Abstract
The important challenge about cancer is diagnosis in primary stages and proper treatment. Although classical clinico-pathological features of the tumor have major prognostic value, the advances in diagnosis and treatment are indebted to discovery of molecular biomarkers and control of cancer in the pre-invasive state. Moreover, the efficiency of available therapeutic options is highly diminished, and chemotherapy is still the main treatment due to lack of enough specific targets. Accordingly, finding the new noninvasive biomarkers for cancer is still an important clinical challenge that is not achieved yet. There are current technologies to screen, diagnose, prognose, and treat cancer, but the limitations of these implements and procedures are undeniable. Liquid biopsy as a noninvasive method has a promising future in the field of cancer, and exosomes as one of the recent areas have drawn much attention. In this review, the potential capability of exosomes is summarized in cancer with the special focus on breast cancer as the second cause of cancer mortality in women all around the world. It discusses reasons to choose exosomes for liquid biopsy and the studies related to different potential biomarkers found in the exosomes. Moreover, exosome studies on milk as a specific biofluid are also discussed. At last, because choosing the method for exosome studies is very challenging, a summary of different techniques is provided.
Keywords: extracellular vesicles, bio-fluids, breast milk, biofluid, exosome, clinical, application
Main Text
Identification of specific biomarkers is required for early detection and cancer screening to conquer cancer. Due to the heterogeneous characteristics of most tumors and different genomic profiles, conventional biopsies cannot reflect the whole nature of primary or secondary tumors (metastasis area). Each tissue biopsy provides a small sample size and fails to reflect tumor heterogeneity, which is essential in the treatment procedure.1, 2 Moreover, to detect the therapy response, repeated cancer cell sampling is needed to identify tumor genetic changes during cancer treatment, but after surgery, the tumor is not accessible to be monitored during treatment.3 Accordingly, new low-cost and noninvasive sampling is needed for early detection, screening, and investigating tumor dynamics as well as the risk of relapses.4 Isolation of genetic materials from bioliquids is a new and minimally invasive method to diagnose different types of cancer. In most cancers, some compartments of tumor cells (e.g., DNA, exosome, etc.) or even the whole cancer cells segregate from the original tumor bulk and enter the bloodstream or any other bioliquid.5 Relative to direct tumor biopsies, the easy-to-obtain nature of bioliquids makes them an attractive alternative source for clinical application. Releasing exosomes from heterogeneous cancer cells in biofluids could provide the potential information of tumors.3, 6
Exosomes were first reported by Pan and Johnstone7 in 1983 at McGill University when culturing sheep reticulocytes. These lipid bilayered vesicles with endocytic origins are released into the extracellular region8 by a variety of mammalian cells, including cancer cells.9 Exosomes from different types of cells enclose different proteins that have important roles in their biogenesis and are used as markers for their recognition in experimental procedures. Some examples of these proteins are Rab GTPase family,10 tetraspanins (CD9, CD81,11 and CD6312), annexins,13 and chaperones (heat shock protein [HSP] 7012 and HSP9014). Exosomes are exciting in a vast range of biofluids, including serum,15 normal and malignant urine,16 plasma, breast milk, saliva,17 malignant pleural effusions,18 bronchial lavage fluid,19 ocular samples, tears,20 nasal lavage fluid,21 semen,22 synovial fluid,23 amniotic fluid, and pregnancy-associated serum.24
The current review discusses advantages of liquid biopsy, especially milk as a specific breast biofluid for exosome-based studies. Then different exosome studies related to diagnosis, treatment, and response to therapy in breast cancer (BC)16 were investigated. Finally, recent studies on exosomal genetic materials in the field of cancer biomarkers were scrutinized, and the study concentrated on different methods related to laboratory works on cancer exosomes.
Encouraging Reasons to Use Exosomes for Liquid Biopsy
Traditional biopsies, such as fine needle aspiration, rely on accessing the tumor cells25, but exosome-based liquid biopsy relies on subcellular particles and their cargos. Compared with the other sources of liquid biopsies, exosomes have superiority in different aspects. First, compared with other subcellular particles such as apoptotic bodies and microvesicles, exosomes are more homogeneous in terms of size,26 and their cup-shaped appearance makes them easily distinguishable through electron microscopy.27 Second, unlike circulating tumor cells (CTCs) thought to be a new source for cancer biomarkers,6 many isolation and characterization protocols are developed to use exosomes in research and therapy.28 There are many commercial kits that let us quickly and efficiently isolate exosomes from a small amount of human body liquid.29 Third, exosomes express specific markers including HSP70 and Alix, which can be applied to separate exosomes from other subcellular vesicles.30 Moreover, exosomes can mirror the original cell markers by presenting specific surface proteins31 and even their target cells.32 These features cause easy isolation of both the tissue and target cell-specific exosomes. Fourth, exosomes are stable in the circulation33 and are found in almost every potential body fluid; therefore, they can be used as diagnostic tools for many diseases including BC. One of the BC clinical antibody-targeted receptors carried by exosomes, HER2,34 might trigger the idea to use exosomal HER2 as a prognostic tool through the liquid biopsy. Fifth, the nucleic acid content of exosomes is a practical source for cancer investigation. Double-stranded DNA content of exosomes can show the mutational status of the original cell.35 Moreover, exosomes represent their parental tumor-specific RNA and protein profiles,33 and their architecture protects circulating RNA and microRNA (miRNA) from RNase catalytic function.36 Circular RNAs in exosomes are more various than that of the cell of origin.37 Therefore, exosomal nucleic acids can be utilized to find genetic signatures in patients with cancer.38 Databases such as ExoCarta39 and Urinary Exosome Protein Database40 facilitate the sharing of data about the nucleic acid and protein content of exosomes.
Breast Milk- and Breast Fluid-Derived Exosomes
The majority of biomarker-based studies on BC focus on blood-derived exosomes. Most of the cells in the human body, including breast tissue cells, release their exosomes into the blood.41 Therefore, the concentration of breast-specific exosomes (BSEs) seems to decrease in blood samples. However, breast fluids, especially milk, might be full of BSEs. Thus, breast milk may be a more reliable source for exosome studies and finding specific biomarkers. Hypothetically, BSEs may have breast tissue-specific markers that let them bind specifically to the BC cells and could be used in nanoengineering and targeted therapy. In addition, collecting human milk in large scales for commercial purposes is more practical and less invasive than that of human serum. The yield of milk-derived exosomes (335 mg/L) is enough for commercial purposes.42 In this regard, scientists traced human milk exosomes using their protein profile (including CXCL5, MIA, and KLK6).43 One study showed that drug-loaded bovine milk exosomes were used to inhibit human BC cells (MDA-MB-231 and T47D) proliferation. It concluded that bovine milk-derived exosomes increased the stability and cellular uptake of the drug. The regulatory effect of human milk on the immune system is already demonstrated;44 researchers presented the anti-cancer impact of milk exosomes by NF-KB pathway in H1299 cells.42 Furthermore, in 2016, Yassin et al.45 proposed that camel milk exosomes can be utilized as potentially safe nanocarriers. Interestingly, human milk exosomes contain high levels of transforming growth factor-β2 (TGF-β2) and could promote epithelial-to-mesenchymal transition (EMT) in MCF7 and MCF10A breast cells. Hence, secretion of TGF-β2 in breast milk might increase the risk of BC.46 In the opposite case, bovine milk exosomes decreased BC cell viability in vitro.47 The stability of bovine milk exosomes under the digestive system acidic condition makes it ideal for oral application. Loading bovine milk exosomes with paclitaxel (PTX) raised therapeutic efficacy and reduced systemic toxicity compared with free PTX in an animal model for lung cancer.48 A recent study assessed the capacity of milk-derived exosomes to optimize curcumin delivery to cancer cells. The mentioned study proved that milk exosomes encapsulated with curcumin had higher solubility in hydrophilic solutions, which elevated the curcumin delivery to cancer cells. Exosomal curcumin was stable under digestive system conditions, as well as endocytosis by human intestinal cells in vitro.49 In conclusion, milk exosomes might be cost-effective drug carriers and suitable for oral application. Further investigation on human breast milk exosomes may discover new roles for them in drug delivery.
Exosomes’ Role in BC Diagnosis, Treatment, and Resistance to Therapy
Diagnosis
Similar to other cancers, BC tumor-related exosomes exert multiple functions in tumor growth, metastasis, and chemoresistance. Exosomes and their components (DNA, RNA, and proteins) may influence immune escape, tissue invasion, metastasis, and angiogenesis.50 Existence of circulating exosomal miRNAs might be a diagnostic biomarker for breast malignancies. Triple-negative BC (TNBC) cells produce exosomes containing certain proteins and miRNAs (Figure 1), which result in malignant transformation. In comparison with malignant cells, normal cells release exosomes that pack neutral miRNAs.51 Several investigations showed correlation between exosomal miRNA (miR-195,52 miR-21,53 miR-484/19154) and tumorigenesis and pathological stages. Hannafon et al.41 indicated that levels of miR-21 and miR-1246 were higher in plasma exosomes of patients with BC compared with those of the control samples. Moreover, interestingly, the result of the study by Palma et al.51 showed that exosomes released by BC cells can be classified depending on their miRNAs content. Analyzing small RNA content of the serum exosomes of five patients with BC by RNA sequencing (RNA-seq) technique showed that BC diagnosis was associated with changes in the levels of specific subtypes of miRNAs.55 In 2016, Fiskaa et al.56 assessed the whole small RNA content of nine BC cell lines and indicated exosomal small RNA signatures to identify BC cell lines from each other and also from those of other non-BC cell lines. Based on the abundant studies focused on the role of exosomes in BC, Park et al.57 designed a diagnostic kit. This useful equipment measures the amount of 10 miRNAs in breast tumor-derived exosomes. The aggregation of some miRNAs indicates BC samples, and a decreased amount of them directs to normal samples.57 To sum up, the miRNA content of bioliquid exosomes can be potentially applied in early diagnosis and staging of patients with BC.
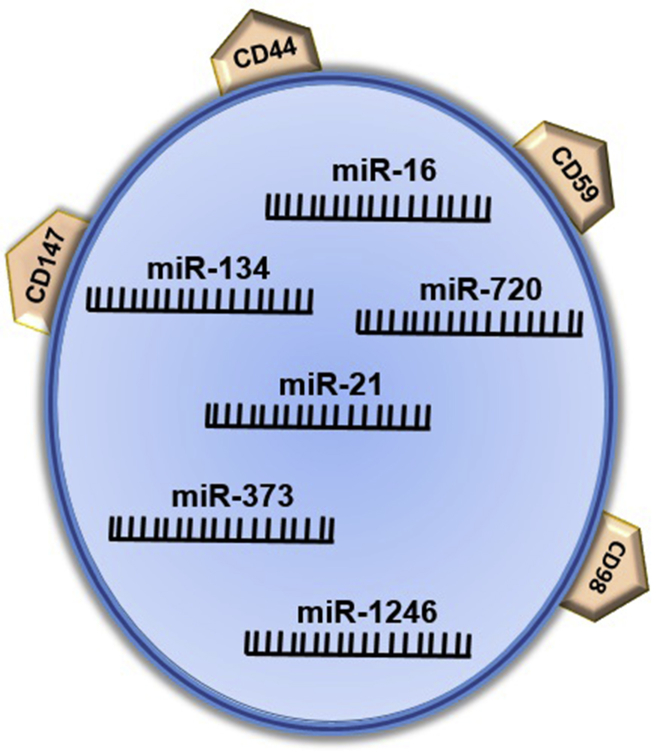
Figure 1.
Schematic Representation of TNBC Exosomes
TNBC exosomes represent some surface proteins, including CD98, CD147, and CD59, and some overexpressed miRNAs (miR-134, miR-21, miR-373, and miR-1246).
As well as miRNAs, some proteins are differentially expressed in certain stages and types of BC, which are found in the extracted exosomes. For example, CD24, likely as a late-stage BC biomarker,58 exists in seral exosomes.59 In 2005, Ryan et al.60 showed that survivin and its splice variants were differentially expressed in BC tissues and had different roles in the apoptosis of BC cells. Accordingly, Khan et al.61 considered that BC cells released survivin packed in the exosomes. Then, they found survivin-2B as an anti-apoptotic marker in the serums of patients with BC. Therefore, expression analysis of survivin-2B may serve as a diagnostic and prognostic marker in early BC stages.62
Treatment
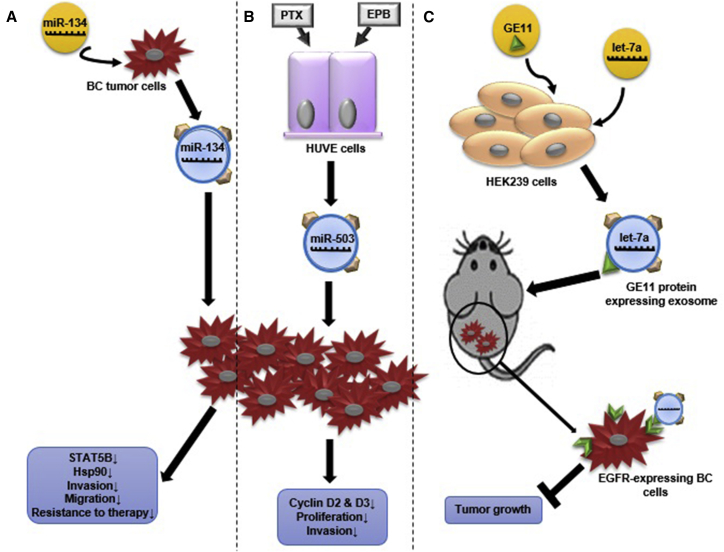
Recent research presented exosomes as novel therapeutic targets. Intrinsic and engineered exosomes can be applied as therapeutic agents to lay off the progression of the disease. It was indicated that engineered exosomes loaded with specific elements such as miRNAs can be utilized as a potential therapeutic option. O’Brien et al.,63 in 2015, found that miR-134-loaded exosomes can decrease the migration and invasion of BC cells. Additionally, it enhances the sensitivity of BC cells to anti-HSP90 agents, 17-AAG and PU-H7163 (Figure 2A). miR-503 inhibits the proliferation of BC cells and their invasive capacities by CCND2 and CCND3 knockout. By miRNA profiling, Bovy et al.64 identified that miR-503 was specifically upregulated in exosomes released from endothelial cells after treatment with PTX and epirubicin (EPB). Based on their study, endothelial exosomes loaded with miR-503 might obstruct the proliferation of tumor cells, and in that way contribute to the direct effect of taxanes and anthracyclines therapy64 (Figure 2B). An in vivo study on RAG2−/− mice showed that exosomes were potent to deliver let-7a to epidermal growth factor receptor (EGFR)-expressing BC cells. This study suggested that exosomes can be used to target EGFR-expressing BC cells by carrying off nucleic acid drugs65 (Figure 2C).
Figure 2.
Proposed Exosomes’ Role in BC Treatment
(A) miR-134 transfected Hs578Ts cells released miR-134-carrying exosomes that can downregulate STAT5B and HSP90 expression. Also, these exosomes reduce migration and invasion, and increase anti-HSP90 drug sensitivity in secondary Hs578Ts cells. (B) Human umbilical vein endothelial (HUVE) cells released miR-503-overexpressing exosomes after PTX and EPB treatment. These exosomes had the potential to reduce BC invasion and cyclin D2 and D3 expression that led to decline in BC cells proliferation. (C) Human embryonic kidney cells (HEK239) were transfected with GE11 protein (specifically binds to EGFR-expressing cells) and let-7a miRNA. HEK239 cells released GE11-expressing and let-7a-overexpressing exosomes, which bind specifically to EGFR-expressing xenograft BC tissues, and inhibited tumor development in animal model.
For the first time in 2015, Jenjaroenpun et al.66 characterized the whole RNA content of exosomes secreted by two human metastatic BC cell lines. They suggested that exosomal RNA analysis might distinguish low metastatic BC cell line (MDA-MB-436) from highly metastatic BC cell line (MDA-MB-231).66 By the RNA-seq technique, miRNA expression profiles of metastatic BC, as well as normal mammary cell lines, are identified. Based on the multiple algorithms, miR-105 is selected for in vivo and in vitro analyses. It indicated that exosomes mediate the transfer of miR-105, which efficiently breaks the cell-cell tight junctions and induces metastasis. In addition, overexpressed miR-105 in non-metastatic BC cell line (MCFDCIS)-derived exosomes induced metastasis and vascular permeability in null mice.67 In conclusion, exosomes can be used in drug delivery and targeted therapy of BC cells, or target the inhibition of the cancer signaling pathways.
Resistance to Therapy
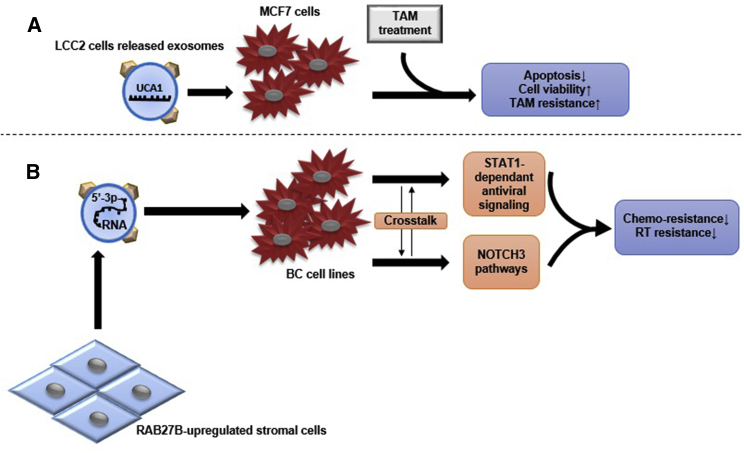
The role of exosomes in the resistance of BC to treatment was assessed in some studies. Urothelial carcinoma-associated 1 (UCA1) protein exerts a regulative effect on chemoresistance of different cancer cells (e.g., gastric, bladder, and colorectal cancer).68, 69, 70 In 2016, Xu et al.71 compared the UCA1 mRNA amount in exosomes released from tamoxifen-sensitive (MCF7) and -resistant (LCC2) BC cells. Significantly higher levels of UCA1 were detected in both LCC2 cells and their exosomes. Interestingly, LCC2-released exosomes had higher UCA1 expression than those of the LCC2 parent cells. They concluded the role of exosomal transfer of UCA1 in the induction of tamoxifen resistance in MCF-7 cells71 (Figure 3A). Boelens et al.72 demonstrated that upregulation of RAB27B protein resulted in over-release of exosomes in stromal cells. These exosomes induce the STAT1 protein in adjacent BC cells and activate NOTCH3 pathways. All of these processes lead to chemoresistance and radiation resistance in BC cells72 (Figure 3B). In this regard, using RNA-seq data, scientists assessed BC patient-derived xenograft models to predict the chemotherapy response.73 In summary, BC cells’ exosomes and stromal cells’ exosomes, which are affected by cancer niche, transfer intercellular messages in order to maintain cancer cell’s niche.
Figure 3.
Proposed Exosomes’ Role in BC Resistance to Therapy
(A) Tamoxifen-resistant BC cells (LCC2 cell line) secrete UCA1-overexpressed exosomes, which can cause resistance to tamoxifen (TAM) treatment of the MCF7 cell line and decrease apoptosis through reduction of cleaved caspase-3 expression. (B) RAB27B-upregulated stromal cells release exosomes that contain 5′-triphosphate RNAs and activate STAT1-dependent antiviral signaling and NOTCH3 pathways in adjacent BC cells. The crosstalk between these two signaling pathways results in reduction of chemo-resistance and radiotherapy (RT) resistance in different BC cell lines.
Exosomal Genetic Materials in Other Cancers
Researchers studied other cancer exosomes in different biofluids. Some of the reputable studies focusing on non-BC cells are summarized in Table 1. The study on serum samples illustrated the advantages of these vesicles as potential biomarker sources in different cancers. As an example, the study by Taylor et al.74 showed that the signature on circulating exosomal miRNAs accurately reflected the tumor profiles. Therefore, exosomal mRNA profiling can be performed as an alternative diagnostic procedure.74 Moreover, a study on glioblastoma multiforme in patients’ serum samples indicated that exosomal miR-320, miR-574-3p, and RNU6-1 could serve as diagnostic biomarkers for early detection and monitoring of the disease.15 Another study on seral exosomes of patients with melanoma showed an association between circulating miR-125b downregulation and disease progression.75 The selected miR-1246, miR-3976, miR-4644, and miR-4306 were significantly upregulated in 83% of pancreatic adenocarcinoma seral exosomes, compared with those of the control group.76 Similar studies on esophageal cancer, meningioma, and prostate cancer revealed evidence in favor of using exosomes in cancer diagnosis or investigation of cell-to-cell communication.38, 77
Table 1.
Non-BC Studies on Biofluid-Derived Exosomes
| Primary Sample | Type of Cancer | Genes | Detection Method | References |
|---|---|---|---|---|
| Urine | prostate cancer |
ERG PCA3 |
qRT-PCR | 101 |
| Bronchial lavage samples | lung cancer | hsa-miR-19b-1 hsa-miR-1285 hsa-miR-1289 hsa-miR-1303 hsa-miR-217 hsa-miR-29a-5p hsa-miR-548-3p hsa-miR-650 U6 snRNA |
qRT-PCR | 102 |
| Serum | glioblastoma multiforme |
RNU6-1 miR-320 miR-574-3p |
qRT-PCR | 15 |
| advanced melanoma | miR-125b | qRT-PCR | 75 | |
| pancreatic cancer | miR-1246 miR-4644 miR-3976 miR-4306 |
qRT-PCR | 76 | |
| adenocarcinoma of the esophagus | miR-223-5p miR-223-3p miR-483-5p miR-409-3p miR-196b-5p miR-192-5p miR-146a-5p miR-126-5p |
qRT-PCR | 77 | |
| prostate cancer | miR-200c miR-605 miR-135a miR-433 miR-106a |
Scano-miR bioassay and qRT-PCR | 38 | |
| meningioma | miR-106a-5p miR-219-5p miR-375 miR-409-3p miR-197 miR-224 |
qRT-PCR | 103 | |
| colorectal adenomas | miR-21 miR-29a miR-92a miR-135b |
qRT-PCR | 104 | |
| Serum and tumor cells | ovarian cancer | miR-21 miR-141 miR-200a miR-200c miR-200b miR-203 miR-205 miR-214 |
microarray | 74 |
| Patients and healthy sera: cell lines (NPC and NP69) | nasopharyngeal carcinoma | miR-24-3p | qRT-PCR | 105 |
| Cell line AZ-P7a | metastatic gastric cancer | let-7 miRNAs family | qRT-PCR | 80 |
| PC-3 | prostate cancer | 364 miRNAs profile | microarray and qRT-PCR | 79 |
| LIM1215 | colorectal cancer | GPA33 CDH17 CEA EPCAM PCNA EGFR MUC13 MINK1 KRT18 CLDN1, CLDN3, and CLDN7 CEP55 EFNB1 and EFNB2 |
immunoaffinity capture | 81 |
In addition to the serum studies, research on urine exosomes showed practical significant results to predict cancer status. A novel triple-RNA signature in prostate cancer can discriminate score 7 from other scores in the first biopsy and reduce unnecessary sampling.78
As well as the studies on different biofluids, there are some evaluations on the exosomes derived from cell lines. For instance, the comparison of normal and prostate cancer cell lines showed the presence of a specific miRNA pattern in the cancer exosomes.79 The results of another study suggested that metastatic gastric cancer cell line released let-7 family miRNAs via exosomes into the extracellular environment to maintain their oncogenesis.80
The specific exosomal proteins in a colorectal cancer cell line may provide the understanding of colon cancer biology and potential screening of biological markers for cancer.81 In 2016, researchers reported 570 proteins as an exosomal protein profile consisting of several cancer-related signaling proteins, tumor antigens, and secreted regulators. The functional value of tumor exosomes in the promotion of angiogenesis and cell migration was also demonstrated.82
Exosome Isolation Protocols
Similar to every newfound area, exosomes need to be validated in different aspects, especially clinical significance. The prerequisite for this level is to develop standard methods to isolate, characterize, and extract biological materials. Based on what was discussed heretofore, these techniques are explained (Table 2).
Table 2.
Comparison of the Current Exosomes Isolation Methods
| Method | Principle | Advantages | Disadvantages | Yield | Purity | References |
|---|---|---|---|---|---|---|
| Ultracentrifugation | centrifugation and ultracentrifugation steps | cost-effective | time consuming | low | high | 83, 84 |
| large primary sample size | ||||||
| low accuracya | ||||||
| contamination with media proteins | ||||||
| Ultrafiltration | centrifugation and filtration | cost-effective | time consuming | low | high | 87 |
| large primary sample size | ||||||
| low accuracy | ||||||
| Density gradient | density | cost-effective | time consuming | high | high | 88, 89 |
| large primary sample size | ||||||
| low accuracy | ||||||
| Immunoaffinity purification | magnetic beads | low primary sample volume | – | high | high | 90, 91, 92 |
| high accuracy | ||||||
| Microfluidically isolation | microfluidic devices | simple | high-priced | high | high | 93 |
| low primary sample volume | ||||||
| high accuracy | ||||||
| Commercial reagents | chemical reagent | rapid | high-priced in large sample size | high | high | 94, 95 |
| low primary sample volume | low accuracy | |||||
| high accuracy |
Contamination with non-exosome particles.
There are different exosome isolation protocols based on the types of starting samples and preferred downstream experiments with their own advantages and disadvantages. The starting sample could be cell cultured medium or one of the biological fluids, and isolated exosomes are assessed in terms of function or content, including proteins, DNAs, mRNAs, and non-coding RNAs such as miRNAs. Although differential centrifugation technique is the gold standard method to purify exosomes,83, 84 different sample viscosities and requirement of special equipment make this method low-efficient and restricted, respectively.84 In high-viscosity fluids, the rate and duration of centrifugation should be increased, or the fluids should be diluted to decrease the viscosity, because samples with high viscosity have lower sedimentation efficiency.85 Because plasma is more viscous than serum, the modified ultracentrifugation protocol could be used to purify exosomes by replacing the single filtration with the first step of centrifugation in plasma.86 Overall, plasma has higher viscosity than serum, serum has higher viscosity than cultured media, and PBS has the least viscosity.
Although this method improves the recovery rate, the contamination with media proteins and a large amount of starting sample was not practical for proteomics analysis and clinical use. To omit large particles and debris from samples, addition of filtration step to ultracentrifugation can improve exosomal extraction, especially in studies that plan to analyze RNAs.87 Sucrose gradient centrifugation for more extraction purity and yield was introduced based on different flotation densities of exosomes in 1997.88 Cantin et al.,89 in 2008, modified this method using the iodixanol (OptiPrep) gradient to separate exosomes from viruses that overlapped in density and size range.
The discovery of specific exosome protein markers in different biological status, from normal to ill, especially in cancer, led to the development of immunoaffinity-based techniques, which can isolate exosomes from small sample volumes.28 Coated magnetic or latex beads are of the specific antigens used to purify the so-called exosomes. This method operates faster with more efficiency, which is important in clinical settings.90, 91, 92
Microfluidic devices were fabricated in order to cover the defects of the current exosome isolation methods; approaches such as immunological separation, sieving, and trapping the exosomes are some examples of the functions of such devices. Simplicity, specificity, efficiency, and high purity of such few-step isolation techniques, which can work with slight volume of starting samples and reagents, make them intriguing for research and clinical settings.93 Moreover, commercial kit is another option that rapidly and simply isolates exosomes from small volumes of different starting samples by chemical reagents, which makes them ideal for pathological purposes. Commercial reagents are recommended if the starting sample volume is limit; this technique is suitable to isolate exosomes from less than 500 μL starting volume.94 Some purification reagents are efficient for future downstream analyses except for protein analysis, unless successive ultracentrifugation and filtration steps are operated to eliminate the non-specific proteins.95 Therefore, the best exosomes isolation method should be employed based on the size and type of the starting sample volume and planned downstream analysis.
The main objective of the studies on the exosomes isolation methods is to take a step forward in their clinical application. There are some challenges on the clinical application methods of exosomes as biomarker sources. First, based on the type of cancer, the proper biofluid should be selected. The selected biofluid must be accessible through a noninvasive or minimally invasive procedure, and the intended biomarkers detectable in medically safe quantity of the biofluid. For example, Cheng et al.36 showed that exosomes from different fractions of blood sample represented different miRNA profiles. Hence, a detectable biomarker in a certain bioliquid may not be identified in another one. Second, a standard procedure should be developed to isolate exosomes accurately and specifically. In other words, an ideal procedure should accurately isolate exosomes from the other extracellular vesicles and particles, and the BSEs from other exosomes specifically. For instance, the study by Caradec et al.96 investigated the possible contamination of seral exosomes with albumin protein. Third, the process of exosome isolation in the clinic had to be repetitive, rapid, easy to handle, cheap, and applicable for different types of tissue-specific exosomes. Fourth, a perfect exosome isolation method should have a low error rate and higher recovery yield.97 To date, no approved exosome isolation procedure is introduced for the clinical setting. Some companies recommended a preclinical exosome isolation kit (like Exoquick-CG; SBI), which is necessary to be validated through clinical trials, and the upcoming results should be compared and compatible with those of the pathology reports.
Exosome Characterization
Several methods are available to distinguish extracted exosomes from other vesicles (Table 3). Liquid chromatography, mass spectrometry, immuno-blot analysis, flow cytometry, western immuno-blotting, and dot blot assay are common to verifying exosome markers. Characterizing based on shape and size of particles is done by electron microscopy, atomic force microscopy (AFM),98, 99 nanoparticle tracking analysis (NTA), dynamic light scattering (DLS) analysis, qNANO GOLD,100 and ELISA approaches. Recently, lateral flow immunoassay (LFIA) was developed to detect exosomes, targeting tetraspanins CD9, CD63, and CD81 on their membranes. Some studies run western blot on exosome samples to indicate the lack of microvesicle molecular markers (CD29, CD40, and p-selectin) and endoplasmic reticulum molecular markers (calnexin), and confirmed that they specifically isolated exosomes.42
Table 3.
Different Exosomes Characterization Methods
| Exosome Features | Exosome Quantification Tool |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM | WB | Chro. | MS | NTA | DLS | Dot Blot | ELISA | qNano | AFM | FACS | LFIA | |
| Shape | X | X | ||||||||||
| Size | X | X | X | X | X | |||||||
| Distribution | X | X | ||||||||||
| Morphology (structure) | X | X | X | X | X | X | X | |||||
AFM, atomic force microscopy; Chro., chromatography; DLS, dynamic light scattering; EM, electronic microscope; FACS, fluorescence-activated cell sorting; LFIA, lateral flow immunoassay; MS, mass spectrophotometry; NTA, nanoparticle tracking analysis; WB, western blot.
Conclusions
Nowadays, exosomes draw attention as a potential source to discover new biomarkers for different diseases including cancer. A perfect cancer biomarker can show the existence of tumor mass and its molecular features in the early stages. Exosomes have special properties, which make them an ideal tool for minimally invasive liquid biopsy. These subcellular particles are detectable in almost every biofluid; therefore, in accordance with the type of cancer, researchers can select a special biofluid to detect patients’ exosomes. Proper isolation protocol should be applied based on the downstream analysis, type, and volume of starting sample. Exosomes contain proteins, RNAs, and DNA, which might indicate the biological and pathological features of the tumor mass in real-time status. Up to now, many candidate exosomal biomarkers are suggested for BC, but none of them are approved yet. The role of exosomes to inhibit proliferation and elevation of response to chemotherapy in BC cells is proved. In addition to the repressive effects of exosomes on BC cells, engineered exosomes specifically target BC cells, conclusively reduce the side effects of chemotherapy on normal cells, and increase the chemotherapy response and half-life of drug in circulation. On the other hand, BC cells exosomes induce oncogenic features in normal mammary cells and resistance to chemotherapy and radiotherapy in chemosensitive BC cells. These exosomes activate signaling pathways, which lead to migration and metastasis in noninvasive BC cells, but related mechanisms are not validated yet. There is still a long way for scientists to discover reliable procedures to diagnose, treat, and monitor BC through cancer-specific exosome-based liquid biopsy. To eliminate the effect of normal cell exosomes, isolation and characterization methods should be developed, and based on the liquid of origin, a consensus should be achieved. Experts in biology and bioengineering should cooperate to advance exosome-based technologies.
Author Contributions
Conceptualization and Validation, K.M.-A. and R.E.; Writing – Original Draft, S.H., S.D., Z.E.-S., T.S., N.J.-I., and T.O.B.; Writing – Review & Editing, R.E. and S.H.; Visualization, S.H., Z.E.-S., S.D., and T.O.B.; Supervision, R.E.; Project Administration, S.H.; Funding Acquisition, R.E.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This review was supported under Iran National Science Foundation Projects funding scheme (project number 95849123). We thank Iran National Science Foundation for their financial support.
References
- 1.Esposito A., Criscitiello C., Locatelli M., Milano M., Curigliano G. Liquid biopsies for solid tumors: understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol. Ther. 2016;157:120–124. doi: 10.1016/j.pharmthera.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feller S.M., Lewitzky M. Hunting for the ultimate liquid cancer biopsy—let the TEP dance begin. Cell Commun. Signal. 2016;14:24. doi: 10.1186/s12964-016-0147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izzotti A., Carozzo S., Pulliero A., Zhabayeva D., Ravetti J.L., Bersimbaev R. Extracellular microRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am. J. Cancer Res. 2016;6:1461–1493. [PMC free article] [PubMed] [Google Scholar]
- 5.Perakis S., Speicher M.R. Emerging concepts in liquid biopsies. BMC Med. 2017;15:75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alix-Panabières C., Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 7.Pan B.-T., Johnstone R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 8.Simpson R.J., Jensen S.S., Lim J.W. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 9.Suetsugu A., Honma K., Saji S., Moriwaki H., Ochiya T., Hoffman R.M. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev. 2013;65:383–390. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Savina A., Fader C.M., Damiani M.T., Colombo M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6:131–143. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 11.Demory Beckler M., Higginbotham J.N., Franklin J.L., Ham A.J., Halvey P.J., Imasuen I.E., Whitwell C., Li M., Liebler D.C., Coffey R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharaziha P., Chioureas D., Rutishauser D., Baltatzis G., Lennartsson L., Fonseca P., Azimi A., Hultenby K., Zubarev R., Ullén A. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget. 2015;6:21740–21754. doi: 10.18632/oncotarget.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welton J.L., Khanna S., Giles P.J., Brennan P., Brewis I.A., Staffurth J., Mason M.D., Clayton A. Proteomic analysis of bladder cancer exosomes. Mol. Cell. Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buschow S.I., van Balkom B.W., Aalberts M., Heck A.J., Wauben M., Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010;88:851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]
- 15.Manterola L., Guruceaga E., Gállego Pérez-Larraya J., González-Huarriz M., Jauregui P., Tejada S., Diez-Valle R., Segura V., Samprón N., Barrena C. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-oncol. 2014;16:520–527. doi: 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryzgunova O.E., Zaripov M.M., Skvortsova T.E., Lekchnov E.A., Grigor’eva A.E., Zaporozhchenko I.A., Morozkin E.S., Ryabchikova E.I., Yurchenko Y.B., Voitsitskiy V.E., Laktionov P.P. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS ONE. 2016;11:e0157566. doi: 10.1371/journal.pone.0157566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lässer C., Alikhani V.S., Ekström K., Eldh M., Paredes P.T., Bossios A., Sjöstrand M., Gabrielsson S., Lötvall J., Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamperl H., Plattfaut C., Freund A., Quecke T., Theophil F., Gieseler F. Extracellular vesicles from malignant effusions induce tumor cell migration: inhibitory effect of LMWH tinzaparin. Cell Biol. Int. 2016;40:1050–1061. doi: 10.1002/cbin.10645. [DOI] [PubMed] [Google Scholar]
- 19.Gregson A.L., Hoji A., Injean P., Poynter S.T., Briones C., Palchevskiy V., Weigt S.S., Shino M.Y., Derhovanessian A., Sayah D. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am. J. Respir. Crit. Care Med. 2015;192:1490–1503. doi: 10.1164/rccm.201503-0558OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkumas K.M., Hoffman E.A., McKay B.S., Allingham R.R., Stamer W.D. Myocilin-associated exosomes in human ocular samples. Exp. Eye Res. 2007;84:209–212. doi: 10.1016/j.exer.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lässer C., O’Neil S.E., Shelke G.V., Sihlbom C., Hansson S.F., Gho Y.S., Lundbäck B., Lötvall J. Exosomes in the nose induce immune cell trafficking and harbour an altered protein cargo in chronic airway inflammation. J. Transl. Med. 2016;14:181. doi: 10.1186/s12967-016-0927-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison M.N., Roller R.J., Okeoma C.M. Human semen contains exosomes with potent anti-HIV-1 activity. Retrovirology. 2014;11:102. doi: 10.1186/s12977-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skriner K., Adolph K., Jungblut P.R., Burmester G.R. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 24.Keller S., Rupp C., Stoeck A., Runz S., Fogel M., Lugert S., Hager H.D., Abdel-Bakky M.S., Gutwein P., Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 25.Wu M., Burstein D.E. Fine needle aspiration. Cancer Invest. 2004;22:620–628. doi: 10.1081/cnv-200027160. [DOI] [PubMed] [Google Scholar]
- 26.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Luga V., Zhang L., Viloria-Petit A.M., Ogunjimi A.A., Inanlou M.R., Chiu E., Buchanan M., Hosein A.N., Basik M., Wrana J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Lobb R.J., Becker M., Wen S.W., Wong C.S., Wiegmans A.P., Leimgruber A., Möller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawakubo-Yasukochi T., Morioka M., Hayashi Y., Nishinakagawa T., Hazekawa M., Kawano S., Nakamura S., Nakashima M. The SQUU-B cell line spreads its metastatic properties to nonmetastatic clone SQUU-A from the same patient through exosomes. J. Oral Biosci. 2016;58:33–38. doi: 10.1016/j.job.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross J.C., Chaudhary V., Bartscherer K., Boutros M. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 32.Chavez-Muñoz C., Morse J., Kilani R., Ghahary A. Primary human keratinocytes externalize stratifin protein via exosomes. J. Cell. Biochem. 2008;104:2165–2173. doi: 10.1002/jcb.21774. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Yuan X., Shi H., Wu L., Qian H., Xu W. Exosomes in cancer: small particle, big player. J. Hematol. Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koga K., Matsumoto K., Akiyoshi T., Kubo M., Yamanaka N., Tasaki A., Nakashima H., Nakamura M., Kuroki S., Tanaka M., Katano M. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25(6A):3703–3707. [PubMed] [Google Scholar]
- 35.Thakur B.K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng L., Sharples R.A., Scicluna B.J., Hill A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3:23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M., Prasad N., Levy S., Coffey R.J., Patton J.G., Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alhasan A.H., Scott A.W., Wu J.J., Feng G., Meeks J.J., Thaxton C.S., Mirkin C.A. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl. Acad. Sci. USA. 2016;113:10655–10660. doi: 10.1073/pnas.1611596113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao C. Urinary protein biomarker database: a useful tool for biomarker discovery. Adv. Exp. Med. Biol. 2015;845:195–203. doi: 10.1007/978-94-017-9523-4_19. [DOI] [PubMed] [Google Scholar]
- 41.Hannafon B.N., Trigoso Y.D., Calloway C.L., Zhao Y.D., Lum D.H., Welm A.L., Zhao Z.J., Blick K.E., Dooley W.C., Ding W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munagala R., Aqil F., Jeyabalan J., Gupta R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371:48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larssen P., Wik L., Czarnewski P., Eldh M., Löf L., Ronquist K.G., Dubois L., Freyhult E., Gallant C.J., Oelrich J. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol. Cell. Proteomics. 2017;16:502–511. doi: 10.1074/mcp.M116.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrado C., Raimondo S., Chiesi A., Ciccia F., De Leo G., Alessandro R. Exosomes as intercellular signaling organelles involved in health and disease: basic science and clinical applications. Int. J. Mol. Sci. 2013;14:5338–5366. doi: 10.3390/ijms14035338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yassin A.M., Abdel Hamid M.I., Amer H., Warda M., Farid O.A. Dromedary milk exosomes as mammary transcriptome nano-vehicle: their isolation, vesicular and phospholipidomic characterizations. J. Adv. Res. 2016;7:749–756. [Google Scholar]
- 46.Qin W., Tsukasaki Y., Dasgupta S., Mukhopadhyay N., Ikebe M., Sauter E.R. Exosomes in human breast milk promote EMT. Clin. Cancer Res. 2016;22:4517–4524. doi: 10.1158/1078-0432.CCR-16-0135. [DOI] [PubMed] [Google Scholar]
- 47.Reinhardt T.A., Lippolis J.D., Nonnecke B.J., Sacco R.E. Bovine milk exosome proteome. J. Proteomics. 2012;75:1486–1492. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Agrawal A.K., Aqil F., Jeyabalan J., Spencer W.A., Beck J., Gachuki B.W., Alhakeem S.S., Oben K., Munagala R., Bondada S., Gupta R.C. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine (Lond.) 2017;13:1627–1636. doi: 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Vashisht M., Rani P., Onteru S.K., Singh D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl. Biochem. Biotechnol. 2017;183:993–1007. doi: 10.1007/s12010-017-2478-4. [DOI] [PubMed] [Google Scholar]
- 50.Khan S., Jutzy J.M.S., Aspe J.R., Valenzuela Malyn M.A., Park J.S., Turay D., Wall N.R. The application of membrane vesicles for cancer therapy. In: Gali-Muhtasib H., editor. Advances in Cancer Therapy. InTech; 2011. pp. 21–52. [Google Scholar]
- 51.Palma J., Yaddanapudi S.C., Pigati L., Havens M.A., Jeong S., Weiner G.A., Weimer K.M., Stern B., Hastings M.L., Duelli D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heneghan H.M., Miller N., Kelly R., Newell J., Kerin M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 54.Hu Z., Dong J., Wang L.E., Ma H., Liu J., Zhao Y., Tang J., Chen X., Dai J., Wei Q. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33:828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 55.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Martin D.I. Deep sequencing of serum small RNAs identifies patterns of 5′tRNA half and YRNA fragment expression associated with breast cancer. Biomark. Cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiskaa T., Knutsen E., Nikolaisen M.A., Jørgensen T.E., Johansen S.D., Perander M., Seternes O.M. Distinct small RNA signatures in extracellular vesicles derived from breast cancer cell lines. PLoS ONE. 2016;11:e0161824. doi: 10.1371/journal.pone.0161824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park K.-h., Yong -Y.-r., Kang H.-j., Kim G.-h., Park D.-H., Lee M.-y. Composition and kit for diagnosing breast cancer including polynucleotide within vesicle, and method of diagnosing breast cancer using the same. U.S. Patent. December 2013 20170145519. [Google Scholar]
- 58.van’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 59.Rupp A.K., Rupp C., Keller S., Brase J.C., Ehehalt R., Fogel M., Moldenhauer G., Marmé F., Sültmann H., Altevogt P. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 2011;122:437–446. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 60.Ryan B., O’Donovan N., Browne B., O’Shea C., Crown J., Hill A.D., McDermott E., O’Higgins N., Duffy M.J. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br. J. Cancer. 2005;92:120–124. doi: 10.1038/sj.bjc.6602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan S., Jutzy J.M., Aspe J.R., McGregor D.W., Neidigh J.W., Wall N.R. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16:1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan S., Bennit H.F., Turay D., Perez M., Mirshahidi S., Yuan Y., Wall N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176. doi: 10.1186/1471-2407-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Brien K., Lowry M.C., Corcoran C., Martinez V.G., Daly M., Rani S., Gallagher W.M., Radomski M.W., MacLeod R.A., O’Driscoll L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget. 2015;6:32774–32789. doi: 10.18632/oncotarget.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bovy N., Blomme B., Frères P., Dederen S., Nivelles O., Lion M., Carnet O., Martial J.A., Noël A., Thiry M. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–10266. doi: 10.18632/oncotarget.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohno S., Takanashi M., Sudo K., Ueda S., Ishikawa A., Matsuyama N., Fujita K., Mizutani T., Ohgi T., Ochiya T. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenjaroenpun P., Kremenska Y., Nair V.M., Kremenskoy M., Joseph B., Kurochkin I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., Yu Y., Chow A., O’Connor S.T., Chin A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shang C., Guo Y., Zhang J., Huang B. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother. Pharmacol. 2016;77:1061–1067. doi: 10.1007/s00280-016-3029-3. [DOI] [PubMed] [Google Scholar]
- 69.Pan J., Li X., Wu W., Xue M., Hou H., Zhai W., Chen W. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382:64–76. doi: 10.1016/j.canlet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 70.Bian Z., Jin L., Zhang J., Yin Y., Quan C., Hu Y., Feng Y., Liu H., Fei B., Mao Y. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu C.G., Yang M.F., Ren Y.Q., Wu C.H., Wang L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016;20:4362–4368. [PubMed] [Google Scholar]
- 72.Boelens M.C., Wu T.J., Nabet B.Y., Xu B., Qiu Y., Yoon T., Azzam D.J., Twyman-Saint Victor C., Wiemann B.Z., Ishwaran H. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lewis M.T., Dobrolecki L.E., Hilsenbeck S.G., Simon L.M., Shaw C.A. Identification of molecular predictors of differential chemotherapy response using patient-derived xenografts. Clin. Cancer Res. 2016;22(Suppl 16):B04. [Google Scholar]
- 74.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 75.Alegre E., Sanmamed M.F., Rodriguez C., Carranza O., Martín-Algarra S., González A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 2014;138:828–832. doi: 10.5858/arpa.2013-0134-OA. [DOI] [PubMed] [Google Scholar]
- 76.Madhavan B., Yue S., Galli U., Rana S., Gross W., Müller M., Giese N.A., Kalthoff H., Becker T., Büchler M.W., Zöller M. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 77.Warnecke-Eberz U., Chon S.H., Hölscher A.H., Drebber U., Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36:4643–4653. doi: 10.1007/s13277-015-3112-0. [DOI] [PubMed] [Google Scholar]
- 78.Donovan M.J., Noerholm M., Bentink S., Belzer S., Skog J., Brown G.A., Cochran J.S., O’Neill V. Interim performance of a non-DRE urine exosome gene signature to predict Gleason ≥7 prostate cancer on initial prostate needle biopsy from patients enrolled in a prospective observational trial. J. Clin. Oncol. 2015;33(Suppl 15):5064. [Google Scholar]
- 79.Hessvik N.P., Phuyal S., Brech A., Sandvig K., Llorente A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim. Biophys. Acta. 2012;1819:1154–1163. doi: 10.1016/j.bbagrm.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 80.Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., Muramatsu K., Fukuda Y., Ogura S., Yamaguchi K., Mochizuki T. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mathivanan S., Lim J.W., Tauro B.J., Ji H., Moritz R.L., Simpson R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greening D.W., Ji H., Chen M., Robinson B.W., Dick I.M., Creaney J., Simpson R.J. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci. Rep. 2016;6:32643. doi: 10.1038/srep32643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Momen-Heravi F., Balaj L., Alian S., Mantel P.Y., Halleck A.E., Trachtenberg A.J., Soria C.E., Oquin S., Bonebreak C.M., Saracoglu E. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013;394:1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Momen-Heravi F., Balaj L., Alian S., Trachtenberg A.J., Hochberg F.H., Skog J., Kuo W.P. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 2012;3:162. doi: 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 87.Cecilia L., Maria E., Jan L. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012;2012:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bess J.W., Jr., Gorelick R.J., Bosche W.J., Henderson L.E., Arthur L.O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 89.Cantin R., Diou J., Bélanger D., Tremblay A.M., Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J. Immunol. Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 90.Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 91.Greening D.W., Xu R., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 92.Clayton A., Court J., Navabi H., Adams M., Mason M.D., Hobot J.A., Newman G.R., Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 93.Liga A., Vliegenthart A.D., Oosthuyzen W., Dear J.W., Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15:2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 94.Schageman J., Zeringer E., Li M., Barta T., Lea K., Gu J., Magdaleno S., Setterquist R., Vlassov A.V. The complete exosome workflow solution: from isolation to characterization of RNA cargo. BioMed Res. Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamada T., Inoshima Y., Matsuda T., Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. J. Vet. Med. Sci. 2012;74:1523–1525. doi: 10.1292/jvms.12-0032. [DOI] [PubMed] [Google Scholar]
- 96.Caradec J., Kharmate G., Hosseini-Beheshti E., Adomat H., Gleave M., Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin. Biochem. 2014;47:1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 97.Tang Y.-T., Huang Y.Y., Zheng L., Qin S.H., Xu X.P., An T.X., Xu Y., Wu Y.S., Hu X.M., Ping B.H., Wang Q. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017;40:834–844. doi: 10.3892/ijmm.2017.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palanisamy V., Sharma S., Deshpande A., Zhou H., Gimzewski J., Wong D.T. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharma S., Rasool H.I., Palanisamy V., Mathisen C., Schmidt M., Wong D.T., Gimzewski J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garza-Licudine E., Deo D., Yu S., Uz-Zaman A., Dunbar W.B. Portable nanoparticle quantization using a resizable nanopore instrument—the IZON qNano™. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2010;2010:5736–5739. doi: 10.1109/IEMBS.2010.5627861. [DOI] [PubMed] [Google Scholar]
- 101.Donovan M.J., Noerholm M., Bentink S., Belzer S., Skog J., O’Neill V., Cochran J.S., Brown G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015;18:370–375. doi: 10.1038/pcan.2015.40. [DOI] [PubMed] [Google Scholar]
- 102.Schmidt B., Rehbein G., Fleischhacker M. Liquid profiling in lung cancer – quantification of extracellular miRNAs in bronchial lavage. Adv. Exp. Med. Biol. 2016;924:33–37. doi: 10.1007/978-3-319-42044-8_7. [DOI] [PubMed] [Google Scholar]
- 103.Zhi F., Shao N., Li B., Xue L., Deng D., Xu Y., Lan Q., Peng Y., Yang Y. A serum 6-miRNA panel as a novel non-invasive biomarker for meningioma. Sci. Rep. 2016;6:32067. doi: 10.1038/srep32067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Uratani R., Toiyama Y., Kitajima T., Kawamura M., Hiro J., Kobayashi M., Tanaka K., Inoue Y., Mohri Y., Mori T. Diagnostic potential of cell-free and exosomal microRNAs in the identification of patients with high-risk colorectal adenomas. PLoS ONE. 2016;11:e0160722. doi: 10.1371/journal.pone.0160722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ye S.B., Zhang H., Cai T.T., Liu Y.N., Ni J.J., He J., Peng J.Y., Chen Q.Y., Mo H.Y., Jun-Cui Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J. Pathol. 2016;240:329–340. doi: 10.1002/path.4781. [DOI] [PubMed] [Google Scholar]