Abstract
Species from lower invertebrates to a spectrum of mammals show antiaging health benefits of phytochemical(s). Here, we explored the pro-longevity effects of a natural triterpenoid, ursolic acid (3β-hydroxy-urs-12-en-28-oic acid; UA) in Caenorhabditis elegans with maximal life span being evident at 25 µM UA. Similar to eat-2 mutants, UA uptake by worm results in reduced fat storage and attenuation of reactive oxygen species (ROS), independent of superoxide dismutase(s) activation. The genetic requirements for UA-mediated longevity are quite similar to dietary restriction (DR) achieved through SKN-1/NRF-2 exhibiting upregulation of downstream target genes gcs-1 and daf-9. Longevity mechanism was independent of PHA-4/FOXA and attributed to partial dependence on sir-2.1. Altogether, our study suggests differential use of UA-elicited signaling cascades in nutrient sensing for longevity. Both the redox state and the proteostasis of an organism play critical role in aging and disease resistance. Interestingly, we observed a reduction of toxic protein aggregation in transgenic polyglutamine (polyQ) C. elegans model and UA-mediated JNK-1 (c-Jun-NH2-terminal kinase) activation in wild-type animals. Thus, our study demonstrates a small extent of prevention against proteotoxic stress by UA coupled with positive aspects of DR-mediated longevity.
Keywords: Healthy aging, Ursolic acid, Polyglutamine, ROS
Aging is marked by physical decline hence affected by the nutritional state of an organism and its ability to restore redox imbalance. Consistent evidence for the pro-longevity effects of phytochemical(s) has been shown to delay age-linked maladies in various model systems tested with safer toxicity profile (1). So far, diverse pharmacological interventions have been documented to modulate gerontogene(s) taking part in the biological course of aging such as pharmacological suppression of the GH/IGF-1 axis and TOR-S6K pathway, restrictive protein intake, activators of conserved protein deacetylase SIR2, and the use of spermidine, metformin, and other epigenetic modulators (2).
Ursolic acid (3β-hydroxy-urs-12-en-28-oic acid; UA) is a pentacyclic ursane type of triterpene usually present in the stem bark, leaves, or fruit peel of plants in the form of free acid or aglycones. It exhibits a wide range of pharmaceutical properties, including anti-inflammatory, hepatoprotection, antitumor, anti-HIV, antiangiogenic, and antibacterial activity (3). In our earlier studies, we have identified UA as an antiaging phytochemical, with enhanced health parameters in C. elegans (4). UA has also proved to have broad-spectrum antitumor effects tested in various cancer models (5–7), with a lead in preclinical and clinical studies where liposomal ursolic acid was used to determine the toxicity and pharmacokinetics in healthy adult human volunteers and in patients with advanced solid tumors (8, 9). Moreover, a unified model integrating cancer to aging has been documented in several studies (10,11). This study sought to understand healthy aging; therefore, UA was used to delineate the molecular profile for longevity as healthy aging, prevents many age-associated malignancies and cancer is one of the major risk factors for aging.
Aging studies in mammals are challenging owing to relatively long life span and a limited set of genetic tools that are capable of assessing the genome-wide changes associated with disease. By contrast, C. elegans has a shorter life span of only 2 to 3weeks, easily propagated in culture, facilitating rapid genetic analyses. Genetic resources for C. elegans include hundreds of publicly available mutant strains; whole-genome RNA interference (RNAi) libraries etc. Interestingly, there are substantially more protein similarities between C. elegans and Homo sapiens than in any other cross-species comparison.
Since aging and disease shoot from common mechanisms (12), delaying disease by delaying aging is the rationale behind the ongoing research. To this end, we used transgenic polyglutamine (polyQ) C. elegans model typically associated with Huntington’s disease (HD). The mutation prone for HD has been recognized as a CAG expansion located at the 5’ end of the gene htt, translated into a polyQ stretch at protein level (13). Opportunities remain in understanding how therapeutic potential of phytochemical(s) accomplish healthy aging with disease resistance. Henceforth, this study has been conducted taking the safer toxicity profile of UA, exploring its role in nutrient-response pathways and implication in age-associated pathologies.
Results and Discussion
UA Considerably Affects Life span and Health Span
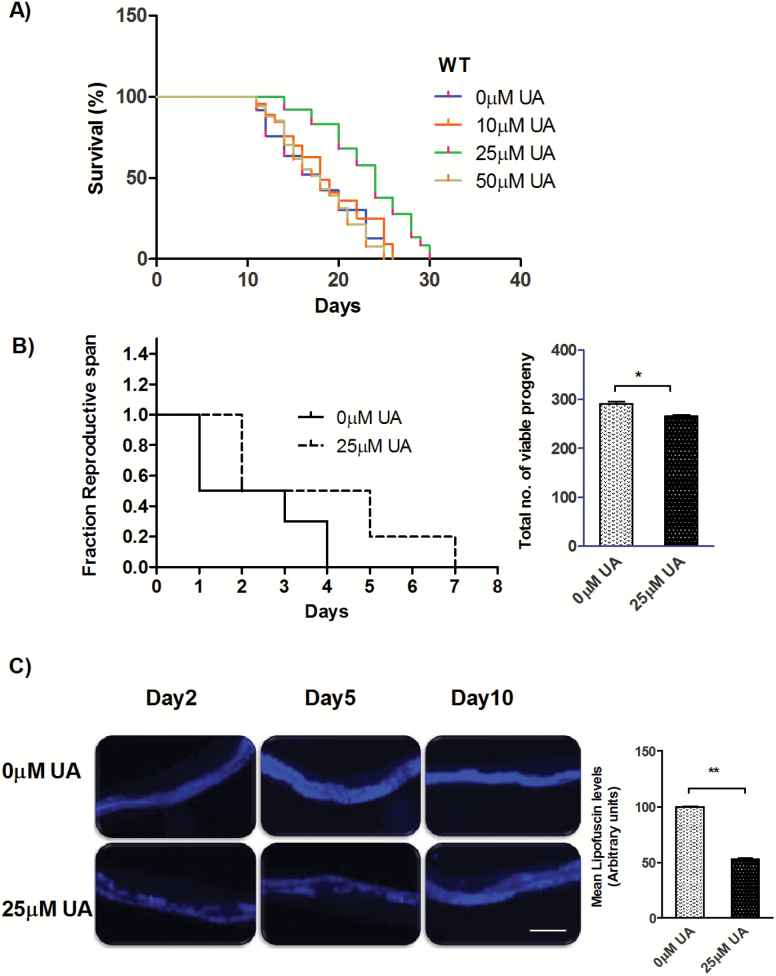
We observed a significant increase ~31.3% in the mean life span of wild-type (WT) N2 worms treated with UA [MLS ± SEM on 0 µM UA is 17.74 ± 0.01 days, while on 25 µM UA is 23.3 ± 0.03 days, p < .0001; Figure 1A, Supplementary Table 1]. UA has poor water solubility. As we move from concentration 25 µM to 50 µM and above, the solution was not homogenous showing cloudiness hence poor bioavailability. There might be chances that it could not be assimilated by C. elegans. Therefore, we observed normal life span at 50 µM and 100 µM UA. We did not observe any undesirable effects on all life-span endpoint parameters for these concentration(s). UA treatment also enhanced health parameters; it lowered as well as delayed accumulation of age pigment lipofuscin to a significant extent (47.07%, p = .0018; Figure 1C). We also observed a noteworthy protection against heat shock (31.1%, p = .0006; Supplementary Figure 1A), healthier chemotaxis index and mobility impairment (Supplementary Figure 2), when compared to control worms. Taken together, UA positively influences life span with health assurance in C. elegans.
Figure 1.
Life span of wild-type (WT) C. elegans strain exposed to various conc. of UA. The 25 µM test concentration showed the most significant increase in life span (31.3%, p < .001). DMSO (0.05%) was taken as control (0 µM UA) for all the experiments. Life span assay was performed at 20°C. Survival plots were drawn by Kaplan–Meier survival assay. (B) UA treatment resulted in longer reproductive span (MRS) on control is 2.3 ± 0.44 days on 25 µM UA is 3.9 ± 0.67 days; p = .0247 with small brood size (inset; brood size for 0 µM UA is 290.5 ± 2.5 while for 25 µM UA is 266.0 ± 2.0; p = 0.0381) (C) The lower and delayed accumulation of lipofuscin in the intestine indicates better health span; scale bars, 100 µm. Right panel represents quantification of the mean fluorescence (47.07%, p = 0.0018 in adult day 5 animals) using imageJ.
UA Works Independently of Insulin/IGF-1 (Insulin-Like Growth Factor 1) Signaling
To elucidate the mechanistic basis for UA-dependent life-span extension, we tested some of the pathway mutant(s) starting from the canonical GH/IGF-1 axis. In this pathway, a mutation in daf-2 [insulin/IGF-1 (insulin-like growth factor 1) signaling (IIS) receptor] results in life-span extension under the control of conserved FOXO transcription factor DAF-16 and heat shock transcription factor (HSF)-1 (14). When daf-2 (e1370) worms were supplemented with UA, we observed a significant life-span extension compared to control worms [MLS ± SEM on 0 µM UA is 25 ± 0.11 days, while on 25 µM UA is 32.35 ± 0.09 days; p < .0001; Supplementary Figure 3A, Supplementary Table 1] suggesting that UA may act through a parallel pathway. In addition, the UA-mediated life-span extension requires neither daf-16 nor hsf-1 [MLS ± SEM of daf-16 (mgDf50) on 0 µM UA is 12.77 ± 0.11 days, on 25 µM UA is 16.35 ± 0.07 days, p < .0001; Supplementary Figure 3B]. hsf-1(sy441) mutants have an impaired protein-folding capacity and we investigated whether UA could rescue the short life span by reducing proteotoxic stress. UA failed to extend life span of hsf-1(sy441) mutant worms compared to control worms at 20°C [hsf-1(sy441) on 0 µM UA is 12.20 ± 0.37 days and on 25 µM UA is 12.57 ± 0.56 days, p = .066; Supplementary Figure 3C, Supplementary Table 1]. Higher temperature is unfavorable for hsf-1(sy441) so, we further tested its life span at 15°C to support our hypothesis. We observed an extended life span of UA-treated worms over control worms [hsf-1(sy441) on 0 µM UA is 14.78 ± 0.42 days and on 25 µM UA is 20.27 ± 0.21 days, p < .0001; Supplementary Figure 3D, Supplementary Table 1]. Altogether, this genetic data show that UA influence longevity independent of the IIS pathway possibly using a different mechanism.
UA Supplementation Mimics a dietary restriction-Like State
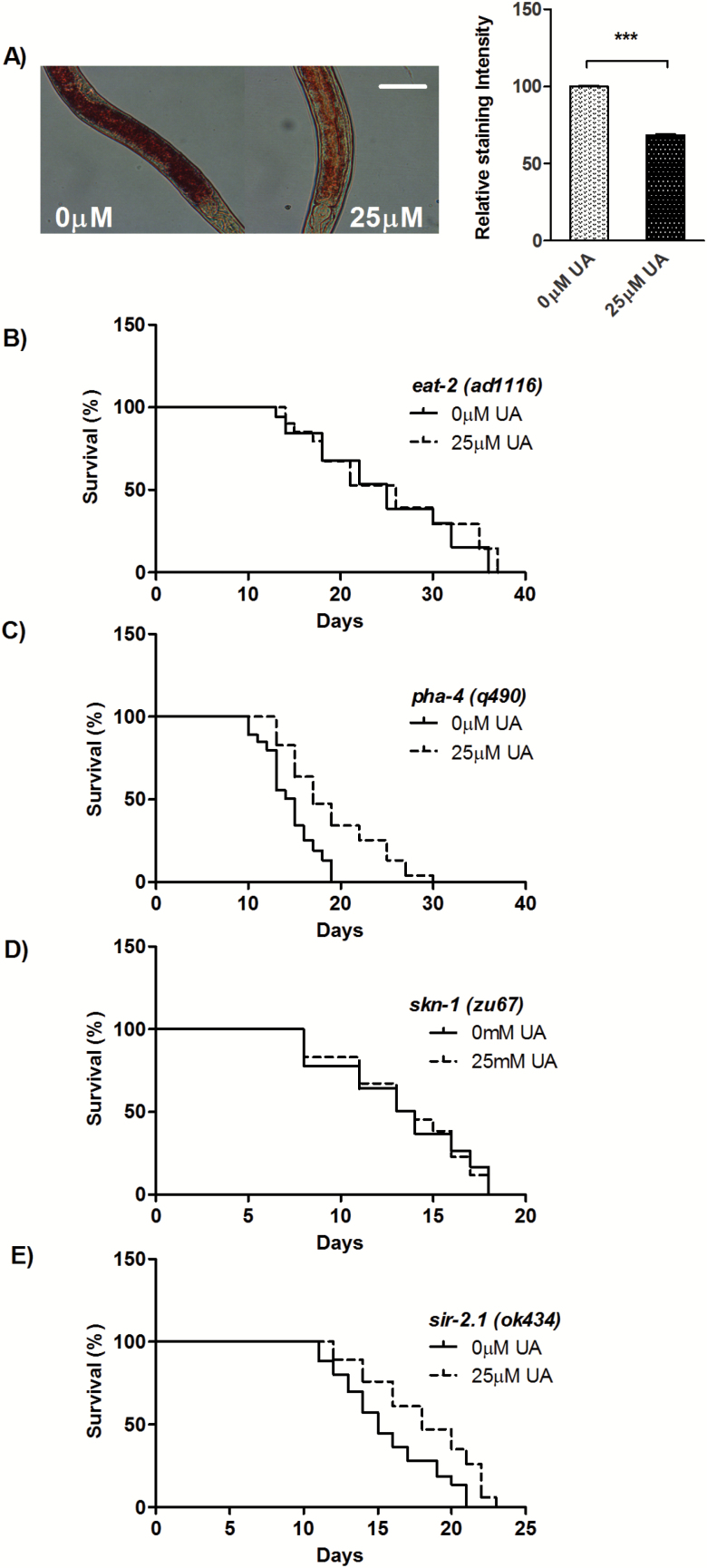
Since UA acts independently of the IIS pathway for longevity, we next examined its role in other longevity pathways. The eat-2 mutants have been used as a genetic surrogate of the dietary restriction (DR); characterized by long life span and reduced fat storage (15,16). Accordingly, we quantified the levels of stored fat in UA-supplemented worms using Oil Red O (17). We observed significantly reduced fat storage in the intestine (31.52%, p = .0006; Figure 2A) compared to control worms. Consequently, we assessed genetic interactions of UA with eat-2. We found that UA failed to increase life span of eat-2 (ad1116), compared to the control [MLS ± SEM on 0 µM UA is 24.80 ± 0.17 days, while on 25 µM UA is 25.41 ± 0.06 days; p = 0.119; Figure 2B, Supplementary Table 1], indicating that they may function in similar way for life-span modulation. Quite similar to eat-2 mutants (18,19), UA-treated worms have small brood size [inset; brood size on 0 µM UA is 290.5 ± 2.5, while on 25 µM UA is 266.0 ± 2.0, p = .0381] and longer reproductive span (mean reproductive span on 0 µM UA is 2.3 ± 0.44 days, while on 25 µM UA is 3.9 ± 0.67 days; p = .0247; Figure 1B) compared to control worms. One of the characteristics of DR is low pharyngeal pumping rate and small body size compared to well-fed animals (15). Interestingly, we found UA-fed worms were having regular body size and feeding rate (Supplementary Figures 4A and B).
Figure 2.
UA regulates longevity in response to dietary restriction. A) Oil Red staining of fixed WT worms grown on Control (0 µM UA) or 25 µM UA shows significant decrease in fat storage by UA (31.52%, p = .0006), right panel represents quantification of staining. B) UA failed to extend life span eat-2 (ad1116) to a significant extent whereas it increased the life span of C) pha-4 (q490), suggesting no role for it. It also failed to increase life span of D) skn-1(zu67), to a significant extent. E) UA-mediated longevity was attributed to partial dependence on sir-2.1(ok434). Life-span assay were performed at 20°C. ***p < .0001.
FOXA transcription factor, PHA-4 is specifically required for genetic and nongenetic models of DR (20) for life-span extension and it does not have any requirement in the longevity governed by IIS pathway. Subsequently, next, we asked whether UA-mediated longevity mechanism requires pha-4. We evaluated whether UA supplementation can increase life span in the absence of pha-4. Under this condition, UA significantly increased the life span [MLS ± SEM on 0 µM UA is 14.51 ± 0.23 days, while on 25 µM UA is 19.05 ± 0.08 days; p < .0001; Figure 2C, Supplementary Table 1]. Hence, unlike eat-2 mutant UA does not require PHA-4 for longevity. The stress-protective transcription factor SKN-1/NRF-2 regulate DR-induced longevity in the ASI neurons, whereas, in the intestine, it acts downstream of the IIS pathway to modulate oxidative stress tolerance (21–23). We asked whether UA-mediated longevity requires SKN-1. In UA-supplemented plate, it failed to increase the life span [MLS ± SEM on 0 µM UA is 13.29 ± 0.03 days, while on 25 µM UA is 13.49 ± 0.13 days; p = .77; Figure 2D, Supplementary Table 1] to a significant extent over control worms. Thus, UA-mediated longevity depends on skn-1, in a way similar to eat-2 mutant (23). Altogether, multiple lines of evidence suggest that UA supplementation extends longevity and initiate a DR-like state.
DR may have cross-talk with other genetic pathways (24). Therefore, we further asked if sir-2.1 (encodes a protein deacetylase of Sirtuin family) has a role in the pathway underlying the effect of UA. The role of sir-2.1 in longevity course has been fiercely debated; in some of the studies, it is shown to have life-span–promoting effects (25,26) but not in others (27). We assessed life span of null mutant sir-2.1(ok434) and observed significant increase over control worms [MLS ± SEM on 0 µM UA is 15.65 ± 0.30 days, while on 25 µM UA is 18.13 ± 0.41 days; p < .0001; Figure 2E, Supplementary Table 1] but not to the same extent as WT control worms, symptomatic of partial dependent on sir-2.1. Altogether, our findings show different ways of life span extension by UA acting either independently or in nonlinear genetic pathways in congruence with the earlier reports (28).
UA Regulates Expression of a Subset of SKN-1 Target Genes
Genes downstream of SKN-1 coordinately affect the rate of aging in all the cells in the body. In the qRT-PCR expression analysis (Supplementary Figure 5), we observed a significant upregulation of gcs-1 (1.52-fold, p = .02) for conferring oxidative stress resistance along with the upregulation of daf-9 (encodes for cytochrome P450; 1.36-fold, p = .049) involved in xenobiotic detoxification. In addition to this, we also observed increase in mRNA transcript levels of sir-2.1 (1.34-fold, p = .043) confirming its positive role in pro-longevity.
UA Directly Mitigates Reactive Oxygen Species and Depends on the Activity of Mitochondrial Complex II
The most probable underlying cause of aging is damage to cellular macromolecules rendered by potent oxidizing agents (29,30). Reduced reactive oxygen species (ROS) levels have always been a remarkable feature of DR. In line with this supposition, we quantified the total intracellular ROS in UA-supplemented worms and found significantly lower ROS levels over control worms (51.15%, p ≤ 0.001; Supplementary Figure 6A). This was supported by gene expression profiling of UA-fed worms in qRT–PCR analysis as we observed no significant upregulation in superoxide dismutase(s) genes (Supplementary Figure 5). Also, in the biochemical analysis, the total cellular superoxide dismutase activity along with core antioxidant enzymes glutathione (GST) and catalase (CAT) activities remained unchanged (Supplementary Figure 7). We propose that antioxidant potential of UA directly mitigates oxidative damage, thus reducing the need for the activation of intrinsic antioxidant defense. This reduced expression of key antioxidants in UA-mediated longevity, suggests evolutionarily conserved effects of DR. Also, in UA-fed worms, we observed greater stress resistance against paraquat that induces severe oxidative stress (18.33%, p = .003; Supplementary Figure 1B). To correlate these phenotypic manifestations of redox status with molecular response, we assessed life span of short-lived mev-1 mutants in which a mutation in cytochrome b of the mitochondrial Complex II results in overproduction of superoxide and increased oxidative stress. UA failed to rescue the short life span of these animals significantly [MLS ± SEM on 0 µM UA is 10.87 ± 0.18 days, while on 25 µM UA is 10.46 ± 0.16 days; p = .327; Supplementary Figure 6B, Supplementary Table 1]. This genetic analysis suggests that UA reduces oxidative stress and requires an endogenous detoxification pathway to support life-span extension. However, recent human studies have documented that antioxidant supplementation failed to show any betterment in health span; provoking debates on the oxidative stress theory of aging (31). Moreover, other relevant evidence have reported a beneficial role for ROS in life span under stress conditions which in turn, induce protective responses that slow aging (32–37).
We cannot purge all stress. And in fact, that is not desirable because with moderate stress comes survival behavior, motivation, and positive striving dealing with various aspects of aging on a population level (33). However, it cannot be ruled out that specific targets (proteins, lipids, or nucleic acids) accumulate damage over time and accelerate aging rate. Therefore, the stringent molecular details are required to regulate ROS and validate the latest aging theories postulating a linkage between ROS and aging. To this end, we have further measured toxic protein aggregation (a new aging biomarker) at organismal level in transgenic polyglutamine (polyQ) C. elegans model for HD. The preclinical and clinical studies have indicated a link between oxidative stress and toxic protein aggregation. The use of antioxidants including essential fatty acids, coenzyme Q10, and creatine, has been reported as potential therapeutic strategies for such detrimental effects (38–41).
UA Modulates JNK-1 to Prevent Polyglutamine Protein Aggregation
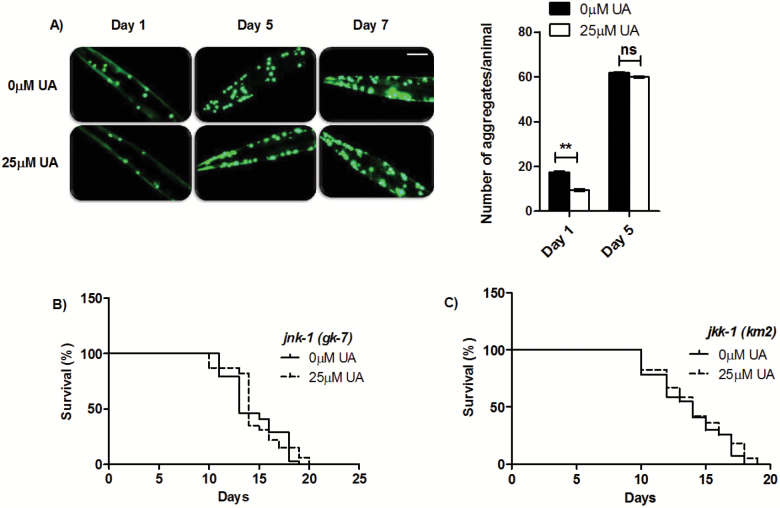
DR has the remarkable ability to protect against multiple diseases including diabetes, cancer, cardiovascular, and neurodegenerative disorders (42). One of the hallmarks of aging is the gradual loss of proteostasis over time, and multiple neurological disorders exhibit symptoms of impaired proteostasis. In a study, it has been documented that aging process itself, in the absence of disease, leads to the insolubilization and increased aggregation of toxic proteins (43). In addition, it was reported that higher levels of inherent protein aggregation aggravated toxicity in a C. elegans HD model and genetic pathway regulating longevity can alter the time course of age-associated polyglutamine‐mediated phenotypes (44). Taking UA as a lead antiaging phytochemical, we next evaluated whether UA improves protein homeostasis. We quantified aging-induced polyQ protein aggregation in unc-54p:Q40::YFP transgenic worms (45). These transgenic animals show a soluble Q40::YFP distribution in body wall muscle cells immediately after hatching and reach adulthood with an entirely Q40::YFP-aggregated phenotype. We observed that the UA-fed Day 1 Q40::YFP animals had approximately less than half as many aggregates as control animals (No. of aggregates in control = 17.5 ± 0.5 and in 25 µM UA is 9.5 ± 0.5; p = .0035; Figure 3A). Although the average number of aggregates increased with age, the UA treatment had no effect on Q40::YFP aggregation in Day 5 and Day 7 adults in which number of aggregates were increased along with diffused fluorescence for polyQ foci. This result suggests that UA treatment prevents the aggregation at the young adult stage and preserves the impaired proteostasis.
Figure 3.
UA reduces toxic protein aggregates in C. elegans transgenic polyQ model. A) Fluorescence micrographs of transgenic animals expressing Q40::YFP in body wall muscle cells with or without UA treatment. Scale bars, 100 μm. Right panel shows quantification of Q40::YFP aggregates. Results are expressed as mean + SEM numbers of aggregates/animal from three biological repeats. B) UA failed to extend life span of JNK-1 deletion mutant jnk-1(gk7) and B) JNK-1 upstream kinase JKK-1 deletion mutant jkk-1(km2), to a significant extent, suggesting a positive interaction with UA.
To further strengthen our hypothesis, the α-synuclein model for Parkinsons’ disease was examined employing transgenic C. elegans strain NL5901 strain, constitutively expressing YFP-fused human α-syn protein in the body wall. As it is well-established that the over-expression of α-synuclein correlates with increased ROS levels and UA mitigates ROS levels, it might also be able to reduce the toxicity of α-synuclein (46). The α-syn aggregation in the head area was comparable between the tested treatment groups. A significant reduction in the accumulation of α-syn in UA-fed Day 1 worms was observed by up to 36.52%; p < .0001; Supplementary Figure 8. The UA treatment had no effect on α-syn levels in Day 5 and Day 7 adult animals, suggesting efficacy of UA treatment at early stages of age-associated neurological disorder(s).
Research conducted on the cellular and animal models of HD have shown that expanded polyQ proteins induce a stress response associated with the c-Jun-NH2-terminal kinase (JNK) pathway (47). In order to assess the importance of this pathway in UA-mediated longevity, we examined life span of the mutants for JNK pathway, including, jnk-1/JNK and upstream kinase jkk-1 (48). In both the mutants, UA treatment failed to considerably increase the life span; jnk-1 (gk7) [MLS ± SEM on 0 µM UA is 14.53 ± 0.17 days, while on 25 µM UA is 14.69 ± 0.76 days; p = . 2182; Figure 3B, Supplementary Table 1] and in the upstream kinase jkk-1(km2) [MLS ± SEM on 0 µM UA is 13.74 ± 0.09 days, while on 25 µM UA is 14.18 ± 0.21 days; p = .114; Figure 3C, Supplementary Table 1]. In addition, we also observed increased mRNA transcript levels of jnk-1 (3.14-fold, p = .048; Supplementary Figure 5) in qRT-PCR gene expression profiling. Altogether, these results suggest that UA mediates JNK-1 activation to elicit its positive effects and are congruent with our earlier findings (4).
In summary, we present results where UA regulates the longevity of C. elegans in DR-dependent manner (Figure 4) and prevents toxic protein aggregation by inducing JNK-1 in WT animals. HD is more amenable to early pharmacological interventions because of its monogenic nature making it fully penetrant compared to other more prevalent neurodegenerative disease(s), including Alzheimer’s disease and Parkinson’s disease, which share such features as impaired proteostasis, selective neuronal vulnerability, and delayed onset (49). Understanding the shared genetic network of aging and associated diseases implies in harnessing the full benefits of phytochemical(s) on life span and health span. It remains interesting to study whether the positive effects of UA can delay onset of age-related morbidities in mammals.
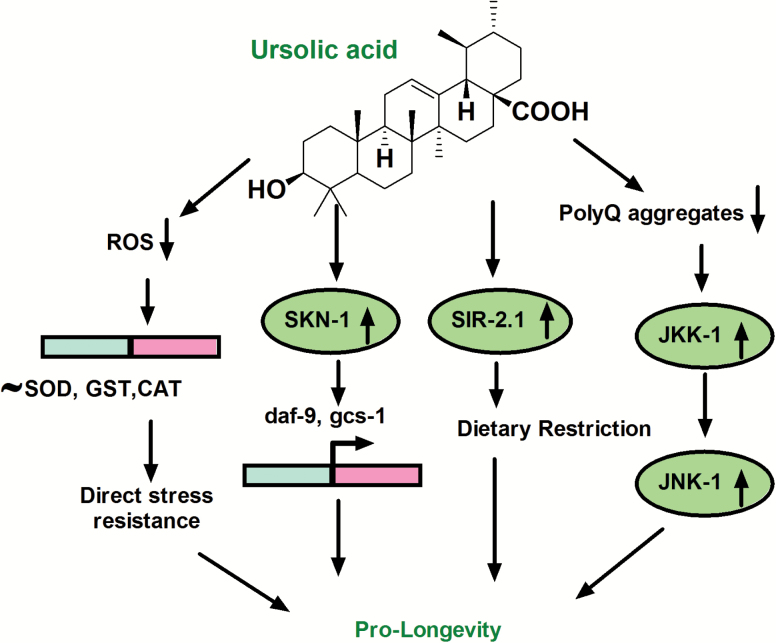
Figure 4.
UA in dietary restriction (DR) mediated longevity. Owing to its own antioxidant potential UA attenuates ROS directly independent of intrinsic antioxidant enzymatic system. As observed in DR worms, SKN-1 and eat-2 mediating longevity in partial dependence of sir-2.1. Moreover, this natural triterpenoid restores proteostasis in transgenic C. elegans polyQ model and mediates JNK-1 activation in wild-type N2 worms. It makes UA a much favored life-span–extending intervention which may have substantial implications in aging and associated pathologies.
Supplementary Material
Supplementary data is available at Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Experimental Procedures
Detailed experimental procedures and associated references are provided as supplementary information.
Funding
This work was supported by MLP 02 Research Grant (to R.P.). First author H.N. gratefully acknowledges the financial support from Indian Council of Medical Research, New Delhi (3/1/3/JRF-2012/HRD-120 (80061)) for fellowship.
Conflict of Interest
None declared.
Supplementary Material
Acknowledgments
We are grateful to the Director, CSIR–Central Institute of Medicinal and Aromatic Plants, Lucknow, India, for his invariable support. We extend our sincere gratitude toward Dr Arnab Mukhopadhyay and lab members (NII, New Delhi, India) for protocol sharing, providing strains, and helpful suggestions. We also thank Dr Gautam Kao and lab members (University of Gothenburg, Sweden.) for providing some of the C. elegans strains and critical comments on data analysis part. Authors wish to thank the Caenorhabditis Genetics Centre, MN, USA, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for reagents.
References
- 1. Leonov A, Arlia-Ciommo A, Piano A, et al. Longevity extension by phytochemicals. Molecules. 2015;20:6544–6572. doi:10.3390/molecules20046544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longo VD, Antebi A, Bartke A, et al. Interventions to slow aging in humans: are we ready? Aging Cell. 2015;14:497–510. doi:10.1111/acel.12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J. Pharmacology of oleanolic acid and ursolic acid. J Ethnopharmacol. 1995;49:57–68. [DOI] [PubMed] [Google Scholar]
- 4. Negi H, Shukla A, Khan F, Pandey R. 3β-Hydroxy-urs-12-en-28-oic acid prolongs lifespan in C. elegans by modulating JNK-1. Biochem Biophys Res Commun. 2016;480:539–543. doi:10.1016/j.bbrc.2016.10.073 [DOI] [PubMed] [Google Scholar]
- 5. De Angel RE, Smith SM, Glickman RD, Perkins SN, Hursting SD. Antitumor effects of ursolic acid in a mouse model of postmenopausal breast cancer. Nutr Cancer. 2010;62:1074–1086. doi:10.1080/01635581.2010.492092 [DOI] [PubMed] [Google Scholar]
- 6. Shanmugam MK, Dai X, Kumar AP, Tan BK, Sethi G, Bishayee A. Ursolic acid in cancer prevention and treatment: molecular targets, pharmacokinetics and clinical studies. Biochem Pharmacol. 2013;85:1579–1587. doi:10.1016/j.bcp.2013.03.006 [DOI] [PubMed] [Google Scholar]
- 7. Kim SH, Ryu HG, Lee J, et al. Corrigendum: ursolic acid exerts anti-cancer activity by suppressing vaccinia-related kinase 1-mediated damage repair in lung cancer cells. Sci Rep. 2015;5:15864. doi:10.1038/srep15864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang XH, Zhou SY, Qian ZZ, et al. Evaluation of toxicity and single-dose pharmacokinetics of intravenous ursolic acid liposomes in healthy adult volunteers and patients with advanced solid tumors. Expert Opin Drug Metab Toxicol. 2013;9:117–125. doi:10.1517/17425255.2013.738667 [DOI] [PubMed] [Google Scholar]
- 9. Both DM, Goodtzova K, Yarosh DB, Brown DA. Liposome-encapsulated ursolic acid increases ceramides and collagen in human skin cells. Arch Dermatol Res. 2002;293:569–575. [DOI] [PubMed] [Google Scholar]
- 10. Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi:10.1038/nature05949 [DOI] [PubMed] [Google Scholar]
- 11. Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi:10.1038/nature05985 [DOI] [PubMed] [Google Scholar]
- 12. Johnson SC, Dong X, Vijg J, Suh Y. Genetic evidence for common pathways in human age-related diseases. Aging Cell. 2015;14:809–817. doi:10.1111/acel.12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeats that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993; 72: 971–983. [DOI] [PubMed] [Google Scholar]
- 14. Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi:10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- 15. Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 2009;4:e7545. doi:10.1371/journal.pone.0007545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yen K, Le TT, Bansal A, Narasimhan SD, Cheng JX, Tissenbaum HA. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS ONE. 2010; 5, e12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crawford D, Libina N, Kenyon C. Caenorhabditis elegans integrates food and reproductive signals in lifespan determination. Aging Cell. 2007;6:715–721. doi:10.1111/j.1474-9726.2007.00327.x [DOI] [PubMed] [Google Scholar]
- 19. Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. [DOI] [PubMed] [Google Scholar]
- 20. Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi:10.1038/nature05837 [DOI] [PubMed] [Google Scholar]
- 21. Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi:10.1038/nature05904 [DOI] [PubMed] [Google Scholar]
- 22. Tullet JM, Hertweck M, An JH, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi:10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park SK, Link CD, Johnson TE. Life-span extension by dietary restriction is mediated by NLP-7 signaling and coelomocyte endocytosis in C. elegans. FASEB J. 2010;24:383–392. doi:10.1096/fj.09-142984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chamoli M, Singh A, Malik Y, Mukhopadhyay A. A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell. 2014;13:641–655. doi:10.1111/acel.12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi:10.1038/nature02789 [DOI] [PubMed] [Google Scholar]
- 26. Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi:10.1016/j.devcel.2005.09.017 [DOI] [PubMed] [Google Scholar]
- 27. Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev. 2007;128:546–552. doi:10.1016/j.mad.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 28. Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi:10.1111/j.1474-9726.2009.00459.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pamplona R, Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Res Rev. 2007;6:189–210. doi:10.1016/j.arr.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 30. Page MM, Robb EL, Salway KD, Stuart JA. Mitochondrial redox metabolism: aging, longevity and dietary effects. Mech Ageing Dev. 2010;131:242–252. doi:10.1016/j.mad.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 31. Ristow M. Unraveling the truth about antioxidants: mitohormesis explains ROS-induced health benefits. Nat Med. 2014;20:709–711. doi:10.1038/nm.3624 [DOI] [PubMed] [Google Scholar]
- 32. Smith SW, Latta LC, 4th, Denver DR, Estes S. Endogenous ROS levels in C. elegans under exogenous stress support revision of oxidative stress theory of life-history tradeoffs. BMC Evol Biol. 2014;14:161. doi:10.1186/s12862-014-0161-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Epel ES, Lithgow GJ. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S10–S16. doi:10.1093/gerona/glu055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chávez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi:10.1534/genetics.107.072587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi:10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- 36. Hernández-García D, Wood CD, Castro-Obregón S, Covarrubias L. Reactive oxygen species: a radical role in development? Free Radic Biol Med. 2010;49:130–143. doi:10.1016/j.freeradbiomed.2010.03.020 [DOI] [PubMed] [Google Scholar]
- 37. Miranda-Vizuete A, Veal EA. Caenorhabditis elegans as a model for understanding ROS function in physiology and disease. Redox Biol. 2017;11:708–714. doi:10.1016/j.redox.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gil-Mohapel J, Brocardo PS, Christie BR. The role of oxidative stress in Huntington’s disease: are antioxidants good therapeutic candidates? Curr Drug Targets. 2014;15:454–468. [DOI] [PubMed] [Google Scholar]
- 39. Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi:10.1089/ars.2006.8.2061 [DOI] [PubMed] [Google Scholar]
- 40. Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45:667–678. doi:10.1016/j.freeradbiomed.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 41. Chen CM, Wu YR, Cheng ML, et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem Biophys Res Commun. 2007;359:335–340. doi:10.1016/j.bbrc.2007.05.093 [DOI] [PubMed] [Google Scholar]
- 42. Speakman JR, Mitchell SE. Caloric restriction. Mol Aspects Med. 2011;32:159–221. doi:10.1016/j.mam.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 43. David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi:10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brignull HR, Morley JF, Garcia SM, Morimoto RI. Modeling polyglutamine pathogenesis in C. elegans. Methods Enzymol. 2006;412:256–282. doi:10.1016/S0076-6879(06)12016-9 [DOI] [PubMed] [Google Scholar]
- 45. Nollen EA, Garcia SM, van Haaften G, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi:10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi:10.1371/journal.pgen.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Merienne K, Helmlinger D, Perkin GR, Devys D, Trottier Y. Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. J Biol Chem. 2003;278:16957–16967. doi:10.1074/jbc.M212049200 [DOI] [PubMed] [Google Scholar]
- 48. Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci USA. 2005;102:4494–4499. doi:10.1073/pnas.0500749102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi:10.1016/S1474-4422(10)70245-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.