Abstract
Progress in gene editing research has been accelerated by utilizing engineered nucleases in combination with induced pluripotent stem cell (iPSC) technology. Here, we report transcription activator-like effector nuclease (TALEN)-mediated reincorporation of Arg1 exons 7 and 8 in iPSCs derived from arginase-1-deficient mice possessing Arg1Δ alleles lacking these terminal exons. The edited cells could be induced to differentiate into hepatocyte-like cells (iHLCs) in vitro and were subsequently used for transplantation into our previously described (Sin et al., PLoS ONE 2013) tamoxifen-inducible Arg1-Cre arginase-1-deficient mouse model. While successful gene-targeted repair was achieved in iPSCs containing Arg1Δ alleles, only minimal restoration of urea cycle function could be observed in the iHLC-transplanted mice compared to control mice, and survival in this lethal model was extended by up to a week in some mice. The partially rescued phenotype may be due to inadequate regenerative capacity of arginase-1-expressing cells in the correct metabolic zones. Technical hurdles exist and will need to be overcome for gene-edited iPSC to iHLC rescue of arginase-1 deficiency, a rare urea cycle disorder.
Keywords: TALEN, gene editing, homology-directed repair, arginase-1 deficiency, hepatocyte-like cell transplant
Introduction
Engineered sequence-specific nucleases, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPRs have been exploited for modeling diseases and developing novel therapeutic applications. These programmable nucleases can be broadly classified into two categories based on their mode of DNA recognition: ZFNs and TALENs achieve specific DNA binding via protein-DNA interactions, whereas CRISPR/Cas9 is directed to specific target sites by a single guide RNA that base-pairs directly with the target DNA sequence, as well as by protein-DNA interactions between Cas9 protein and the protospacer-adjacent motif (PAM).1, 2, 3, 4
Among these gene-editing technologies, TALENs exhibit a high degree of targeting specificity and flexibility.5 They recognize and cleave nearly any given DNA sequence with high efficiency in a broad range of organisms, including human cells, rodents, zebrafish, and plants.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Moreover, the low off-target effects and reduced nuclease-associated cytotoxicities make TALENs great tools for gene engineering in cells.10, 16 These site-specific nucleases are engineered fusion proteins of the catalytic domain of the endonuclease FokI with the TALE DNA-binding domain specifically designed to target a desired genomic site for gene modifications.3, 5 TALEs bind specifically to DNA targets via a central repeat domain and activate transcription of their targets by means of a C-terminal transcriptional activation domain.17 The binding domain is a series of customized 34-residue TALE repeat arrays in “two amino acids for one base” (“repeat variable di-residue”; RVD) recognition code, providing DNA-binding specificity.18, 19 Double-strand breaks (DSBs) occur when two independent TALENs, working in heteromeric pairs, bind to opposite strands of the target site separated by a 12- to 20-bp spacer region, thereby allowing dimerization of FokI and cleavage of the target locus.3, 5 This cleavage can then be resolved by cellular DNA repair, either by error-prone non-homologous end joining (NHEJ), which has the potential to introduce mutations at the site of the DSB, or by template-dependent homologous recombination (also termed homology-directed repair [HDR]) for precise genetic modification. Depending on the donor DNA template design, the repair mechanism can be initiated for gene replacement, single-nucleotide substitutions, or large-scale deletions.1
Arginase deficiency is a rare urea cycle disorder with hyperargininemia and profound neurological impairment as hallmark features.20 Arginase-1-deficient mouse models have been created, and they exhibit a profound lethal phenotype approximately 2 weeks after birth in the global knockout (KO) mice or after induction of tamoxifen-induced gene KO in adult mice (about 2 weeks after induced KO).21, 22, 23 Recently, we generated induced pluripotent stem cells (iPSCs) from our inducible arginase-1-deficient mouse model carrying a deletion of Arg1 exons 7 and 8 (Arg1Δ), which results in defective function of arginase-1.24 We showed that the deletion was repaired using CRISPR/Cas9, in combination with an excisable piggyBac transposon system to target corrective sequences to the endogenous Arg1 locus. In the current study, we report on TALEN-mediated correction of the Arg1Δ alleles in iPSCs and their successful differentiation to hepatocyte-like cells (HLCs) coupled with piggyBac transposon selection technology for seamless genetic manipulation, followed by secondary expansion to propagate the repaired iPSC-derived HLCs (named iHLCs hereafter). The genetically repaired iHLCs were transplanted to restore arginase-1 expression in Arg1Δ mice, and survival of the arginase-1-deficient mice was extended by up to a week in some mice. Although an elevation in arginase-1 expression was observed in the iHLC-transplanted mice, urea cycle function was still considerably lower than in wild-type control mice, thus necessitating further refinement in gene-edited iHLC transplantation cell therapy.
Results
Design of TALEN-Targeting Strategy for Correcting Arg1-Deficient (Arg1Δ) iPSCs
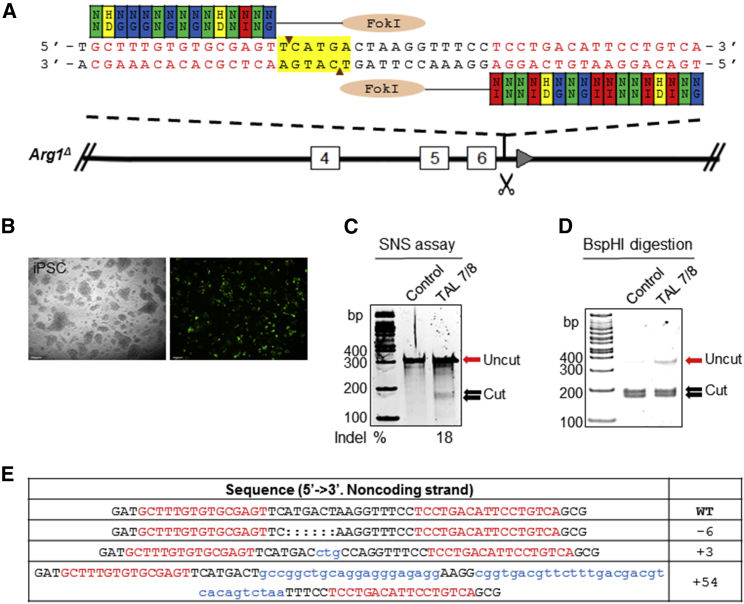
We designed a TALEN pair targeting intron 6 of the Arg1 locus at a region adjacent to the position targeted previously by CRISPR/Cas9.24 Each monomer contains an array of RVDs to bind the target DNA sequences (Figure 1A). The TALEN expression constructs showed good transfection efficiency when introduced into mouse iPSCs by electroporation (Figure 1B). Surveyor nuclease cleavage to detect NHEJ events of PCR products from the target region produced two bands in the 162- to 186-bp range, with insertion or deletion (indel) frequencies up to 18% in mouse iPSCs (Figure 1C), similar to that with CRISPR/Cas9.24 The TALEN pair target sites were selected to utilize a BspHI restriction site located within the spacer region to determine editing efficiency. The loss of the BspHI recognition sequence was demonstrated by the presence of uncleaved PCR products compared to the control (Figure 1D). Sequencing results confirmed that the TALENs induced DSBs at the predetermined position in the Arg1 locus (Figure 1E).
Figure 1.
TALEN-Mediated Gene Targeting in Arg1Δ Mouse iPSCs
(A) Schematic diagram showing the site of Arg1 gene modification using TALEN set 7/8. The TALEN 7/8 pair was designed to target intron 6 of Arg1. Each TALEN arm consists of a DNA-binding domain with repeat variable di-residues (RVDs) corresponding to DNA binding sequence proceeded by a 5′ T nucleotide and a 17-bp spacer region containing a BspHI recognition site (highlighted in yellow) to assay activity. TALE repeat domains are colored to indicate the identity of the RVD. Site-specific double-stranded breaks (DSBs) are generated upon dimerization of fused FokI endonucleases. (B) Electroporation to deliver 7 μg of each TALEN and pMax-GFP into cells to assess transfection efficiency. Images were acquired 24 hr post-electroporation. Scale bars, 95 μm. (C) Surveyor nuclease assay for detection of NHEJ-induced indels resulting from DSBs. The cleavage products were shown as extra bands (between 162 and 186 bp) indicated by the arrows. Mutation frequencies (indels %) were calculated by measuring the band intensities. (D) BspHI digestion results. The cut products were shown as extra bands (169 bp + 183 bp). (E) Sequencing data from PCR amplicons of TALEN-modified genomic DNA showing a few examples of NHEJ-mediated indel mutations at the desired location. The wild-type sequence is shown above with the TALEN-binding sites in red. Deleted bases are indicated by colons, and inserted bases are shown by lowercase letters in blue. The net change in length caused by each indel mutation is to the right of each sequence.
TALEN-Mediated Reincorporation of Deleted Exons via HDR
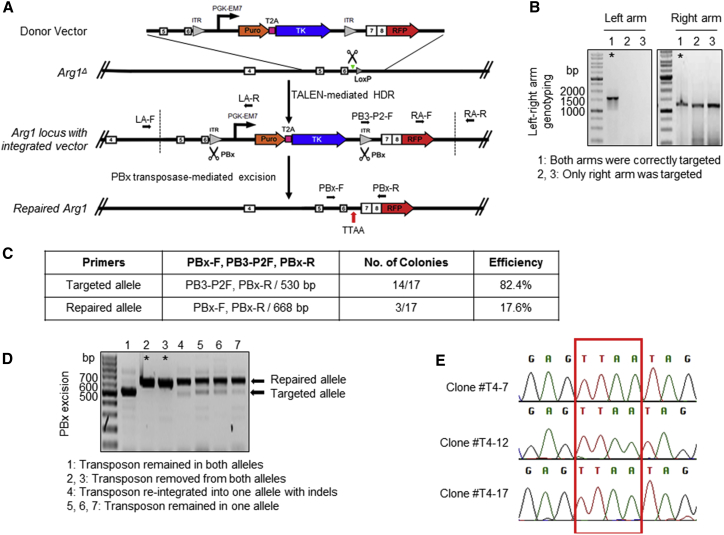
By taking advantage of iPSCs and their propensity for unlimited self-renewal capacity and their differentiation potential, we examined a TALEN-mediated targeted knockin approach for deleted exons 7 and 8 of Arg1 in Arg1Δ iPSCs. TALEN-expressing vector delivery, together with a linearized targeting vector, which consists of exons 7 and 8 cDNA fused to an RFP monomeric-encoding segment with piggyBac inverted terminal repeats (ITRs) flanking a PGK-Puro-TK cassette, was performed (Figure 2A). An HDR stimulatory compound, L-755,507, was added into the culture to enhance TALEN-mediated HDR efficiency.25 Upon cleavage by TALENs and subsequent HDR with the repair template, exons 7 and 8 should be reincorporated into the Arg1 locus. Eleven puromycin-resistant iPSC colonies were obtained and screened for proper targeting by PCR. We designed two primer pairs spanning the junction between the insert and endogenous sequence in both homologous arms as indicated in Figure 2A to examine correct integration. Of the 11 clones analyzed, two were correctly targeted (clones #T4 and #T10; efficiency of 18.2%) for both PCR primer sets (Figure 2B). These two clones were subjected to piggyBac-mediated excision of the integrated selection cassette. Upon ganciclovir selection, PCR screening of 17 ganciclovir-resistant colonies revealed three with biallelic repair (Figure 2C). All three completely repaired clones were from the #T4 line (Figure 2D). Our TALEN-mediated targeting experiments demonstrated HDR knockin efficiency comparable to that of the CRISPR/Cas9 system.24 Sequence analysis showed the presence of a single TTAA sequence26 at the site of transposon excision in the repaired cell lines (clones #T4–7, #T4–12, #T4–17), an evidence for scarless removal of selectable marker cassettes following successful HDR (Figure 2E).
Figure 2.
Footprint-free TALEN-Mediated Correction of Arg1Δ Mouse iPSCs
(A) Schematic of strategy used for TALEN-mediated reincorporation of Arg1 exons 7 and 8 via homology-directed repair in combination with piggyBac transposon methodology. Green triangle denotes the target site of TALEN 7/8 in intron 6 of Arg1. Black arrows indicate primers for PCR-based screening to confirm the selected clones. The remnant LoxP left from the initial Cre-excision of exons 7 and 8 would be removed upon targeting vector integration. The characteristic “footprint” TTAA sequence at the site of transposon excision is shown. PBx, piggyBac transposase; T2A, viral sequence for ribosomal skipping; Puro, puromycin; ITR, inverted terminal repeat; TK, thymidine kinase. (B) Representative gel images showing integration-specific PCR of puromycin-resistant single-cell clones derived from Arg1Δ iPSC after TALEN treatment. Each homology arm was amplified independently by PCR. Amplicon sizes of the left and right arm were 1,530 and 1,233 bp, respectively. The clones indicated by an asterisk are correctly targeted with integrated selection cassette and corrective sequence in the desired position at the Arg1 locus. (C) PCR-based excision screening. A table shows primer combination to uniquely identify different alleles after piggyBac excision and negative selection by ganciclovir. (D) A representative gel of different banding patterns and corresponding genotypes. Amplicon sizes of the repaired and targeted alleles were 668 bp and 530 bp, respectively. PBx-excised clones are indicated by asterisks. (E) Footprint sequencing analysis of transposon-free repair clones. TTAA target sites are boxed.
Transplantation of Repaired iHLCs Extends Survival in Some Mice after Induced Arginase-1 Loss of Expression
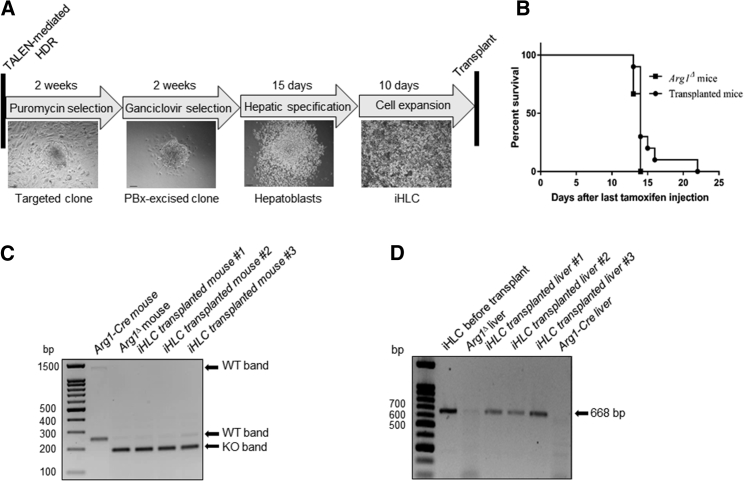
To facilitate iPSC-mediated ex vivo gene therapy, efficient differentiation into the proper cell type after gene repair is essential. Considering the challenges in coaxing iPSCs to mature functional hepatocytes in culture in our recent work,24 we attempted to test whether iHLCs can mature after transplantation and if Arg1 gene editing could lead to the restoration of functional arginase in the liver. Repaired clones (#T4–7 and #T4–12) were subjected to hepatic differentiation using a stepwise protocol, which includes hepatic specification, hepatoblast formation, and iHLC expansion (Figure 3A). At the end of the 25-day differentiation protocol, 2 million iHLCs were transplanted into Arg1-Cre mice via splenic administration. More importantly, two cycles of retrorsine treatment followed by partial hepatectomy was performed prior to transplantation to stimulate engraftment and regenerative capacity.27 Mice were then allowed to recover for 15 weeks, at which time the 5-day sequential tamoxifen-induced global Arg1 gene disruption was carried out. Kaplan-Meier survival curves of Arg1Δ mice versus iHLC-transplanted mice began to diverge on day 14 after the last dose of tamoxifen. In fact, the survival of the iHLC-transplanted mice was extended up to an extra week (day 22 post-tamoxifen) compared to the Arg1Δ mice, which died at the usual time point (around 14 days post-tamoxifen) from a wasting phenotype21, 22, 23 (Figure 3B). Successful excision of exons 7 and 8 of Arg1 induced by tamoxifen administration in experimental mice was verified by PCR genotyping of tail samples for the diagnostic band of 195 bp (Figure 3C). Using primers flanking the Arg1 and RFP domains, liver tissue samples obtained from iHLC-transplanted mice yield bands at 668 bp, indicating the presence of the repaired allele (Figure 3D).
Figure 3.
Generation of Hepatocyte-like Cells from Repaired Mouse iPSCs for Transplantation
(A) Schematic diagram showing time allocated for TALEN-mediated correction, clone screening, and hepatic differentiation prior to transplantation. Scale bars, 95 μm. (B) Kaplan-Meier survival curves of tamoxifen-induced Arg1Δ mice (n = 3) and Arg1Δ mice injected with repaired iHLCs (n = 10). (C) Representative agarose gel of PCR genotyping using genomic DNA from tail biopsies to confirm the deletion of exons 7 and 8. Arg1-Cre mice exhibited two bands at 1.2 kb and 252 bp (indicative of intact exons 7 and 8), while Arg1Δ mice only showed a single band at 195 bp. (D) PCR to confirm the presence of the repaired allele in liver samples obtained from iHLC transplanted (668 bp). Cultured repaired iHLCs were used as a positive control, while liver from an untreated Arg1-Cre mouse served as negative control.
Recovery of Arginase Expression in Livers Repopulated with Transplanted Repaired iHLCs
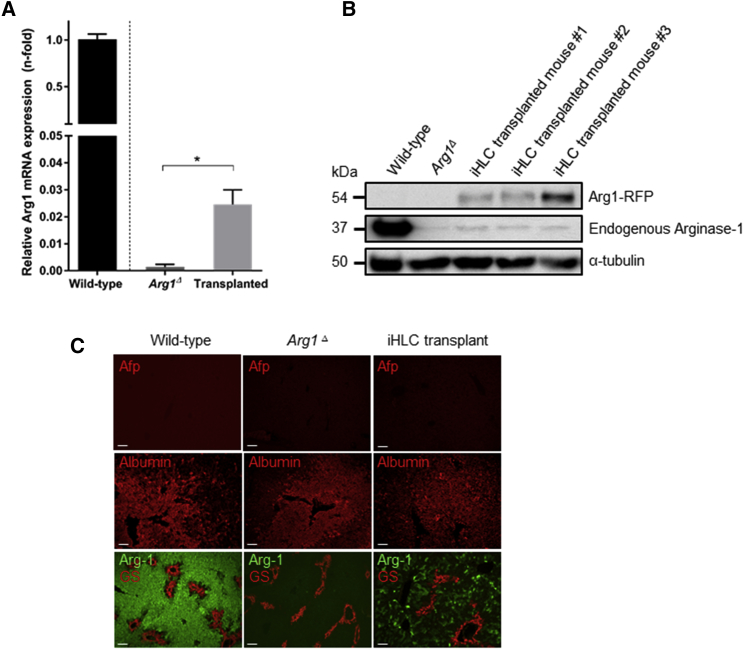
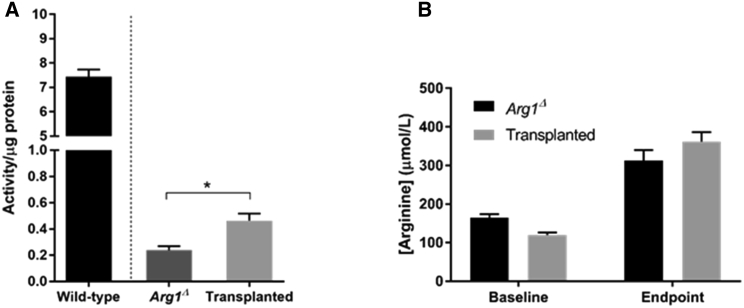
Real-time qPCR analysis was carried out to evaluate Arg1 mRNA in liver using primers in the region encoded by exons 7 and 8. Our data show significantly increased Arg1 transcripts in liver tissues of iHLC-transplanted mice compared to non-transplanted mice, confirming the restoration of full-length Arg1 mRNA expression after gene correction (Figure 4A). However, the level was still distinctly lower than in livers obtained from wild-type mice. After excision of the ITR-flanked PGK-Puro-TK cassettes by piggyBac transposase (PBx), the C-terminal Arg1-RFP should be expressed under the control of the endogenous Arg1 promoter. Hence, hepatocytes expressing fusion protein Arg1-RFP were analyzed by western blot analysis. While wild-type liver samples showed a strong immunoreactive 37 kDa Arg1 band, appreciable amounts of Arg1-RFP fusion product were detected in the iHLC-transplanted mice (Figure 4B). Next, we performed immunohistochemical staining in liver sections to detect restored arginase-1 protein and examine repopulation efficiency. As shown in Figure 4C, the results clearly demonstrate that transplantation of the edited iHLCs improves arginase-1 expression as compared to Arg1Δ mice. Moreover, the transplanted iHLCs did not develop into hyperplastic nodules. Interestingly, scattered distribution of arginase-1 expression was observed throughout the regenerated liver parenchyma in the iHLC-transplanted mice with no signs of colocalization with glutamine-synthetase surrounding the central veins. As in sections from wild-type and Arg1Δ mice, immunostaining of liver slices obtained from iHLC-transplanted mice showed only the hepatic marker albumin (Alb) expression and lack of immature hepatocyte-specific marker alpha-fetoprotein (Afp) expression, providing evidence of in vivo maturation of iHLCs. Together, our data indicate that genetically corrected mouse iPSCs can express the Arg1 protein once they differentiate into mature hepatocytes. Finally, we assessed the potential of restored Arg1 for improving functional enzyme activity. Although iHLC-transplanted mice showed significantly higher arginase-1 activity when compared to Arg1Δ mice, the level is still considerably low compared to normal activity (Figure 5A). No significant differences were observed in blood arginine levels in the iHLC-transplanted mice compared to Arg1Δ mice at both baseline (4 days post-tamoxifen administration) and humane endpoint, when arginine levels are substantially increased (Figure 5B).
Figure 4.
Arg1 Expression Profile in Liver Tissues
(A) Real-time qPCR analysis of Arg1 gene expression in liver tissues. The Ct values of all genes were normalized to the Ct values of GAPDH. The y axis represents the fold change of gene expression compared with wild-type liver using the comparative threshold method (2−ΔΔCt). Values are mean ± SEM for n = 3–10. Statistical significance between groups was determined by Student’s t test (*p < 0.05). (B) Western blot analysis of total liver extracts (20 μg/well). Arg1 protein expression was evaluated by immunoblotting with an anti-Arg1 antibody (C-terminal) and anti-red fluorescent protein (RFP) antibody. α-tubulin was used as loading control. (C) Immunostaining for Arg1 (Alexa Fluor 488, green), alpha-fetoprotein (Afp), albumin, and glutamine synthetase (GS) (Alexa Fluor 594, red) in serial liver sections from representative mice. Scale bars, 95 μm.
Figure 5.
Analysis of Hepatic Function of iHLC-Transplanted Mice
(A) Arg1 enzyme activity in livers of iHLC-transplanted mice measured at endpoint compared to wild-type and non-transplanted Arg1Δ mice. (B) Measurement of blood arginine levels at baseline (4 days after tamoxifen administration) and humane endpoint. Values are mean ± SEM for n = 3–10. Statistical significance between groups was determined by Student’s t test (*p < 0.05).
Discussion
In this study, we demonstrate TALEN-mediated gene editing to repair the dysfunctional Arg1Δ allele in iPSCs, in concert with transplantation-based studies. Our data reveal that TALEN-mediated site-specific genome modification in mouse iPSCs was similar in efficiency with the CRISPR/Cas9 system.24 Our application of TALEN-mediated gene repair highlights the feasibility and potential for gene-editing strategies using engineered cell therapies, albeit with modest improvement in survival after iHLC transplantation in the arginase-1 KO mouse model. qPCR and western blot analyses verified the presence of repaired cells expressing arginase-1. Nevertheless, transplantation of repaired iHLCs only resulted in about 5% repopulation of livers, and the lifespan of the transplanted mice could only be modestly extended by up to a week in some mice. The transplanted cells failed to fully recapitulate the normal liver distribution of arginase-1 in the correct metabolic zones, hence leading to marginal urea cycle function and elevation of blood arginine.
The major challenges highlighted by this study are the low rates of engraftment and the lack of hepatocyte repopulation in the correct liver zones. There are two main metabolic zonations in the liver. Metabolic activities such as glycolysis, lipogenesis, and xenobiotic disposal preferentially localized in perivenous (PV) areas encode glutamine synthetase, GLT1 (a glutamate transporter), and RHBG (an ammonia transporter).28 In contrast, key enzymes of the urea cycle such as ARG1 and carbamoyl-phosphate synthase (CPS1) are preferentially expressed in the periportal (PP) regions.
Intrasplenic transplantation of hepatocytes, which we employed in this study, was reported to facilitate cell integration in PP locations.29 We also performed partial hepatectomy to create a growth advantage for transplanted cells.27 However, we could not achieve optimum engraftment and functional regeneration for long-term therapeutic effects. We are not certain as to why transplanted edited cells were distributed in a scattered pattern and not in clusters as seen in our previous work30 and by others,31 since similar protocols were used for transplantation studies. Further work is required to understand these phenomena. Taking our current study and previous observations30 together, we reasoned that proper integration of transplanted cells in the correct metabolic zone of liver parenchyma is critical for enhancing the metabolic urea cycle capacity of the liver. The success of the hepatocyte infusion protocol is likely to depend on engraftment of sufficient numbers of hepatocytes in the PP loci for optimum arginase-1 enzyme activity. There are several questions to be addressed: (1) What is the optimal differentiation stage of iHLCs to achieve the highest level of engraftment and urea cycle function? (2) As the remaining hepatocytes after partial hepatectomy still have extensive proliferative potential, will they constrain competitive advantage for the transplanted cells? (3) Is there senescence or any immunogenic response to the newly corrected gene product of transplanted iHLCs? One of the major concerns of iPSC development is the potential tumorigenicity of iPSCs and their progeny.32 However, recent findings using in vitro differentiated iPSC-derived cells have sparked optimism over their therapeutic potential. There was no evidence of immune rejection of iPSCs that have matured to an adult fate, including endothelial cells, hepatocytes, and neuronal cells upon transplantation into syngeneic mice.33, 34 The discrepancies between those studies may be attributable to different iPSC lines used in their experiments. In this regard, it should be noted that in vitro differentiated gene-edited cells may have distinctive immunogenicity due to genetic manipulation and their long culture time. It remains to be determined whether iHLCs derived from gene-edited iPSCs used in our transplantation studies were immunogenic and if transient immunosuppression is required following transplant. (4) Can zonal regeneration of gene-edited hepatocytes be manipulated to improve therapeutic efficacy? Several signaling pathways have been identified to direct zonal organization including Wnt/β-catenin35, 36 and its antagonistic pathway, Ras/MAPK/Erk.37 Further investigation is needed to refine our current understanding in shaping desired metabolic zonation.
In summary, this report demonstrates another successful application of gene editing in iPSCs. Despite the lack of efficient metabolic functional repair after transplantation, the application of gene correction into HLCs might still be a realistic goal for ex vivo gene therapy of liver diseases with further experimental optimization. To our knowledge, these results represent the first description of transplantation using cells derived from HDR-mediated repair of iPSCs for arginase-1 deficiency. The development of efficient targeted gene editing using TALENs and CRISPR/Cas9 systems could open exciting new avenues for arginase-1 gene therapy.
Materials and Methods
Mice and Cell Source
The inducible Arg1-deficient mouse strain (herein referred to as Arg1-Cre mice), derived from parental strains Arg1flox (JAX strain 008817, C57BL/6-Arg1tm1Pmu/J) and CreERT2 (JAX strain 008463, B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J) were injected intraperitoneally (i.p.) on 5 sequential days with tamoxifen to induce global Arg1 deficiency (herein referred to as Arg1Δ mice) as previously described.21 All procedures were reviewed and approved by the Queen’s University Animal Care Committee (approval #Funk-2011-048-R1-A4) and conformed to the Guidelines of the Canadian Council on Animal Care. Arg1Δ mouse iPSCs were generated from Arg1Δ PMEFs as described in a previous study.24
Design and Assembly of TALENs Targeting the Mouse Arg1 Gene
A pair of TALENs (left TALEN 5′-GCTTTGTGTGCGAGT-3′, right TALEN 5′-TCCTGACATTCCTGTCA-3′) were designed and assembled to target a region close to exon 6 in the Arg1 gene using the GoldyTALEN scaffold method.38 The TALEN target site comprises binding sites for two TALE-FokI fusion proteins that are each preceded by a T at the 5′ end and a unique restriction enzyme (BspHI) recognition site within the spacer region. Specificity of TALENs was examined using NCBI Primer-BLAST. Potential off-target sites were assessed in silico using the Paired Target Finder tool on the TAL Effector Nucleotide Targeter 2.0 web interface to scan the mouse genome for sequences containing mismatches within the TALEN pair target site.39 The repeat variable di-peptide (RVD)-containing units for TAL-7 (NN HD NG NG NG NN NG NN NG NN HD NN NI NN NG) and TAL-8 (NG NN NI HD NI NN NN NI NI NG NN NG HD NI NN NN NI) were assembled using the Golden Gate approach.40 After assembly, the RVDs were cloned into the pC-Goldy TALEN destination vector for expression in mammalian cells.
Functional Evaluation of TALEN Cutting Efficiency
Arg1Δ iPSCs (3 × 106) were electroporated with equal amounts of each TALEN plasmid DNA (7 μg each) to induce double-stranded breaks (Gene Pulser System, 250 V, 500 μF, 0.4 cm cuvettes) (Bio-Rad, Mississauga, ON, Canada). Genomic DNA from the electroporated cells was isolated 2 days later. The genomic region surrounding the TALEN target site (within intron 6 and 3′ UTR of the Arg1 gene) was amplified with Phusion high-fidelity DNA polymerase (NEB, Ipswich, MA). Primers used were as follows: forward 5′-CTAACCGTCATTAACTTCACTCTG-3′, reverse 5′-GCACTGTCTAAGCCCGAGAGTATC-3′. Purified PCR products were subjected to a re-annealing process using a step gradient (95°C–25°C over 30 min) to enable heteroduplex formation. For mismatch cleavage assays, the annealed products were treated with Surveyor nuclease (Transgenomics, Omaha, NE), and resultant cleavage products (two bands at 162–185 bp and 167–185 bp) were analyzed in 8% Tris-borate-EDTA polyacrylamide gels. The indel efficiency was quantified based on the relative band intensities measured using Quantity One Software (Bio-Rad, Mississauga, ON, Canada). The indel percentage was calculated using the following formula: 100 × (1 − (1 − (b + c)/(a + b + c))1/2), wherein a is the intact band and b and c represent the Surveyor nuclease digestion products. The PCR products were also assessed for gene modification by BspHI (NEB, Ipswich, MA) digestion, which gave rise to 183 and 169 bp fragments. To confirm TALEN-mediated gene modification, PCR amplicons were subcloned into pCR 2.1-TOPO TA vector (Invitrogen, Carlsbad, CA) and individual colonies were subjected to sequence analysis.
TALEN-Mediated Gene Targeting and Excision of Selection Markers Using PBx
A custom-designed repair targeting vector24 with Arg1 homology arms, which consists of exons 7 and 8 cDNA fused to the coding sequence of monomeric RFP, a hybrid PGK-EM7 promoter, a positive-negative drug resistance cassette carrying a puromycin-thymidine kinase resistance gene, and a self-cleaving 2A peptide (T2A), all flanked by the piggyBac transposon ITRs (Transposagen, Lexington, KY) was introduced into the cells together with TALEN plasmids to initiate HDR. The targeting vector was linearized with NotI (NEB, Ipswich, MA) prior to electroporation. L-755,507 (5 μM, Tocris BioScience, Minneapolis, MN) was also added to the cells to increase the efficiency of donor incorporation.25 Puromycin treatment (1 μg/mL) was initiated 4 days after electroporation to identify cells that successfully integrated the targeting vector. The resulting colonies were picked on day 14 and expanded on puromycin-resistant mitotically-inactivated PMEFs (iPMEFs) for further characterization. PCR genotyping was carried out for both left and right arms independently using LongAmp Taq DNA polymerase (NEB, Ipswich, MA), with one of the two PCR primers designed to anneal outside the region spanned by both homology arms to ensure on-target integration. Primers were as follows: left arm forward (LA-F) 5′-GTCTGCAGAGATTCGGAAGGTAAC-3′, reverse (LA-R) 5′-CTGACTAGGGGAGGAGTAGAAGGT-3′; right arm forward (RA-F) 5′- CCGTAATGCAGAAGAAGACC-3′, reverse (RA-R) 5′-GGCTATTGAAGATTTAACATTTGG-3′. The selection cassette was removed by introducing PBx (Transposagen, Lexington KY) into the puromycin-resistant cells. Four days after piggyBac excision, ganciclovir selection (2 μM, Cayman Chemical, Ann Arbor, MI) was carried out to eliminate the cells that have residual vector expression. After a 2-week selection period, colonies were picked and expanded on gelatin-coated dishes without feeder cells. Genomic DNA was extracted and subjected to PCR-based screening of transposon-excised clones using primers as follows: PBx-F 5′-TCACAGGACTTACAGTGATC-3′; PBx-R 5′-CATGAACTCCTTGATGACG-3′ and PB3-P2-Forward 5′-GCGACGGATTCGCGCTATTTAGAAA-3′ (transposon-specific primer). PCR products resulting from the removal of the selection cassette were sequenced to confirm the intended genetic modification and the presence of the TTAA sequence at the excised site.26
Differentiation of Mouse iPSCs into HLCs In Vitro
TALEN-mediated gene-edited cells were differentiated into HLCs using a modified stepwise protocol as previously described.24 In brief, mouse iPSCs were trypsinized into single-cell suspensions and resuspended in advanced RPMI containing 10% fetal bovine serum (FBS) (Wisent, St-Bruno, Quebec, Canada), 100 ng/mL activin A (R&D Systems, Minneapolis, MN), and 50 ng/mL Wnt3a (R&D Systems, Minneapolis, MN). One million cells per well were seeded into 12-well plates coated with 2% Matrigel (BD Biosciences, Mississauga, ON, Canada) and incubated at 37°C/5% CO2 for direct definitive endoderm (DE) induction. FBS was reduced to 0.2% on the following day until day 5. On day 6–10, the medium was replaced with hepatic commitment medium (advanced RPMI, 2% FBS, 50 ng/mL BMP4 (PeproTech, Montreal, Quebec, Canada), 20 ng/mL FGF-2, (PeproTech, Montreal, Quebec, Canada). On day 11-15, the medium was replaced with advanced RPMI containing 2% FBS, and 20 ng/mL hepatocyte growth factor (HGF; PeproTech, Montreal, Quebec, Canada) to promote the expansion of early hepatic progenitor cells. Cells were maintained in advanced RPMI supplemented with 2% FBS, 20 ng/mL HGF, 20 ng/mL Oncostatin M (R&D System, Minneapolis, MN), 50 nM dexamethasone (Sigma-Aldrich, St. Louis, MO), and 1× insulin-transferrin-selenium (Corning, Corning, NY) for at least 10 days prior to transplantation.
iHLC Transplantation Studies
Twelve-week-old female Arg1-Cre mice were used as recipients. The mice were conditioned with retrorsine (70 mg/kg i.p., twice at 2 week intervals) (Sigma-Aldrich, St. Louis, MO) prior to transplantation. Two-thirds partial hepatectomy27 was performed after the last retrorsine dose to create a selective growth advantage for transplanted cells. In brief, mice were anesthetized with isoflurane/O2 and 20 mg/kg subcutaneously (s.c.) of Tramadol for pre-surgery analgesia. The left lobe and median lobes were ligated with a silk filament at the base before resection. Two million iHLCs (donor cells) were suspended in 100 μL of Hanks’ Balanced Salt Solution (HBSS) and injected slowly over 20–30 s into the lower pole of the spleen. All operations were performed on a heating pad with sterile surgical techniques. Immediately after surgery, mice received 1 mL of warmed saline and 2.5 mg/kg Meloxicam. Five hundred microliters of saline, Tramadol, and Meloxicam were administered for the next 3 days. The following experimental groups were defined: group 1-wild-type (n = 3), group 2-Arg1 knockout (n = 3), and group 3-transplantation of repaired iHLCs (n = 10). Fifteen weeks later, allowing sufficient time for donor cell engraftment, the 5-day sequential tamoxifen-induced Arg1 KO regimen was carried out.21 Changes in body weight of mice were carefully monitored daily during the experimental period. Humane endpoints were defined as body weight loss of >15% relative to the weight at the time of final tamoxifen administration, accompanied by hunched posture. The endpoint took place between days +11 to +14 with the mean at day +13.21, 30 Hence, lifespan extension of mice that underwent iHLC transplantation was recorded from this time point (day +13).
PCR Genotyping
Genomic DNA from tail biopsies was extracted by standard protocols and subjected to PCR for genotyping using primer sets (Integrated DNA Technologies, Coralville, IA) as follows: F1 5′-TGCGAGTTCATGACTAAGGTT-3′, R1 5′-AAAGCTCAGGTGAATCGG-3′, and R2 5′-GCACTGTCTAAGCCCGAGAGTATC-3′. Cycle parameters were as follows: denaturation at 94°C for 30 s, annealing at 64.5°C for 1 min, and elongation at 72°C for 1 min for 35 cycles.
Liver Engraftment Analysis by Immunohistochemistry
Liver tissues were collected immediately at the time mice were sacrificed. Paraformaldehyde-fixed and paraffin-embedded mouse liver sections (4 μm thick) were stained following standard immunohistochemical protocols, performing heat-induced antigen retrieval (10 mM citrate buffer [pH 6.0], 0.02% Tween 20) before incubation with primary antibodies. IHC analysis (rather than direct fluorescence detection of red fluorescent protein [RFP]) was preferred as livers exhibit high autofluorescence. Primary antibodies used in this study were as follows: Arg-1 (Abcam, Cambridge, MA, #ab91279, 1:200), glutamine synthetase (Abcam, Cambridge, MA, #ab64613, 1:200), Alb (Abcam, Cambridge, MA, #ab19196, 1:400), Afp (Novus Biologicals, Littleton, CO, #NBP-762755, 1:200). Fluorescent secondary antibodies, Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) (Molecular Probes, Eugene, OR, #A-11068, 1:400), Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes, Eugene, OR, #A-11005, 1:400), and Texas Red goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, #111-075-144, 1:200) were used for primary antibody detection. Slides were then dehydrated and mounted with ProLong Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA). Visualization was performed with a fluorescent microscope (Leica, DM IRB, Richmond Hill, ON).
Gene Expression Analysis
Liver tissues were pulverized in liquid nitrogen prior to RNA extraction. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), followed by RNA cleanup using a GeneJET RNA Purification Kit (Fisher Scientific, Unionville, ON, Canada) and treated with DNase I (1 μg/μL, Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. RNA quality was assessed with the RNA 6000 Nano Kit (Agilent Technologies, Mississauga, ON, Canada). cDNA was synthesized from 1 μg of total RNA using an iScript cDNA synthesis kit (Bio-Rad, Mississauga, ON, Canada). qPCR was performed using a thermal cycler (Applied Biosystems Model 7500) with SYBR Green PCR master mix (Bio-Rad, Mississauga, ON, Canada). Melting curves were performed on completion of the cycles to ensure absence of nonspecific products. Primers (Integrated DNA Technologies, Coralville, IA) were designed to span exon-exon boundaries to avoid the amplification of genomic DNA. Primer sets were as follows: Arginase-1 (accession number, NM_007482), 5′-ACAAGACAGGGCTCCTTTCAG-3′ (sense), 5′-TGAGTTCCGAAGCAAGCCAA-3′ (antisense); and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (accession number: NM_001289726), 5′-CATGGCCTTCCGTGTTCCTA-3′ (sense), 5′-ATGCCTGCTTCACCACCTTCT-3′ (antisense). Relative gene expression was calculated using the comparative threshold method (2−ΔΔCT) and is presented as fold-change of transcripts for target genes normalized to the expression of housekeeping gene GAPDH. Gene expression values in wild-type hepatocytes were set to 1.
Western Blot Analysis
Liver tissues were pulverized in liquid nitrogen and homogenized in ice-cold radioimmunoprecipitation assay (RIPA) buffer (Millipore, Bedford, MA), including protease inhibitor cocktail (Roche, Mississauga, ON, Canada). Twenty-micrograms of centrifuged, clarified protein samples were subjected to western blot analysis and probed with rabbit polyclonal anti-Arg1 antibody (C-terminal) (Abcam, Cambridge, MA, #ab91279, 1:10,000), mouse monoclonal anti-RFP antibody (Rockland Immunochemicals, Limerick, PA, #200-301-3795, 1:2,000), and mouse polyclonal anti-α-tubulin antibody (Sigma-Aldrich, St. Louis, MO, #T5168, 1:5,000) used as loading control. Immunoreactive proteins were detected using horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibody (Sigma-Aldrich, St. Louis, MO, 1:5,000) and visualized by enhanced chemiluminescence detection (GE Healthcare, Mississauga, ON, Canada).
Arginase Activity Assay and Biochemical Analysis
Arginase activities of all samples were assayed as described previously.21, 30 One unit of activity is defined as 10 nmol urea/μg protein. Whole-blood samples were obtained from the submandibular vein on day 4 and 14 following the final tamoxifen (or vehicle) injection and humane endpoint (for mice surviving beyond day 14 post-tamoxifen administration). Drops of blood were collected into microcapillary tubes prior to transfer onto the Whatman 903 filter paper cards (GE Healthcare, Mississauga, ON, Canada). Sample preparation was based on the method described previously30 prior to analysis by mass spectrometry.
Statistical Analysis
All experiments were performed at least in three biological replicates. Survival curves were computed in each group of mice using the Kaplan-Meier method and compared across groups using the log rank test. All results are expressed as mean ± standard error of mean (SEM). Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA). Means were compared using the two-tailed Student’s t test. p values of < 0.05 were considered statistically significant.
Author Contributions
Y.Y.S. and C.D.F. designed the experiments and wrote the manuscript. Y.Y.S. and C.R.R. performed the experiments and analyzed the data. L.L.B. conducted the animal experiments. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
Y.Y.S. is supported by a fellowship from the Urea Cycle Disorders Consortium (UCDC; U54HD061221), which is a part of the NIH Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the Office of Rare Diseases Research (ORDR), the National Center for Advancing Translational Science (NCATS), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). This work was supported by a microgrant from the Rare Disease Foundation and the BC Children’s Hospital Foundation (BCCHF) (#18-19 to Y.Y.S.). C.D.F. is supported by the Canada Research Chairs program and the Canadian Institutes of Health Research (CIHR) (MOP-341036).
References
- 1.Kim H., Kim J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 2.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 3.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 5.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 6.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Q., Lee Y.K., Schaefer E.A., Peters D.T., Veres A., Kim K., Kuperwasser N., Motola D.L., Meissner T.B., Hendriks W.T. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma N., Liao B., Zhang H., Wang L., Shan Y., Xue Y., Huang K., Chen S., Zhou X., Chen Y. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J. Biol. Chem. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakuma T., Hosoi S., Woltjen K., Suzuki K., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013;18:315–326. doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- 10.Park C.Y., Kim J., Kweon J., Son J.S., Lee J.S., Yoo J.E., Cho S.R., Kim J.H., Kim J.S., Kim D.W. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc. Natl. Acad. Sci. USA. 2014;111:9253–9258. doi: 10.1073/pnas.1323941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tesson L., Usal C., Ménoret S., Leung E., Niles B.J., Remy S., Santiago Y., Vincent A.I., Meng X., Zhang L. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 12.Wefers B., Meyer M., Ortiz O., Hrabé de Angelis M., Hansen J., Wurst W., Kühn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl. Acad. Sci. USA. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander J.D., Cade L., Khayter C., Reyon D., Peterson R.T., Joung J.K., Yeh J.R. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T., Liu B., Spalding M.H., Weeks D.P., Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhang F., Li X., Baller J.A., Qi Y., Starker C.G., Bogdanove A.J., Voytas D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161:20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdanove A.J., Schornack S., Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Moscou M.J., Bogdanove A.J. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 19.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 20.Sin Y.Y., Baron G., Schulze A., Funk C.D. Arginase-1 deficiency. J. Mol. Med. (Berl.) 2015;93:1287–1296. doi: 10.1007/s00109-015-1354-3. [DOI] [PubMed] [Google Scholar]
- 21.Sin Y.Y., Ballantyne L.L., Mukherjee K., St Amand T., Kyriakopoulou L., Schulze A., Funk C.D. Inducible arginase 1 deficiency in mice leads to hyperargininemia and altered amino acid metabolism. PLoS ONE. 2013;8:e80001. doi: 10.1371/journal.pone.0080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasten J., Hu C., Bhargava R., Park H., Tai D., Byrne J.A., Marescau B., De Deyn P.P., Schlichting L., Grody W.W. Lethal phenotype in conditional late-onset arginase 1 deficiency in the mouse. Mol. Genet. Metab. 2013;110:222–230. doi: 10.1016/j.ymgme.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrage L.C., Sun Q., Elsea S.H., Jiang M.M., Nagamani S.C., Frankel A.E., Stone E., Alters S.E., Johnson D.E., Rowlinson S.W., Members of Urea Cycle Disorders Consortium Human recombinant arginase enzyme reduces plasma arginine in mouse models of arginase deficiency. Hum. Mol. Genet. 2015;24:6417–6427. doi: 10.1093/hmg/ddv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sin Y.Y., Price P.R., Ballantyne L.L., Funk C.D. Proof-of-concept gene editing for the murine model of inducible arginase-1 deficiency. Sci. Rep. 2017;7:2585. doi: 10.1038/s41598-017-02927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser M.J., Ciszczon T., Elick T., Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell C., Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 28.Gebhardt R., Matz-Soja M. Liver zonation: novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 2014;20:8491–8504. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajvanshi P., Kerr A., Bhargava K.K., Burk R.D., Gupta S. Studies of liver repopulation using the dipeptidyl peptidase IV-deficient rat and other rodent recipients: cell size and structure relationships regulate capacity for increased transplanted hepatocyte mass in the liver lobule. Hepatology. 1996;23:482–496. doi: 10.1002/hep.510230313. [DOI] [PubMed] [Google Scholar]
- 30.Ballantyne L.L., Sin Y.Y., Al-Dirbashi O.Y., Li X., Hurlbut D.J., Funk C.D. Liver-specific knockout of arginase-1 leads to a profound phenotype similar to inducible whole body arginase-1 deficiency. Mol. Genet. Metab. Rep. 2016;9:54–60. doi: 10.1016/j.ymgmr.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espejel S., Roll G.R., McLaughlin K.J., Lee A.Y., Zhang J.Y., Laird D.J., Okita K., Yamanaka S., Willenbring H. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. J. Clin. Invest. 2010;120:3120–3126. doi: 10.1172/JCI43267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao T., Zhang Z.N., Rong Z., Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 33.Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S., Sugiura M., Ideno H., Shimada A., Nifuji A., Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 34.Guha P., Morgan J.W., Mostoslavsky G., Rodrigues N.P., Boyd A.S. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013;12:407–412. doi: 10.1016/j.stem.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Torre C., Perret C., Colnot S. Transcription dynamics in a physiological process: β-catenin signaling directs liver metabolic zonation. Int. J. Biochem. Cell Biol. 2011;43:271–278. doi: 10.1016/j.biocel.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Planas-Paz L., Orsini V., Boulter L., Calabrese D., Pikiolek M., Nigsch F., Xie Y., Roma G., Donovan A., Marti P. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 37.Braeuning A., Menzel M., Kleinschnitz E.M., Harada N., Tamai Y., Köhle C., Buchmann A., Schwarz M. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007;274:4766–4777. doi: 10.1111/j.1742-4658.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- 38.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G., 2nd, Tan W., Penheiter S.G., Ma A.C., Leung A.Y. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., Bogdanove A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]