influence various properties of the GI microbiota (17). Using a 34-item frailty index (FI34) previously validated to predict mortality (18), we hypothesized that diversity in the gut microflora changes with biological aging in community-dwelling adults. We further hypothesized that these changes are more specifically related to one’s estimated biological age rather than chronological age and that changes in the gut microbiota differentiate biological from chronological aging.
Methods
Participant Recruitment and Sample Collection
A total of 85 subjects living in the Greater Metropolitan Area of New Orleans, Louisiana, enrolled in the Healthy Aging Family Study (HAFS) (18) volunteered to additionally participate in the study described herein. These subjects represent a subset of the full HAFS cohort, which is composed of randomly sampled offspring of nonagenarians. Subject age ranged from 43 to 79, FI34 score from 0 to 0.365, and body mass index (BMI) from 18.6 to 41.9 (Table 1). A subset of participants (8%) reported antibiotic usage in the 6 months prior to participation. Information on antibiotic usage was not available for 13 of 85 subjects (15%). The remaining participants (76%) reported no recent history of antibiotic usage. Standardized, freezer-safe stool collection kits were distributed to participants. Samples were collected at home (immediately stored at −20°C) within 24 hours of the testing visit then transferred to −80°C for longer-term storage. The activities of this study and the parent study were approved by local Institutional Review Boards. All participants provided written informed consent prior to enrollment into both studies.
Table 1.
Cohort demographics and clinical data
| Cohort | Female | Male |
|---|---|---|
| Demographics | ||
| n | 53 | 32 |
| Age (y) | 63 ± 6 | 64 ± 8 |
| FI34a | 0.12 ± 0.1 | 0.13 ± 0.1 |
| Body mass index (kg/m2) | 26 ± 5 | 29 ± 5 |
| White | 53 (100%) | 32 (100%) |
| Nonagenarian parent | ||
| None | 2 (4%) | 0 (0%) |
| One | 44 (83%) | 28 (88%) |
| Both | 7 (13%) | 4 (12%) |
| Medical history | ||
| Obstructive pulmonary disease | 15 (28%) | 5 (16%) |
| Cardiovascular disease | 24 (45%) | 20 (62%) |
| Cerebrovascular disease | 2 (4%) | 2 (6%) |
| Diabetes | 4 (8%) | 2 (6%) |
| Neurodegenerative disease | 0 (0%) | 0 (0%) |
| Labs | ||
| Triglycerides | 121 ± 66 | 133 ± 68 |
| Cholesterol | 200 ± 38 | 184 ± 30 |
| Hospital nights past 12 mo | ||
| None | 1 (2%) | 4 (12%) |
| 1 | 47 (89%) | 25 (78%) |
| 2–3 | 2 (4%) | 2 (6%) |
| 4–6 | 3 (6%) | 1 (3%) |
| Antibiotics past 6 mo | ||
| Yes | 4 (8%) | 3 (9%) |
| No | 40 (75%) | 25 (78%) |
| Unknown | 9 (17%) | 4 (13%) |
Note: Eighty-five subjects participated in this study. All subjects were community-dwelling adults recruited from the greater metropolitan area of New Orleans, Louisiana.
aDimensionless ratio.
Frailty Index
Frailty was estimated using a FI34 of lifetime health history and function variables previously validated to predict time to death (18). Questionnaire items have been previously published (18).
DNA Purification and 16S rRNA Sequencing
DNA was extracted from 200 mg of bead-homogenized stool using the Qiagen QIAamp DNA Stool MiniKit (Germantown, MD). Purified DNA was subjected to polymerase chain reaction amplification using previously published universal primers for the 16S rRNA hypervariable region V3–V4 (19). Sequencing was performed on an Illumina MiSeq. A minimum sample sequencing depth of 9,097 reads was achieved. Data in this study are derived from a single sequencing run. See Supplementary Methods for further details.
Sequence Variants, Operational Taxonomic Units, and Sample Library Normalization
Raw sequences were truncated to 250 base pairs, denoised, chimera filtered, and clustered into sequence variants using DADA2 v1.1.5 (20). For taxonomically informed analyses, operational taxonomic units (OTU) were generated in DADA2 by taxonomic classification of sequence variants using the Greengenes 97% reference database v13.8 and a minimum bootstrap confidence level of 80. OTUs with matching Greengenes database identifiers were clustered to single OTUs prior to OTU-based analyses. OTUs unresolved for taxonomy were omitted from analyses. Where indicated in the text, variance-stabilized OTU counts were generated using DESeq2. For a more detailed description of the analytical methods used in this study, refer to the Supplementary Methods.
Data Analyses and Statistics
All analyses were performed using the R platform. Beta-diversity p values were generated via distance-based redundancy analysis (dbRDA) and permutational multivariate analysis of variance. Taxon differential abundance p values were generated by the Wald test between levels of age or FI34 in DESeq2. All other p values were generated via multivariate linear mixed-effects modeling in R. Model variables included age, FI34, sex, BMI, antibiotic usage in the past 6 months, sample read count, and subject family membership. Family membership was modeled as a random effect. See Supplementary Methods for a more detailed description of analytical methods used in this study.
Results
Biological Age, Not Chronological Age, Is Associated With Changes in Gut Microbiota Diversity
Alpha- and beta-diversity model two distinct but related aspects of ecological diversity: “within-sample” diversity and “between-sample” diversity. Sample alpha-diversity may be subdivided into two properties: (a) richness and (b) evenness. Sample richness is the number of unique denoised sequenced read clusters known as “sequence variants” present in a single biological sample. Sample evenness is a measure of the uniformity in relative abundance of sequence variants within a sample. Sample richness and evenness were evaluated separately using indices tailored to either property.
Biological Age Correlates With Decreased Subject Stool Richness or Within-Sample Diversity
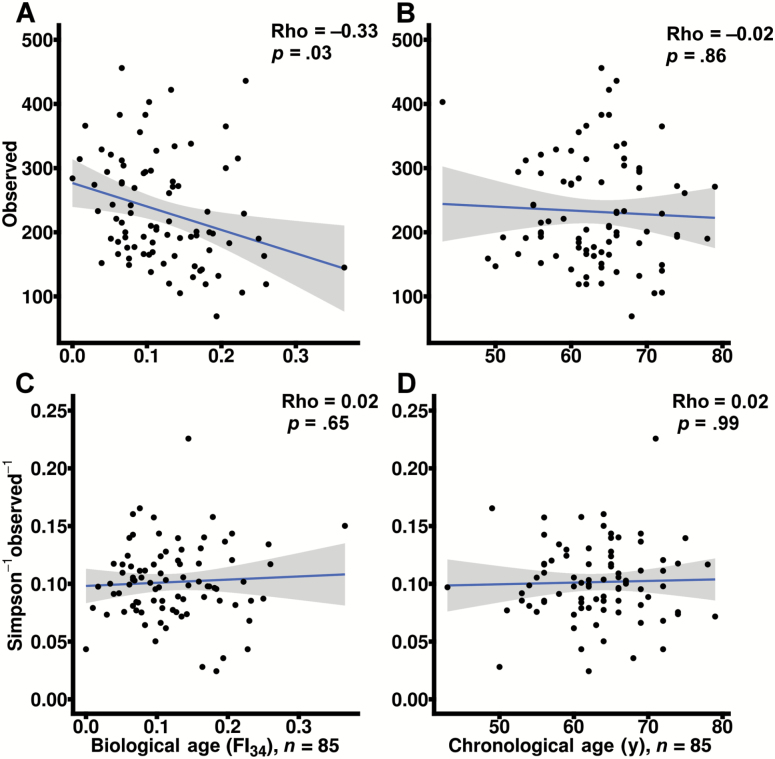
Observed sample richness was calculated on sequence variants generated from subject stool. Subject biological age as measured by the FI34 significantly correlated with decreased observed richness (Figure 1A) after correction for BMI, sex, antibiotic usage, chronological age, sample read count, and family membership modeled as a random effect (p = .03). Subject chronological age failed to correlate with changes in richness after similar correction for clinical and technical covariates (Figure 1B). Correlation statistics between observed richness and FI34 or chronological age (rho = −0.33 and −0.02, respectively) were significantly different by co-correlation analysis (p < .05) (21). Removal of a potential outlying subject (FI34 = 0.365, Figure 1A) yielded no change in findings (p < .05, data not shown).
Figure 1.
FI34 score negatively correlates with stool sample richness. The total number of observed unique sequence variants (denoised sequenced read clusters) in each sample (observed richness) and the evenness of sequence variants in each sample based on the inverse Simpson evenness index were calculated and plotted against FI34 scores and chronological age (A−B and C−D, respectively). Biological age (A and C) and chronological age (B and D) p values are corrected for subject body mass index, sex, sample read depth, family membership, antibiotic usage, and chronological age or FI34. Data are derived from raw sequence variant counts. Spearman rho correlation statistics are additionally provided as indicated. Linear best-fit lines are plotted with shaded 95% confidence intervals.
In contrast to observed richness, true richness requires infinitely large sequencing depth. Richness indices such as the objective Bayes negative binomial (OBNB) estimate the observed and unobserved sample richness, providing for a closer approximation of true richness in the absence of exhaustive sampling. Like observed richness, OBNB richness inversely correlated with biological age after correction for covariates (p = .03; Supplementary Figure 1A). OBNB did not correlate with subject chronological age before or after correction for covariates (Supplementary Figure 1B). Correlation statistics between OBNB richness and FI34 as well as OBNB and chronological age were significantly different (p < .05).
Two indices of evenness were evaluated and compared for their potential association with chronological or biological age. Neither the Pielou (data not shown) nor the inverse Simpson evenness index (Figure 1C and D) correlated with FI34 or chronological age after correction for clinical and technical covariates. Additional correction for observed sample richness did not alter these findings on evenness, which may be confounded by richness in some indices for purely mathematical reasons (22).
Previous studies have explored the relationship between stool alpha-diversity and frailty with combined measures of richness and evenness (15). We likewise confirmed this observation in our cohort using the same mixed-methodology. Shannon’s diversity index in subject stool samples significantly and inversely correlated with biological age but not chronological age (Supplementary Figure 1C and D).
Lastly, these analyses were repeated to compare random- and fixed-effect modeling of family membership. Fixed-effect modeling yielded identical findings to random-effect modeling (Supplementary File 1A). Family membership in our cohort failed to significantly correlate with stool alpha-diversity in both random- and fixed-effect models (p > .05, data not shown). In addition, subject BMI, sex, and antibiotic usage failed to correlate with alpha-diversity (p > .05, data not shown). Sample read counts significantly correlated with alpha-diversity (p < .05, data not shown).
Biological Age Correlates With Subject Stool Beta-Diversity or Between-Sample Diversity
The UniFrac index estimates the phylogenetic similarity of OTUs between a pair of samples (23). Relative similarities are calculated for all sample pairs and then plotted as distances in a co-ordinate plane to identify clusters of samples. These distances may be additionally weighted for similarity in OTU abundance between samples.
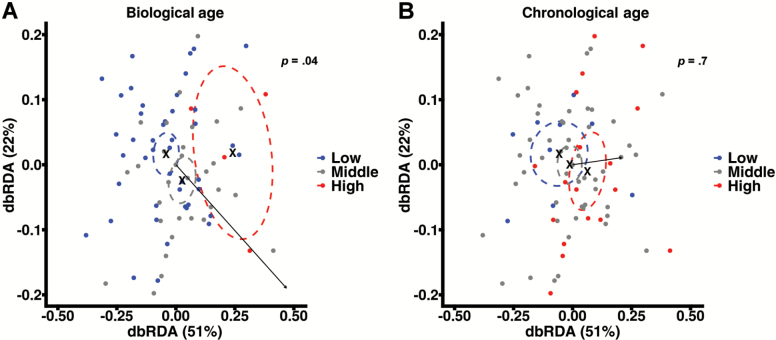
OTUs were generated by assigning taxonomy to sequence variants. OTUs with identical taxonomic assignments were clustered prior to analysis. Both unweighted and weighted UniFrac distances were calculated and tested for association with subject FI34 score or chronological age via dbRDA and permutational multivariate analysis of variance. Biological age significantly correlated with unweighted UniFrac distances after correction for confounders including sample total read counts (p = .04; Figure 2A). Weighted UniFrac distances failed to correlate with biological age in both raw OTU counts and OTU counts normalized to equal variance after corrections (p = .43 and .14, respectively, data not shown). Chronological age failed to correlate with both unweighted (Figure 2B) and weighted (data not shown) UniFrac distances before and after correction for confounders. The effect of biological age on between-subject diversity was similar in direction but greater in magnitude than chronological age as indicated by the dbRDA vectors (black arrows, Figure 2A and B). Subject BMI exhibited a strong but statistically insignificant effect (p > .05; Supplementary Figure 2). Subject sex and antibiotic usage history effects were weak and statistically insignificant (p > .05; Supplementary Figure 2). Sample read count exhibited a modest significant effect (p = .02; Supplementary Figure 2). Overall, these data suggest the relative phylogenetic similarity of stool microbial communities based on subject biological age but not chronological age.
Figure 2.
FI34 score correlates with between-subject similarity in stool taxonomy. Unweighted UniFrac taxonomic similarities were calculated on raw OTU counts between all pairwise comparisons of subject stool samples. Between-subject similarities are plotted via distance-based redundancy analysis (dbRDA). Each point represents a single subject sample. Between-point distances indicate relative taxonomic similarity of subject stool samples. Point colors indicate subject FI34 or chronological age groups separated into low (blue), middle (gray), or upper (red) 33rd percentiles of the full (A) FI34 or (B) chronological age range. Arrows show the magnitude and direction of FI34 or chronological age correlation. Correlation significance (p) is corrected for subject body mass index, sex, antibiotic usage, sample read depth, family membership, and chronological age or FI34. Dashed ovals indicate the 95% confidence interval about the midpoints of low, middle, and high FI34 or chronological age. Midpoints within confidence intervals are indicated by “X”. Supplementary Figure 2 contains dbRDA results of all tested study variables and confounders. Only the FI34 score and sample read count variables significantly correlated with between-subject diversity (p < .05).
Taxon Correlates Distinguish Biological From Chronological Age
Subject FI34 and chronological age were assigned to low, middle, and high tertiles for OTU differential abundance analysis. Low, middle, and high levels contained the lower, middle, and upper 33rd percentiles, respectively, of subject FI34 (low: 0–0.083; middle: 0.091–0.137; high: 0.142–0.365) or chronological age (low: 43–61; middle: 62–66; or high: 67–79) to ensure even group sample sizes for comparison between FI34 and chronological age as correlates of OTU abundance. Change in abundance of genera between low, middle, and high FI34 and chronological age were calculated.
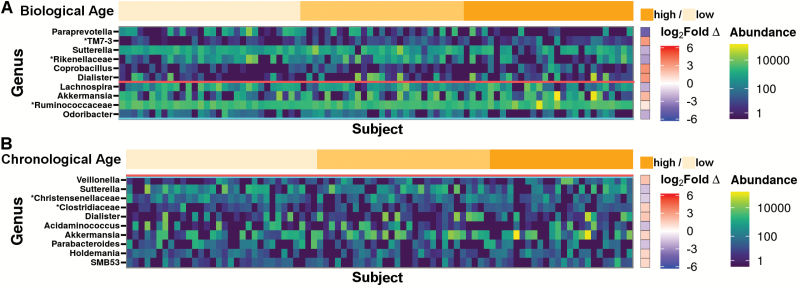
Significant, differentially abundant genera between high and low (Figure 3) and middle and low FI34 scores were identified. Subjects with high FI34 scores, compared with those with low FI34, exhibited increased abundance of Coprobacillus (q = 0.08), Dialister (q = 0.08), and a TM7 candidate-phylum OTU (false-discovery rate q = 0.04; Figure 3A). High FI34 was also associated with decreased Paraprevotella (q = 0.02), Sutterella (q = 0.08), and a Rikenellaceae family OTU (q = 0.08). Middle FI34, compared with low FI34, was associated with greater TM7 candidate-phylum OTU abundance (q = 0.04, log2 fold-change = 3.06). In contrast to FI34 score, all comparisons between levels of chronological age failed to significantly associate with the abundances of individual genera (Figure 3B). The remaining genus-level OTUs failed to significantly correlate in abundance with FI34 score or chronological age (Supplementary Figures 3 and 4).
Figure 3.
Divergent taxa correlate with FI34 score and chronological age. DESeq2-normalized genus-level OTU abundances are expressed as a heatmap. Taxon abundances are sorted along the x-axis by (A) increasing FI34 score or (B) chronological age. Differential abundance of genus-level OTUs between lower (light orange), middle (middle orange), and upper (dark orange) 33rd percentiles of FI34 or chronological age was calculated using DESeq2. Log2 fold-change (Log2 fold ∆) in abundance is expressed as a ratio of the taxon abundance in high-level over low-level FI34 or chronological age. Shades of red fold-change indicate higher abundance in the high group, whereas shades of blue indicate lower abundance in the high group. Group sizes were chosen to normalize sample size between FI34 and chronological age groups (FI34: low n = 30, middle n = 27, high n = 28; Age: low n = 32, middle n = 29, high n = 24). OTUs were filtered for prevalence in at least 25% of subjects. Significant differences in OTU abundances were observed between the lower and upper levels of FI34 scores but not chronological age after correction for multiple comparisons by false-discovery rate (FDR; q < 0.1 above horizontal red bar, p < .05 above the horizontal blue bar). Taxa are sorted down the y-axis by increasing p values. Low FI34 scores ranged from 0 to 0.083, middle scores ranged from 0.091 to 0.137, and high scores ranged from 0.142 to 0.365. Supplementary Figures 3 and 4 contain expanded heatmaps for the results of all tested genera sorted by increasing FI34 score or chronological age, respectively. “*” indicates substitution of higher clade name in lieu of unassigned genus.
Genus-Level Subcommunities Correlate With Biological Age
Dynamic host properties may alter the survival or clearance of groups of interacting, inter-related intestinal microbiota (24). Functional metabolic activities of microbial subcommunities may encourage their formation and conservation in the host and in hosts of similar frailty status (25). Microbial co-occurrence networks are used to identify groups of coabundant organisms. Once identified, the abundance of these groups may then be correlated with clinical variables of interest (26).
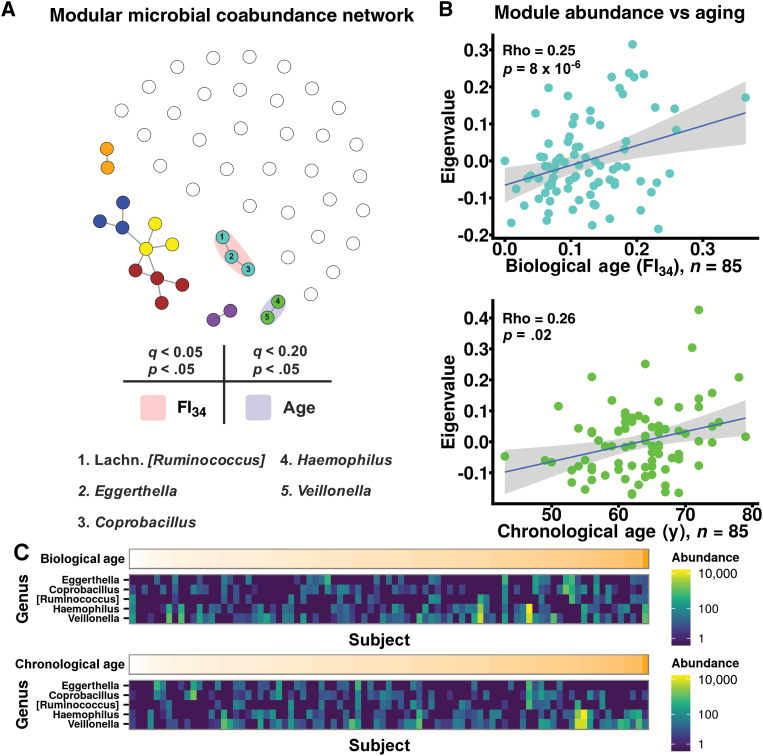
OTUs were collapsed to the genus level, filtered for at least 25% prevalence, and subjected to Sparse Inverse Covariance Estimation for Ecological Association and Statistical Inference (SPIEC-EASI). Coabundant genera were identified by SPIEC-EASI ranging in correlation strength from −0.24 to 0.22 (Supplementary File 1B). Follow-up Weighted Gene Co-expression Network Analysis (WGCNA) on all positive correlations detected seven subcommunities or modules of co-occurring genera (represented as colors) numbering two to four per module (Figure 4A, Supplementary Figure 5 and File 1C). Genera that failed assignment to a module were given the color white and disconnected from other genera (Figure 4A). FI34 significantly and positively correlated with abundance of one module (turquoise; Figure 4A and B, Supplementary Figure 6 and File 1D) after correction for clinical and technical covariates (p = 8 × 10–6) as well as false-discovery rate (q = 6 × 10–5). Omission of a potential outlying subject (FI34 = 0.365) yielded no change in findings (p = 3 × 10–4, q = 2 × 10–3). Chronological age positively correlated with one module after similar correction for covariates (green, p = .02, q = 0.17; Figure 4A and B, Supplementary Figure 7 and File 1D). Comparable results were observed when modeling subject family membership as a random- or fixed-effect (Supplementary File 1A). Family membership as well as BMI, sex, and antibiotic usage failed to significantly associate with turquoise or green module abundances (p > .05, data not shown). Sample read count modestly correlated with turquoise and green module abundances (q < 0.03). Spearman rho correlation coefficients between turquoise module abundance and FI34 as well as green module abundance and chronological age were 0.25 and 0.26, respectively. Turquoise module members consisted of Eggerthella, Coprobacillus, and Lachnospiraceae-family (Lachn.) Ruminococcus. The green module comprised Haemophilus and Veillonella. Biological and chronological-aging-associated module relative abundances were comparable (1.4% ± 0.06% and 1.7% ± 0.09%, respectively, p = .16). The prevalences of the turquoise module genera approximated that of the green module genera (37% ± 7% and 59% ± 6%, respectively, p = .13).
Figure 4.
FI34 score correlates with the abundance of a microbial coabundance module. Genus-level OTUs were filtered for prevalence in at least 25% of subjects and normalized by DESeq2 variance-stabilizing transformation. Correlation coefficients were calculated between all pairs of genera using the Sparse Inverse Covariance Estimation for Ecological Association and Statistical Inference (SPIEC-EASI) algorithm. Positive correlation coefficients were retained for downstream analysis. (A) A microbial coabundance network consisting of positively coabundant genera is plotted. Vertices or points represent a single OTU genus. Edges or lines emanating from nodes connect significantly coabundant genera as inferred by SPIEC-EASI. Relative point positions were plotted according to the competing strengths of their coabundance correlations (Fruchtermann–Reingold projection). Points with similar connectivity were grouped into a module (indicated by color) using Weighted Gene Co-expression Network Analysis (WGCNA). Genera lacking significant coabundance are plotted as empty, white points. Module abundances that correlated with FI34 score or chronological age following correction for clinical and technical confounding (p < .05) and false-discovery rate (FDR; q < 0.05 and q < 0.20) are highlighted in red or blue, respectively. (B) Turquoise and green module abundances (eigenvalue) are plotted against FI34 score or chronological age with respective Spearman rho correlation statistics and p values corrected for clinical and technical confounding. Linear best-fit lines are plotted in blue with shaded 95% confidence intervals. (C) DESeq2-normalized abundances of turquoise and green module OTUs are expressed as heatmaps. OTU abundances are sorted along the x-axis by increasing FI34 score (top panel) or chronological age (bottom panel). Supplementary Figures 6 and 7 contain expanded heatmaps of all modules and member genera sorted by increasing FI34 score or chronological age, respectively.
Age-Associated Subcommunities Possess Distinct Inferred Biochemical Functions
The collective genetic content or “metagenome” of module genera was predicted via Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt; Supplementary Figure 8A). Module biochemical pathways were inferred from the annotated reference genomes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (27). The presence or absence of 6,909 KEGG protein-coding genes was examined in each module (Supplementary Figure 8A). Clustering modules by similarity in KEGG gene content revealed a high degree of variability between modules, suggesting potential module-specific biological activities (vertical dendrogram, Supplementary Figure 8A). Turquoise and green module predicted metagenomes were directly compared. Turquoise and green shared numerous KEGG genes that together made up 64% and 60% of their respective inferred KEGG gene contents (Supplementary Figure 8B). KEGG genes were grouped into canonical biochemical pathways for assessment of higher functionality. Many genes shared between the turquoise and green modules belonged to core biological functions such as amino acid and nucleic acid metabolism, DNA replication and repair, ribosome assembly, and protein translation (Supplementary Figure 9). The remaining unshared genes differentiated the turquoise and green modules (Supplementary Figure 8C). Turquoise and green modules differed primarily in genes associated with “metabolism” and “environmental information processing” followed by “genetic information processing” and “cellular processes” (Supplementary Figure 8C). Ninety-three percentage of “inorganic ion transport and metabolism” genes that were observed in either module were unshared between turquoise and green modules along with 88% of “two-component system,” 62% of “transporters,” 58% of “ABC transporters,” 74% of “signal transduction mechanisms,” 72% of “transcription factors,” and 92% of “protein kinases,” suggesting differences in module sensitivity and response to environmental factors that distinguish biological aging from chronological aging.
Turquoise module genera were enriched over green module genera for the number of unique genes associated with metabolic pathways including “methane metabolism,” “pentose and glucuronate interconversions,” “galactose metabolism,” “glycolysis/gluconeogenesis,” “glycerolipid metabolism,” and “fructose and mannose metabolism” (Supplementary Figure 8C). The turquoise module was further distinguished by genes associated with environmental persistence and host virulence such as “sporulation” (Supplementary Figure 8C) as well as “germination,” “bacterial motility proteins,” “Staphylococcus aureus infection,” and “bacterial toxins” (Supplementary File 1E). “Sporulation” pathway genes (Supplementary File 1F), including the Spo0A master regulator (KEGG ID: K07699; Supplementary File 1G), were enriched in Coprobacillus and Lachn. Ruminococcus (Supplementary Figure 10), whereas threonine aldolase (KEGG ID: K01620; Supplementary File 1G) a source of ethanol-independent acetaldehyde production was found in all three genera. In contrast, the green module was enriched for “nitrogen metabolism” in addition to “glycosyltransferase” genes and “lipopolysaccharide biosynthesis proteins” (Supplementary Figure 8C). These data suggest that biological and chronological age-associated module genera may possess divergent biochemical repertoires and activities.
Discussion
Many health deficits have been individually associated with changes in the GI microbiota (28). The potential interaction between biological aging and gut microbial ecology has been addressed in this study. We have identified both global and specific changes in the GI microbiota that were closely associated with subject biological age but not chronological age. These changes were robust to correction for potential confounders of the gut microbiota that include BMI and recent antibiotic usage. We additionally corrected for participant family relatedness arising from the sampling methodology of this study’s parent study, which has identified a genetic basis of aging and longevity (18). We were unable to identify a relationship between family membership and the gut microbiota in this study. However, a separate, more thorough analysis is needed to draw conclusions on the heritability of gut microbiota community structure within families.
Increasing biological age was associated with pronounced alterations in GI microbiota diversity that were unobserved with chronological age, contributing to the growing body of literature that asserts frailty as a distinct, measurable clinical syndrome that is not otherwise a function solely of chronological age. Frailty has been shown to outperform chronological age as an estimate of risk for age-related adverse health outcomes. These may include GI dysbiosis and subsequent loss of normal gut microbiota function (29).
We examined stool alpha-diversity in subjects with varying biological age and chronological age. We confirmed previously reported observations that frailty correlates with combined measures of richness and evenness in subject stool samples (15). However, we then differentiated between the two domains of alpha-diversity to clarify the impact of each in association with biological aging. This is necessary as changes in microbial dominance or abundance equitability (evenness) is biologically distinct from the introduction or loss of whole species in the GI microbial community (richness). Colonization of the GI tract by a wide range of species may be key for normal GI resilience to changes in environment, including new disability, hospitalization, institutionalization, or other frailty-related complications. Our data suggest that biological age, compared with chronological age, may more accurately reflect the risk of community-dwelling adults for complications due to decreased intestinal richness, which is associated with commensal opportunism, chronic inflammation, and metabolic dysregulation (9,30,31).
Altered environmental and physiological factors associated with aging may influence the selectivity of the gut for particular microbial species. We thus explored how well biological age or chronological age predicted between-subject taxonomic relatedness in gut microbiota. We found that FI34 scores significantly grouped subjects by microbiota phylogenetic similarity. Thus, gut microbial community structure varies according to subject FI34 scores and tends to be similar between subjects of similar FI34 score but not so for those of similar chronological age. Repeating this analysis for abundance-weighted between-sample differences in OTU taxonomy abolished the correlation between biological age and beta-diversity. This suggests that (a) biological aging more strongly influences the taxonomic membership of intestinal microbial communities but not member relative abundance or that (b) our sample size is insufficient to identify a relationship between biological age and a composite index of abundance and taxonomic relatedness. We addressed (b) by normalizing OTU counts to stabilize variance, which has been shown to increase the sensitivity of detection of between-sample differences (32). Variance-stabilized weighted differences improved correlation significance; however, these differences still fell below the threshold of significance for association with biological age.
Global changes in microbial diversity prompted us to investigate specific OTU correlates of biological and chronological age. Biological age but not chronological age was significantly associated with changes in abundance of individual genera. Coprobacillus significantly increased in abundance with increased FI34 as observed via DESeq2 differential OTU abundance analysis. Coprobacillus was additionally assigned to a WGCNA module that independently and positively correlated with subject frailty. The WGCNA workflow groups OTUs into modules based strictly on OTU coabundance patterns unbiased to clinical variables. Modules are then later correlated against clinical variables such as chronological age or FI34 score. This suggests some degree of internal validation of results as obtained by disparate methodologies. Results from both agree that chronological and biological aging are distinguished by their association with the abundances of individual genera and coabundant genera in the gut.
In total, seven coabundance modules were detected by WGCNA topological overlap-clustering of a co-occurrence network generated by SPIEC-EASI. SPIEC-EASI as well as other related network inference algorithms are specifically designed to suppress spurious, false-positive relationships that arise out of artifact in compositional data such as that collected in 16S microbial surveys (33). We additionally denoised sequenced reads using DADA2 and filtered OTUs for at least 25% prevalence in our cohort to focus our analysis on the more represented microbial genera. Together these measures may reveal the most salient and robust coabundance relationships between microbial organisms in human stool. In turn, we identified a frailty-associated coabundance module consisting of Eggerthella, Coprobacillus, and Lachn. Ruminococcus, all of which have been previously reported as individual correlates of frailty (13,15). Our findings suggest that these genera not only increase in abundance with increased frailty but co-occur in the human gut and may co-operate for survival in prefrail and frail individuals.
In silico biochemical pathway reconstitution in microbial subpopulations has been previously performed on cohorts with inflammatory bowel disease (26) but not specifically in those undergoing biological aging. Although this analysis alone does not replace true metagenomic deep-sequencing, it does offer hypotheses and potential targets to guide the design of future metagenome sequencing studies. In our study, the biological age-associated turquoise module was predicted to contain genes responsible for bacterial persistence in the gut such as sporulation and germination and bacterial motility (Supplementary File 1E). Bacterial motility proteins were particularly present in Eggerthella (Supplementary Figure 10) and included those for bacterial ciliary function, which is known to confer adherence to epithelial surfaces in some species (34). Genes for sporulation were sequestered in Coprobacillus and Lachn. Ruminococcus (Supplementary Figure 10 and File 1F). Interestingly, the latter two have been historically reported as nonspore formers based on findings in insult-free, anaerobic, axenic cultures (35) despite the presence of major genetic determinants of sporulation including the Spo0A master regulator (KEGG ID: K07699, Supplementary File 1G) (36). Recent work has found that culturing conditions favoring sporulation are, however, critical to reveal this phenotype, and many previous “nonspore formers” such as Lachn. Ruminococcus gnavus, Blautia producta, and others observed in our cohort, indeed form spores when (a) challenged with ethanol and atmospheric air and (b) grown in polymicrobial culture (37). This study further concluded that spore formation likely promotes survival of strictly anaerobic bacteria in the human gut as well as their transmission between individuals in the environment (37). We speculate that these factors may support microbial growth, transmission, and virulence in the gut of individuals undergoing biological aging.
Knowledge of the pathways encoded in coabundant microbial genomes may illuminate mechanisms of co-operation in potentially virulent organisms. Recent work has revealed novel virulence factors in key species of Eggerthella and Lachn. family Ruminococcus. Ruminococcus species including R. gnavus have been shown to secrete mucolytic alpha- and beta-glucosidases that may sensitize mucous sugars and mucin protein to degradation and consumption by other, co-cultured microbial species (38). Eggerthella is completely dependent on amino acid harvest for energy and may benefit from the activity of Lachn. Ruminococcus (39). In addition, Eggerthella is predicted to produce acetaldehyde from threonine (KEGG ID: K01620, Supplementary File 1G), which is characteristically abundant in mucin proteins (40) and may serve as substrate to increase gut barrier permeability (41). In turn, co-operation may encourage growth, epithelial attachment, and invasion of Eggerthella lenta, which has been recently shown to adhere to the surface of gut epithelial Caco-2 cells in vitro (34) and has been reported in numerous cases of elderly bacteremia (42,43). In our cohort, Eggerthella and Lachn. family Ruminococcus genera were significantly coabundant. One hundred percentage of Eggerthella OTUs were specifically assigned to E. lenta and 99% of Lachn. Ruminococcus were assigned to R. gnavus. Whether Eggerthella spp. co-operate with Lachn. Ruminococcus spp. to achieve virulence in the gut of frail persons through mucolysis remains to be experimentally investigated. If validated, co-operation between virulent symbionts may drive chronic low-grade inflammation and subsequently the aging process and frailty trajectory (44).
Additional activities of Eggerthella have been previously characterized in the GI tract. Eggerthella is a common human GI commensal that has similarly been reported to be increased in relative abundance in prefrail subjects in the United Kingdom as well as mouse models of aging (15,45). Previous studies have demonstrated that Eggerthella directly inactivate cardiac glycosides such as digoxin (46). Increased Eggerthella abundance in frail persons may yet prove as an emerging biomarker of frailty as future studies further characterize its role in frailty syndrome.
The findings in this study are based on microbial communities in stool collected by participants at home. Custom designed stool collection kits were provided to participants designed to minimize variation in sampling effort. It is difficult to completely control for subject-to-subject variation due to at-home sampling. In addition, luminal microbial communities may differ from communities more proximal to the gut epithelium such as those in the outer mucous layer (47). How these differences translate physiologically for the host is a question of on-going research. The outer mucous layer is known to constitutively migrate into the luminal space throughout the GI tract (48), and its accumulation and clearance in stool may mitigate these differences to some degree.
Alternative clinical and molecular measures of biological aging are available. These include the Fried Index (49) and leukocyte telomere length (50), which may also prove informative when examining changes in gut microbial diversity and coabundance and may provide additional understanding of the relationship between the microbiota and aging.
In summary, biological age as estimated by our FI34 was found to be a better indicator of changing gut microbiota structure than chronological age. Biological age correlated with the growth of specific, coabundant organisms in the gut lumen, which differentiated biological from chronological age. Further study of these changes and their role in aging physiology may open the field to the development of novel therapy. Specific interventions reversing or slowing these and other changes in the gut microbiota may promote healthy aging in prefrail or frail persons.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was supported by the National Institutes of Health (grant numbers T32 AA07577, K01 AG027905, P01 AG022064, R24 AA19661, P01 HL076100, P60 AA009803).
Conflict of Interest
The authors declare no conflicts of interest.
Supplementary Material
References
- 1.Population Division, DESA, United Nations. World Population Ageing 1950–2050. New York, NY: United Nations; 2002. [Google Scholar]
- 2. Mitnitski AB, Mogilner AJ, MacKnight C, Rockwood K. The mortality rate as a function of accumulated deficits in a frailty index. Mech Ageing Dev. 2002;123:1457–1460. doi:10.1016/S0047-6374(02)00082-9 [DOI] [PubMed] [Google Scholar]
- 3. Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet. 2013;381:752–762. doi:10.1016/S0140-6736(12)62167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strandberg TE, Pitkala KH. Frailty in elderly people. Lancet. 2007;369:1328–1329. doi:10.1016/S0140-6736(07)60613-8 [DOI] [PubMed] [Google Scholar]
- 5. Matsubayashi K, Okumiya K, Osaki Y, Fujisawa M, Doi Y. Frailty in elderly Japanese. Lancet. 1999;353:1445. doi:10.1016/S0140-6736(05)75972-9 [DOI] [PubMed] [Google Scholar]
- 6. Mitnitski AB, Graham JE, Mogilner AJ, Rockwood K. Frailty, fitness and late-life mortality in relation to chronological and biological age. BMC Geriatr. 2002;2:1. doi:10.1186/1471-2318-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morley JE, Vellas B, van Kan GA, et al. Frailty consensus: a call to action. J Am Med Dir Assoc. 2013;14:392–397. doi:10.1016/j.jamda.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishino R, Mikami K, Takahashi H, et al. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterol Motil. 2013;25:521–528. doi:10.1111/nmo.12110 [DOI] [PubMed] [Google Scholar]
- 9. Grasa L, Abecia L, Forcén R, et al. Antibiotic-induced depletion of murine microbiota induces mild inflammation and changes in Toll-like receptor patterns and intestinal motility. Microb Ecol. 2015;70:835–848. doi:10.1007/s00248-015-0613-8 [DOI] [PubMed] [Google Scholar]
- 10. Bruce-Keller AJ, Salbaum JM, Luo M, et al. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol Psychiatry. 2015;77:607–615. doi:10.1016/j.biopsych.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bindels LB, Delzenne NM. Muscle wasting: the gut microbiota as a new therapeutic target? Int J Biochem Cell Biol. 2013;45:2186–2190. doi:10.1016/j.biocel.2013.06.021 [DOI] [PubMed] [Google Scholar]
- 12. Saraswati S, Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Front Microbiol. 2014;5:764. doi:10.3389/fmicb.2014.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi:10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 14. Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Aging of the human metaorganism: the microbial counterpart. Age (Dordr). 2012;34:247–267. doi:10.1007/s11357-011-9217-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jackson M, Jeffery IB, Beaumont M, et al. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8. doi:10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi:10.1126/science.aac8469 [DOI] [PubMed] [Google Scholar]
- 17. Lee S, Sung J, Lee J, Ko G. Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl Environ Microbiol. 2011;77:7433–7437. doi:10.1128/AEM.05490-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim S, Welsh DA, Cherry KE, Myers L, Jazwinski SM. Association of healthy aging with parental longevity. Age (Dordr). 2013;35:1975–1982. doi:10.1007/s11357-012-9472-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied Environ Microbiol. 2013;79:5112–5120. doi:10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi:10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diedenhofen B, Musch J. cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015;10:e0121945. doi:10.1371/journal.pone.0121945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma M. Species richness vs evenness: independent relationship and different responses to edaphic factors. Oikos. 2005;111:192–198. doi:10.1111/j.0030-1299.2005.13049.x [Google Scholar]
- 23. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi:10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi:10.1038/nature17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol. 2016;428:3230–3252. doi:10.1016/j.jmb.2016.06.021 [DOI] [PubMed] [Google Scholar]
- 26. Tong M, Li X, Wegener Parfrey L, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One. 2013;8:e80702. doi:10.1371/journal.pone.0080702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. doi:10.3402/mehd.v26.26191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langille MG, Meehan CJ, Koenig JE, et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. doi:10.1186/s40168-014-0050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Chatelier E, Nielsen T, Qin J, et al. ; MetaHIT Consortium Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi:10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 31. Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi:10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 32. McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi:10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi:10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho GS, Ritzmann F, Eckstein M, et al. Quantification of Slackia and Eggerthella spp. in human feces and adhesion of representatives strains to Caco-2 cells. Front Microbiol. 2016;7:658. doi:10.3389/fmicb.2016.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2008;58(Pt 8):1896–1902. doi:10.1099/ijs.0.65208-0 [DOI] [PubMed] [Google Scholar]
- 36. Serra CR, Earl AM, Barbosa TM, Kolter R, Henriques AO. Sporulation during growth in a gut isolate of Bacillus subtilis. J Bacteriol. 2014;196:4184–4196. doi:10.1128/JB.01993-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Browne HP, Forster SC, Anonye BO, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi:10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi:10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- 39. Sperry JF, Wilkins TD. Arginine, a growth-limiting factor for Eubacterium lentum. J Bacteriol. 1976;127:780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nichols NL, Bertolo RF. Luminal threonine concentration acutely affects intestinal mucosal protein and mucin synthesis in piglets. J Nutr. 2008;138:1298–1303. [DOI] [PubMed] [Google Scholar]
- 41. Rao RK. Acetaldehyde-induced barrier disruption and paracellular permeability in Caco-2 cell monolayer. Methods Mol Biol. 2008;447:171–183. doi:10.1007/978-1-59745-242-7_13 [DOI] [PubMed] [Google Scholar]
- 42. Venugopal AA, Szpunar S, Johnson LB. Risk and prognostic factors among patients with bacteremia due to Eggerthella lenta. Anaerobe. 2012;18:475–478. doi:10.1016/j.anaerobe.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 43. Gardiner BJ, Tai AY, Kotsanas D, et al. Clinical and microbiological characteristics of Eggerthella lenta bacteremia. J Clin Microbiol. 2015;53:626–635. doi:10.1128/JCM.02926-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao X, Li H, Leng SX. Inflammation and immune system alterations in frailty. Clin Geriatr Med. 2011;27:79–87. doi:10.1016/j.cger.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conley MN, Wong CP, Duyck KM, Hord N, Ho E, Sharpton TJ. Aging and serum MCP-1 are associated with gut microbiome composition in a murine model. PeerJ. 2016;4:e1854. doi:10.7717/peerj.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haiser HJ, Seim KL, Balskus EP, Turnbaugh PJ. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–238. doi:10.4161/gmic.27915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H, Limenitakis JP, Fuhrer T, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi:10.1038/ncomms9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922–G929. [DOI] [PubMed] [Google Scholar]
- 49. Fried LP, Tangen CM, Walston J, et al. ; Cardiovascular Health Study Collaborative Research Group Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:146–156. doi:10.1093/gerona/56.3.M146 [DOI] [PubMed] [Google Scholar]
- 50. Kim S, Bi X, Czarny-Ratajczak M, et al. Telomere maintenance genes SIRT1 and XRCC6 impact age-related decline in telomere length but only SIRT1 is associated with human longevity. Biogerontology. 2012;13:119–131. doi:10.1007/s10522-011-9360-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.