Abstract
Fluorescence-based techniques are widely used to study biomolecular conformations, intra- and intermolecular interactions, and conformational dynamics of macromolecules. Especially for fluorescent-based single-molecule experiments, the choice of the fluorophore and labeling position are highly important. Here, we have studied the biophysical and structural effects that are associated with the conjugation of fluorophores to cysteines in the splicing factor U2AF65 by using single pair Förster resonance energy transfer (FRET) and nuclear magnetic resonance (NMR) spectroscopy. We show that certain acceptor fluorophores are advantageous depending on the experiments performed. The effects of dye attachment on the protein conformation were characterized using heteronuclear NMR experiments. We find that the presence of hydrophobic and aromatic moieties in the fluorophores can significantly affect the conformation of the conjugated protein, presumably by transient interactions with the protein surface. Guidelines are provided for carefully choosing fluorophores, considering their photophysical properties and chemical features for the design of FRET experiments and for minimizing artifacts.
Introduction
Single-molecule experiments provide unique information regarding the conformational states and subpopulations of biological macromolecules in solution. Single-molecule fluorescence offers the advantage that individual molecules can be studied in solution or in their cellular environment upon attaching a fluorescent dye. However, the attachment of any chemical moiety to a protein needs to be carefully considered as it may cause changes to its conformation or dynamics and thereby affect its function. This problem is not unique to labeling of proteins with fluorescent dyes [1,2], but extends to other types of protein tagging methods as well. For example, in NMR and EPR studies, paramagnetic tagging with spin labels (e.g. IPSL, MTSSL), lanthanide chelating tags (e.g. DOTA, DTPA, EDTA), or lanthanide binding tags (LBTs) [3–10], are regularly used for investigation of structure, dynamics, and function of proteins and protein ligand interactions. To date, fluorescent proteins are frequently used in fusion constructs with proteins of interest to be studied due to the straightforward genetic encoding for expression in cells. While potential problems associated with the use of large fluorescent labels, such as GFP mutants have been discussed [11–13], the conjugation of small organic fluorophores is usually thought to have negligible effects. A few studies have already shown the effect of fluorophore attachment to nucleic acids [8–10,14,15] and some deal with the effects of dyes on certain protein characteristics [14,16,17]. For example, Dallmann et al. modified DNA oligomers with 2-aminopurine and investigated their structural changes by 2D NMR spectroscopy. They observed fast water exchange in the proximity of the labels, which affected the adjacent base pairs [18]. Although some of the reports have even used NMR to characterize the effects of fluorophores on the conformation and dynamics of the biological macromolecules studied [7–11,18,19], a detailed characterization of the effect of fluorophores on different properties of proteins is still missing.
Here, we combine NMR and single pair FRET (spFRET) experiments to investigate the effect of various donor-acceptor fluorophore pairs on the structure and conformational dynamics of the splicing factor U2AF65. U2AF65 is an essential splicing factor that recognizes the polypyrimidine tract RNA sequences at the 3′ splice site of human introns for spliceosome assembly. Its minimal RNA binding region consists of two RNA recognition motifs (RRMs), RRM1,2, connected by a flexible linker, which exhibit significant conformational dynamics in the free state and undergo a large conformational transition upon binding to cognate RNA [20–22]. Due to its known conformational change and dynamics as well as its binding affinity for RNA, U2AF65 is an ideal model protein to study the effect of fluorophore attachment on the conformational properties. We used spFRET and NMR to characterize conformational properties and spFRET in solution and surface immobilized to analyze conformational dynamics.
Results
Photophysical and biochemical properties of fluorophores attached to U2AF65
In biophysical studies of biological macromolecules using fluorescence spectroscopy, organic fluorophores are often chosen based on optimal photophysical properties [23]. Fluorophores are characterized regarding extinction coefficients, quantum yields, and photostability (Table S1). Especially for single molecule experiments, a large number of photons (high photostability and molecular brightness) is very important to obtain high signal-to-noise ratios. Typical fluorophores used are rhodamine based, as they have high quantum yields and photostability [24]. Furthermore, FRET requires a combination of two fluorophores with a certain Förster radius that depends on the spectral overlap of the absorption and emission spectra of donor and acceptor dyes (Table S2). Here, we investigate commonly used FRET fluorophores with Förster radii between 57 Å and 69 Å. We compared the effects of two carbocyanine dyes (Alexa647, Atto647N) and one carbopyronin (Cy5) as acceptor fluorophores and two different rhodamine-based dyes as donor fluorophores (Atto532 and Atto565). Due to their advantageous photophysical or biochemical properties, all of these fluorophores are commonly used in fluorescence-based techniques and some of them are particularly valuable for super-resolution microscopy. The fluorescent quantum yield of both donor fluorophores was around 90%, while the quantum yield of the acceptors varied between 30% for Cy5 and 65% for Atto647N (Table S1). Features that should be considered in addition to the photophysical properties of a fluorophore are its size/molecular mass, water solubility, charge, and pH sensitivity. The molecular mass of the fluorophores used here ranges from 0.7 kDa to 1.3 kDa and their net charge varies from -3 for Alexa647, to -1 for Atto532, to +1 for Atto647N, Cy5, and Atto565.
We performed spFRET experiments with multiparameter fluorescence detection and pulsed interleaved excitation (MFD-PIE) to gather information on the behavior of the various fluorophores[25]. Figure S1 shows how experiments using MFD-PIE can be used to gain information on the fluorescence lifetime of the donor and acceptor molecules and thus fluorophore quenching, and how the isomerization of cyanine-based dyes can be detected, as discussed previously [23,26]. We observed quenching of the acceptor fluorophore Alexa647 and isomerization of Cy5 bound to U2AF65, while Atto647N did not show differences in its photophysical characteristics upon protein attachment. The lifetime of the Alexa647 bound to U2AF65 in the presence of RNA was found to depend on the proximity ratio (Figure S1A–B) indicating that RNA binding to the protein leads to quenching of the acceptor fluorophore to shorter lifetimes. Alexa647 is partially quenched at this attachment position whereas no quenching is observed with Atto647N or Cy5. Quenching of the acceptor fluorophore leads to a decrease of the apparent FRET efficiency. Using the stoichiometry and acceptor lifetime information obtained from MFD-PIE experiments, however, we are able to account for this effect by adjusting the different populations with different correction factors for the detection efficiency γ and can thus obtain correct FRET efficiency values(Figure S1A–C).
The cyanine dye Cy5 undergoes isomerization between cis- and trans-configurations [26], which leads to a bright and a dark state of the fluorophore, respectively. We can observe this isomerization in histograms of stoichiometry versus FRET efficiency as a population trailing between the donor-only and the dual-labeled as well as the acceptor-only populations (Figure S1D). Often, trailing between donor-only and dual-labeled populations occurs due to photobleaching of the acceptor fluorophore or events in which multiple molecules traverse through the focal volume simultaneously. These photobleaching or multi-molecule events however, were removed by several filtering steps on the data displayed [25,27]. In one-dimensional FRET efficiency histograms, the cis-trans-isomerization of the fluorophores appears as a low FRET population with a FRET efficiency around 0 (Figure S1D, inset). It is thus important to consider the effects described above for the choice of fluorophore, as will be further detailed in the following.
Effects of different conjugated fluorophores on FRET efficiency histograms ofU2AF65
To assess the potential influence of different fluorophores on FRET experiments, U2AF65 was labeled with different donor-acceptor fluorophore combinations in each of the domains (Figure 1); the labeling positions were optimized to obtain the maximal achievable difference in distance (and thus FRET efficiency) for the free and RNA-bound conformations of RRM1,2 as described previously [20]. FRET efficiency histograms from MFD-PIE experiments were compared for different donor fluorophores (Figure S2) and different acceptor dyes (Figure 1). The donors Atto532 and Atto565, in combination with the same acceptor fluorophore, exhibited comparable FRET efficiency distributions; keeping in mind the expected differences in Förster radii and associated shifts in the FRET efficiency histograms (Figure S2B, C, upper panels). Distance distributions calculated from the FRET efficiency histograms show that the shifts in the FRET efficiency histograms reflect the difference in Förster radius of the two pairs of fluorophores (Figure S2B, C, lower panels). The calculated mean distances are 46.5 (±0.5) Å for RRM1,2-Atto532-Alexa647 and 45.7 (±0.2) Å for RRM1,2-Atto565-Alexa647 in the free form and 63.8 (± 0.7)Å and 64.9 (±0.5) Å in the RNA-bound form, respectively. We therefore continued our investigation using Atto532 as donor fluorophore, given its lower hydrophobicity compared to Atto565.
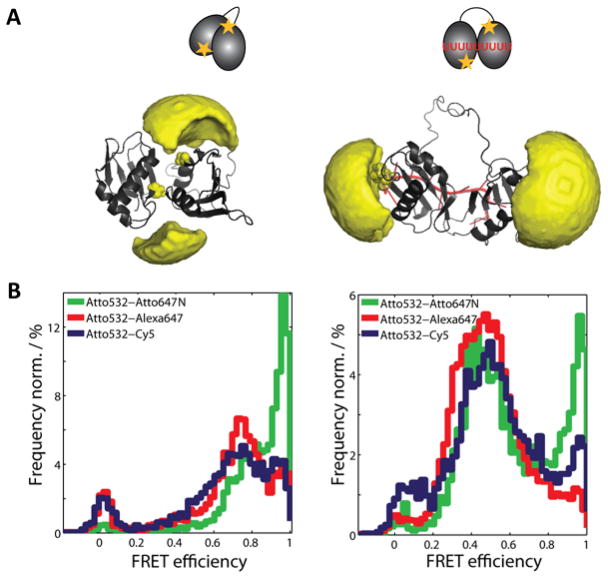
Figure 1. The effect of the acceptor fluorophore on the conformation of U2AF65 RRM1,2-C187-C318 studied by spFRET in solution.
(A) Structure and accessible volume calculations of the fluorophores attached to positions C187 and C318 of RRM1,2 in its closed state in the absence of RNA (PDB accession code: 2YH0) (left panel) and in the presence of U8 RNA (PDB accession code: 2YH1) (right panel). (B) FRET efficiency histograms of RRM1,2 labeled with Atto532 and the acceptor fluorophores Atto647N (green), Alexa647 (red), and Cy5 (blue) in the absence (left panel) and presence (right panel) of U9 RNA.
The FRET efficiency distributions with different acceptor fluorophores (Atto647N, Alexa647 and Cy5) show a consistent general trend for all three combinations of FRET dye pairs. A high FRET state (FRET efficiency 80%) is observed for the free form of U2AF65 for all the different combinations of fluorophores (Figure 1B, left). This FRET state corresponds to a population of molecules dynamically switching between a closed and an open conformation [22]. When bound to RNA, U2AF65 adopts an open conformation, which is visible with a FRET efficiency of ~45% for all combinations of fluorophores (Figure 1B, right). A second population of molecules with a FRET efficiency of 95% is apparent for U2AF65 for all three acceptor fluorophores tested. However, the fraction of this population strongly varies depending on the dye used (Figure 1B). We did not observe any significant change in the residual anisotropy of the three acceptor fluorophores suggesting a similar amount of rotational freedom of the fluorophores in all cases. Thus, we obtain similar results from the spFRET experiments when labeling U2AF65 with different combinations of dye. However, an additional high FRET population is observed whose amplitude depends on the acceptor fluorophore used. This suggests that the dyes have an additional influence on the spFRET spectra. To further investigate the structural effects of the fluorophores on the protein structure, we used heteronuclear NMR spectroscopy.
Attachment site of fluorophores affects the protein conformation
We used heteronuclear 1H, 15N NMR experiments to study the effects of dye attachment on the conformation of U2AF65 (RRM1,2). The chemical shifts of the free and dye-conjugated RRM1,2 were assigned using standard approaches [28]. As the chemical shift is a sensitive reporter of the chemical environment around a nuclear spin, chemical shift changes between the free and dye-labeled RRM1,2 (Figure S3, Figure S4, left) are expected in the vicinity of the dye attachment site, while perturbations farther away may indicate effects on the conformation of the protein. The intensity of backbone amide NMR signals was used as a qualitative assessment of dynamic effects, where a decrease in intensity due to line-broadening may indicate weak, non-specific interactions of the dye with the protein surface. To identify sites for fluorophore attachment with little effects on the protein conformation 1H, 15N NMR correlation spectra were compared for U2AF65 RRM1,2 unlabeled and conjugated with the Atto647N dye at different positions. The Atto647N dye was chosen here as it has the best photophysical properties for FRET experiments but is challenging from a biochemical perspective as it is hydrophobic and often exhibits a high anisotropy when attached to biomolecules, suggesting non-specific interactions [29]. Cysteine residues were engineered at positions 318 (located within helix α2 of RRM2, 4 residues from the carboxy terminal end), 322 (first residue following helix α2), and 326 (located in a loop connecting α2 and β4) in the RRM2 domain. (Figure 2, Figure S6). Residue 318 was originally chosen as a relatively rigid attachment site for paramagnetic relaxation enhancement studies by NMR [30,31], where minimizing the flexibility of the relatively small paramagnetic tag (for example, proxyl spin labels have a molecular weight of ~300 Da) is desirable. The other two locations, 322 and 326, were chosen to be further away from the helix and inmore flexible regions.
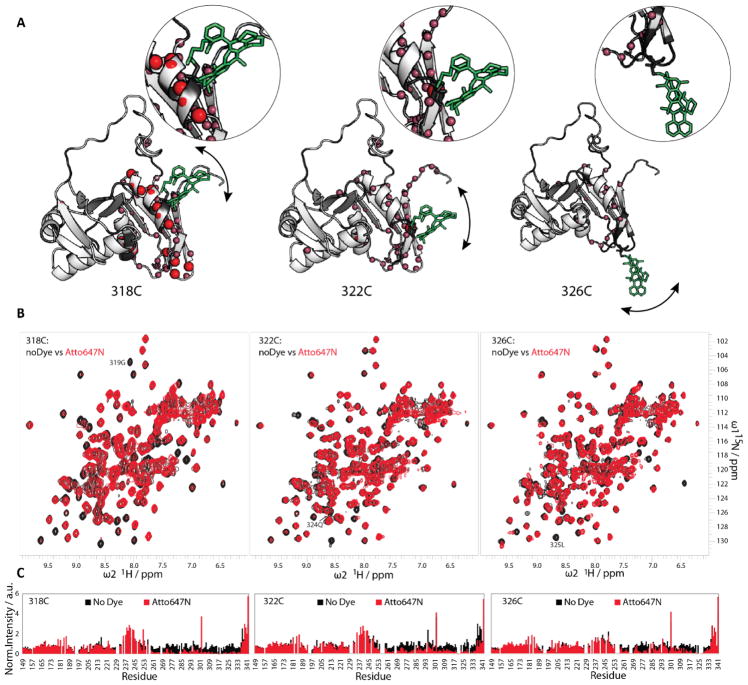
Figure 2. Attachment of the acceptor fluorophore Atto647N to different positions of U2AF65 RRM1,2 studied by NMR.
(A) Structural representations of RRM1,2 with the Atto647N fluorophore placed at positions C318 (left), C322 (middle), and C326 (right). The dye was modelled on every position performing energy minimization. Red spheres indicate the residues with loss of up to 90% of peak height upon fluorophore labeling and the pink spheres represent the residues with a loss between 90–50%. (B) 1H,15N-HSQC spectra of unlabeled RRM1,2 (black) compared to RRM1,2 fluorescently labeled with Atto647N (green) on the positions C318 (left), C322 (middle), and C326 (right). (C) Intensity vs. residue plots of the labeled and unlabeled U2AF65 in the respective positions (318, 322, and 326).
A comparison of NMR spectra for unlabeled and dye-labeled protein shows that attachment of Atto647N to C318 affects many amide signals that are in helix α2 or nearby regions (V275, E277, T280, S281, I310, V312, Q315, G319, L325, K328). NMR signals for many of these residues exhibit an up to 90% decrease of peak heights relative to the unlabeled protein (Figure 2B–C, left), indicating effects to the protein conformation in these regions. When Atto647N is conjugated to C322, intensities of NMR signals are generally less affected, but several residues still show strongly (50–90%) reduced signal intensities (S251, I263, L270, V275, K276, G283, F288, A303, I310, D314, Q315, L325, G328) (Figure 2B–C, middle). In contrast, attachment of Atto647N to position 326 leads to an NMR spectrum with signal intensities comparable to the unlabeled U2AF65 protein with only few residues exhibiting minor line-broadening(L266, N271, S281, F288, S294, M323, R334).
The attachment of the dye at positions 318 and 322 causes more extensive chemical shift changes of amide signals in the NMR spectra when compared to dye conjugation to position 326 (Figure 2B, Figure S3). The larger chemical shifts changes observed when the fluorophore is attached at positions 318 and 322 (which are located in helix α2) may reflect changes to the secondary structure, while residue 326 is found in a flexible loop (Figure 2B, Figure S3). Mapping of intensity and chemical shift differences observed between unlabeled and dye-labeled U2AF65 to the structure of the protein indicates that the dye mainly affects residues in spatial proximity to the dye (Figure 2A–C, Figure S3). The NMR spectral changes induced by dye attachment decrease with increasing distance of the dye attachment from helix α2. Thus position 326 is a preferred position for dye attachment. Moving the attachment sites for the fluorophores farther away from the α-helical region of RRM2 reduces the conformational effects due to the presence of the dye as indicated by the NMR fingerprint spectra.
Chemical properties of fluorophores affect the conformation of U2AF65
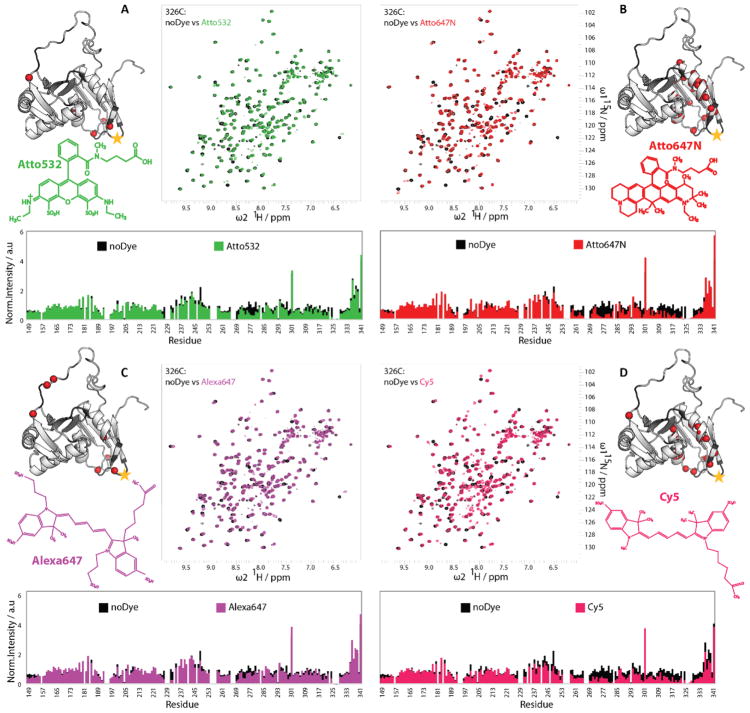
Next, we compared the influence of the four different fluorophores (Atto532, Atto647N, Alexa647, and Cy5) at different positions on the protein. We first started with our optimal attachment site, position C326. Superimposed NMR spectra of 15N-labeled protein conjugated with the different dyes and of the unlabeled protein were analyzed for changes in chemical shifts and signal intensities (Figure 3 and Figure S4). Atto532 and Alexa647 conjugation at position C326 showed very little changes in signal intensity. Those that were detected were concentrated to flexible parts of the protein (Figure 3A, C). Attachment of Cy5 and Atto647N induced a slight reduction of the maximum NMR signal intensities when compared with Atto532 and Alexa647, especially near the dye attachment site and surrounding structured region (Figure 3B, D).
Figure 3. Effect of different fluorophores at position C326 on the conformation of U2AF65 RRM1,2.
1 H,15N-HSQC spectra and intensity versus residue plots of unlabeled RRM1,2 (black) compared to (A) RRM1,2-C326 fluorescently labeled with Atto532 (green), (B) Atto647N (red), (C) Alexa647 (magenta), or (D) Cy5 (pink). Next to each spectrum are the structural formulas of the respective dyes and the structural representations of RRM1,2, where the red spheres indicate the residues with loss of more than 60% of peak height upon fluorophore labeling.
Interestingly, for several of the signals in the Cy5-labeled RRM1,2 spectra, peak doubling is observed, indicating the presence of two conformations that are slowly converting on the NMR chemical shift time scale (>μs-ms). These double-peaks thus indicate distinct but still dynamic contacts of the fluorophore with the surface of the protein. Similarly, the weak intensity of a few NMR signals for Atto647N-labeled U2AF65 may be caused by transient hydrophobic interactions of the highly aromatic dye with neighboring amino acids, among them hydrophobic and aromatic residues(e.g. F262, I263, V275, L278, F282, I317, L330). To investigate the effect of labeling on more sensitive regions of the protein structure, we compared the effect of the fluorophore when attached to the protein at the positions C318 and C322 (Figures S5–S6). Interestingly, the attachment of Atto532 at position C318 shows a strongly reduced effect on the local conformation as compared to Atto647N (Figure S5A). Hence, the attachment of a bulky, hydrophobic and highly aromatic fluorophore within a helix (position C318) can cause distortion even though conjugation of other fluorophores, such as Atto532 (Figure S5A) or the smaller paramagnetic spin labels at this position [30,31] did not significantly alter NMR spectra and signal properties when compared to the unlabeled protein. A further analysis of NMR data with Atto532, Alexa647 and Cy5 labeled at positions C322 (Figure S6) indicates that Atto532 and Alexa64 have a minimal effect on the NMR spectra while a larger effect is observed for Cy5 and Atto647N. Moving the attachment sites for the fluorophores farther away from the α-helical region of RRM2 reduces the conformational effects due to the presence of the dye as indicated by the NMR fingerprint spectra.
We also investigated the effects of the Atto532 at position C187 in RRM1,2, which we used as the second labeling position for the spFRET experiments [22]. The NMR spectrum of Atto532-labeled protein at position C187 is very similar to the unlabeled protein (Figure S5B), indicating minimal effects on the protein conformation. We also performed NMR measurements with double-labeled protein where Atto532, Alexa647 or Atto674N were attached to both positions C187 and C326 (Figure S7). The presence of the label has a minimal effect for Atto532 and Alexa647, whereas the intensity of the NMR signals is reduced for labeling with Atto647N. Thus, it appears that the conjugation of fluorophores at this position preserves the integrity of the protein.
Effects of fluorophore attachment on RNA binding of U2AF65
The U2AF65 RRM1,2 domains bind poly-pyrimidine-rich (Py-tract) RNA, which promotes an open conformation where the two RRM domains are arranged side-by-side [20]. To evaluate the effect of the fluorophores on U2AF65 RRM1,2 when bound to RNA, we labeled the protein at both positions, C187 in RRM1 and C326 in RRM2, with either Alexa647 or Atto647N and collected heteronuclear 1H, 15N NMR spectra. With either dye, the structural integrity of dual-labeled U2AF65 RRM1,2 is maintained (Figure 4C, upper panel). Slightly reduced NMR signal intensities are observed for Atto647N-dual labeled protein when compared to Alexa647-dual labeled protein in the absence of RNA (Figure 4C, upper panels). When titrating a high affinity RNA ligand (U9) to the Alexa647 double-conjugated protein, NMR spectra of RRM1,2 show chemical shift changes and line-broadening for residues involved in RNA binding. This is expected as the presence of the RNA changes the electronic environment of amides in spatial proximity and thus alters the NMR chemical shifts. RNA titration to the Atto647N-dual labeled construct induce severe line-broadening of most NMR signals beyond detection (Figure 4C, lower panel). As this line-broadening is more severe than for the single cysteine mutant RRM1,2-C326, the additional attachment of the hydrophobic Atto647N to C187 in RRM1 apparently leads to significant effects by weak interactions with the protein surface and/or may partially interfere with RNA binding by contacts with RNA bases. Additional RNA titrations were performed on the donor Atto532-dual labeled protein (Figure S7B), which in comparison with the titrations of the unlabeled RRM1,2 (Figure S7A) showed similar changes and almost no intensity loss, thus the attachment of the Atto532 dye at both positions does not affect the integrity and RNA binding capabilityof U2AF65.
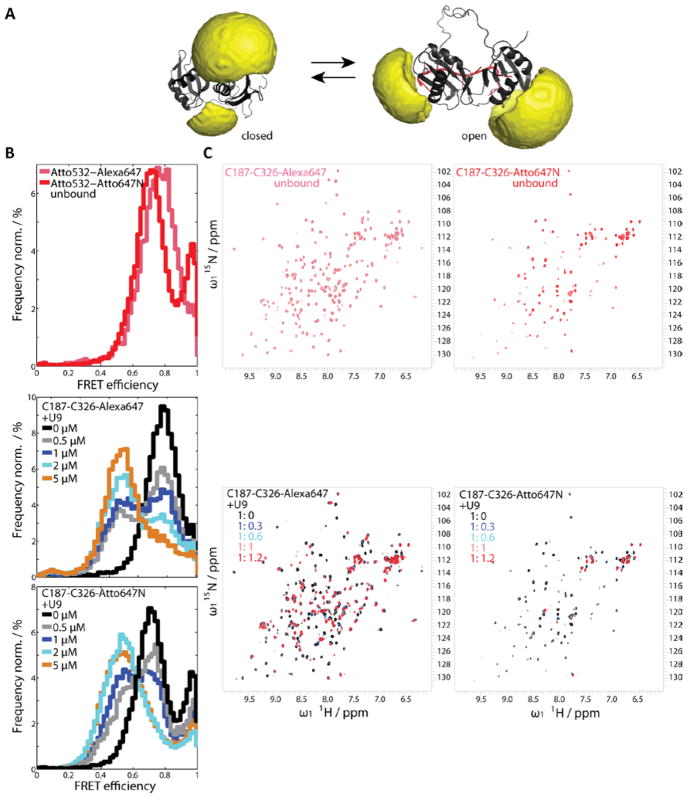
Figure 4. The effect of fluorophores at positions C187 and C326 on the RNA binding of U2AF65 RRM1,2 studied by spFRET in solution and NMR.
(A) Accessible volume calculations of the fluorophores on positions C187 and C326 of RRM1,2 in the closed state in the absence of RNA (PDB accession code: 2YH0) (left) and in the presence of U8 RNA (PDB accession code: 2YH1) (right). (B) FRET efficiency histograms of RRM1,2 labeled with Atto532 and Atto647N (red) or Alexa647 (magenta) in its free form (upper panel). SpFRET efficiency histograms of RRM1,2 labeled with Atto532 and Alexa647 (middle panel) or Atto532 and Atto647N (lower panel) upon addition of RNA at concentration of 0 μM black, 0.5 μM gray, 1 μM dark blue, 2μM light blue, 5 μM orange. (C) Upper panels: 1H,15N-HSQC spectra of unlabeled RRM1,2 (black) and RRM1,2 labeled on positions C187 and C326 with Alexa647 (magenta) or with Atto647N (red). Lower panels: 1H,15N HSQC spectra of a titration series of RRM1,2 fluorescently labeled with Alexa647 (left) or Atto647N (right) withU9 -RNA (black to red). The ratios of RRM1,2 to RNA are indicated in the figure legend, the protein concentration is 120 μM.
Having investigated the influence of the fluorophore on the protein structure and RNA binding, we investigated the conformation changes with spFRET experiments. U2AF65 was labeled stochastically at positions C318 and C326 with the donor fluorophore Atto532 and the acceptors Alexa647 or Atto647N and spFRET measurements were performed in the absence and presence of RNA. Changing the labeling position of the fluorophore from C318 to C326 reduced the amplitudes of the high-FRET state (Figure 4B, upper panel). This subpopulation is even further reduced in the presence of RNA (Figure 4B, middle and lower panel), suggesting that this population of molecules can still actively bind to RNA. SpFRET experiments show that the open conformation of U2AF65 is accessible in the presence of an excess of RNA and that the binding affinity for RNA lies in a similar range, independent of which of the acceptor fluorophores is attached to the positions C187 or C326. Thus, with the exception of the high-FRET state observed for the Atto532-Atto647N FRET pair, we observe the same conformations and RNA binding affinity with the two FRET pairs. Hence, the structural differences we observed with NMR did not have a strong influence on the functionality of the protein.
The influence of different acceptors on the dynamic properties of U 2AF65
Using the relationship between the FRET efficiency and the donor lifetime, we can determine whether a molecule is dynamic, i.e. undergoes a conformational transition during its transit time through the confocal volume [32]. As shown recently, the free U2AF65 protein exhibits dynamics as observed from the deviation from the static FRET line, while U2AF65 bound to RNA is significantly more rigid (Figure S8) [22]. Comparing these two-dimensional histograms of U2AF65 labeled with different acceptor fluorophores, we observe a significantly higher population of the closed state when the protein is labeled with Atto532/Cy5 or with Atto532/Atto647N than with Atto532/Alexa647. This suggests that fewer Atto532/Atto647N- labeled molecules undergo conformational transitions or that the timescale of the conformational dynamics is affected by this combination of fluorophores. Note that free Atto532/Atto647N conjugated U2AF65 shows an additional high FRET state with a FRET efficiency of 95%(Figure 1, Figure S8).
When bound to RNA, dynamic transitions between the closed and the open forms of the protein are still quite pronounced with the acceptor dye Atto647N, while less visible for molecules labeled with different acceptor fluorophores (Figure S8). The additional dynamics observed on a slightly different timescale may be caused by transient interactions of the fluorophores with the protein or with each other.
To investigate dynamics on the milliseconds to second scale, we performed total internal reflection microscopy (TIRF)-based spFRET experiments. U2AF65 labeled with either Atto532/Alexa647 or Atto532/Alexa647 was encapsulated into lipid vesicles and immobilized on the surface of a TIRF prism. For U2AF65-Atto532/Alexa647, only 1% and 5% of the molecules observed in the absence or presence of RNA, respectively, showed a switching between two different FRET states on the timescale of seconds (Table 1). However, 29% and 21% of U2AF65-Atto532/Atto647N molecules (in the absence or presence of RNA, respectively) underwent transitions between different states on the order of seconds (Figure S8C). The FRET efficiencies of these two states correspond to the FRET efficiencies observed for the closed and open conformation of the protein in solution experiments. Dynamic transitions on the second timescale may be due to actual slow dynamics of the protein or due to fluorophore-fluorophore or fluorophore-protein interactions that transiently lock the protein into one of its conformations. In addition, recent studies have described the high propensity of hydrophobic fluorophores to interact with lipids [17], possibly causing additional effects.
Table 1.
Conformational dynamics observed for spFRET on a TIRF microscope.
| Construct | Acceptor fluorophore | Number of dynamic molecules | Number of static molecules | Dynamic molecules [%] |
|---|---|---|---|---|
| RRM1-2-C187-C318 | Atto647N | 129 | 313 | 29 |
| RRM1-2-C187-C318 + U9 |
Atto647N | 35 | 135 | 21 |
| RRM1-2-C187-C318 | Alexa647 | 3 | 234 | 1 |
| RRM1-2-C187-C318 + U9 |
Alexa647 | 5 | 90 | 5 |
Discussion
In single molecule fluorescence spectroscopy studies, it is necessary to collect as many photons as possible. Therefore, whenever possible, organic fluorophores are preferred over fluorescent proteins, which additionally have a high molecular weight and may interfere with the function of the target protein [1]. Every fluorophore comes with its own photophysical and biochemical properties. In this study, common FRET donor-acceptor fluorophore combinations were investigated. Organic fluorophores in the green range of the spectrum (typically used as FRET donors, i.e. Atto532, Atto565) usually perform well with respect to their photophysical properties as well as hydrophilicity and other biochemical characteristics. Finding the optimal fluorophore from the red portion of the spectrum, as FRET acceptors (i.e. Atto647N, Alexa647 and Cy5), is more difficult. While some of the fluorophores show rather good photophysical characteristics, they often come with biochemical properties that are less well suited for experiments on proteins (e.g. high hydrophobicity). Buschmann et al. tested and compared a variety of red fluorophores and described quenching for some of these dyes [33].
Using NMR, we analyzed the conformation of U2AF65 RRM1,2 labeled with three different acceptor fluorophores at positions C318, C322, and C326. The overall behavior of the protein in spFRET experiments and NMR experiments was comparable for all tested fluorophores. However, an additional subpopulation with a high FRET efficiency was observed with different fractional intensities depending on the acceptor fluorophore used. NMR analysis of U2AF65 RRM1,2 labeled with these dyes suggests that this subpopulation may reflect transient interactions of the fluorophore with the protein surface. Additionally, we observed changes in the dynamics of U2AF65 in solution-based MFD-PIE experiments and TIRF measurements of immobilized molecules. Presumably, transient interactions of the fluorophore that are indicated by line-broadening in the NMR spectra affect its conformational flexibility. In TIRF experiments, the dynamics could additionally be influenced by interaction of the fluorophores with the surrounding lipids. This agrees with previous studies in which aromatic rings and negative charges increased the lipophilicity of molecules [34], or where artificial effects of hydrophobic fluorophores on single molecule tracking experiments were described [16].
When potential artifacts are considered and corrected for by using information available from PIE-MFD experiments, consistent results can be extracted from spFRET experiments in solution independent of the pair of fluorophores used. Even artifacts, such as the additional high FRET subpopulation that results from fluorophore attachment to the protein, can be minimized when carefully choosing the labeling position and can even be avoided on the same position when selecting a different fluorophore.
The joint fluorescence and NMR analysis of the conformational effects induced by fluorophore labeling suggest important guidelines for FRET studies: The specific characteristics and effects of each fluorophore should be taken into account when designing in vitro FRET experiments and should be even more carefully considered for in vivo studies where interactions with other proteins or cell components might be influenced by the fluorophore. Especially, when NMR analysis on the studied system is not possible, a number of points should be considered to avoid artifacts and obtain high quality spFRET data:
The position of the dye conjugation within the protein should be evaluated to ensure that this does not affect protein integrity and function, i.e. by avoiding steric clashes with binding ligands or active sites.
The hydrophobicity and electrostatic properties of dyes should be considered to avoid non-specific interactions with the protein or aggregation.
The fluorophore should be attached to conformationally flexible sites such as surface exposed loops, as this may reduce potential effects on the protein structure. In addition, flexible attachment of the fluorophores enables quantitative FRET studies by using the approximation of the orientation factor κ2 as 2/3 for freely rotating dyes. It is interesting to note that a flexible attachment site is not recommended with spin labels for NMR or EPR studies [6].
The fluorophore’s photophysical and biochemical properties have to be considered. While rhodamine-based fluorophores have a high photostability and quantum yields, they sometimes exhibit biochemical properties that are less optimal for the conjugation to certain sites. Depending on the sample under investigation, it is therefore important to consider all the different aspects and analyze the effect of several dye pairs, e.g. by comparing FRET profiles, if little information about the system is available.
In conclusion, great care has to be taken when choosing fluorophores and their attachment sites for single molecule studies. Although certain organic dyes have been described as the best choice for single-molecule tracking [35] or motility assays [36], to date there is no universal answer for the choice of a fluorescent moiety because the requirements for the fluorophore can change for each individual system and experiment. Due to the importance of fluorescence investigations of biological samples, the development of dyes with improved photophysical and biochemical characteristics is ongoing. Especially in the red wavelengths range, new fluorophores are being developed [24,37–39] or known fluorophores are stabilized by additional moieties [40–42].
Here, we have shown the effect of few selected donor and acceptor molecules on the results obtained from spFRET and NMR experiments of U2AF65. We could show that the right choice of labeling position as well as the careful consideration of the different fluorophores properties are highly important. However, by being aware of these effects and verifying results with different fluorophores, one can safely gather the huge wealth of information available from spFRET experiments.
Materials and Methods
Plasmids and constructs
The human U2AF65 (RRM1,2) truncation mutant was designed as described [20]. Cysteines were introduced by site-directed mutagenesis at positions C187, C318, C322, C326 and C187/C318, C187/C326 respectively using a Quick change kit (Agilent Technologies).
Expression and purification of recombinant proteins
U2AF65 mutants were expressed from E. coli BL21(DE3) in M9 minimal media containing 15NH4Cl as only source of nitrogen. Protein purification was done using IMAC (Immobilized Metal Affinity Chromatography) for His-tag purification. Later SEC (Size Exclusion Chromatography) was performed using a Superdex75 Highload 16/60 GE Healthcare column. After verifying their purity with SDS-PAGE, each protein was collected, concentrated up to 1 mM and stored at −80°C until further use. This protocol has being described by Mackereth et al. (2011) [20].
Protein Labeling
Fluorophores were site-specifically conjugated to cysteine residues using sulfhydryl-maleimide chemistry. U2AF65 RRM1,2 harboring single cysteine variants were introduced by site-directed mutagenesis (Supplementary Methods).
Solution-based single-molecule FRET measurements
Single -molecule FRET measurements of fluorescently-labeled proteins in the tens of picomolar range were performed on a custom-build confocal microscope using pulsed interleaved excitation of 532 nm/565 nm and 640 nm lasers and multiparameter fluorescence detection (Supplementary Methods).
Single-molecule FRET measurements of immobilized molecules
Single-molecule FRET measurements of in lipid vesicles immobilized U2AF65 molecules were performed on a custom-build prism-type total internal reflection microscope(Supplementary Methods).
NMR spectroscopy
NMR spectra of 15N-labeled U2AF65(RRM1,2) with and without fluorophores attached were recorded in 20 mM sodium phosphate (pH 6.5), 50 mM NaCl, 1 mM DTT, and 5–10% D2O. The protein concentrations were between 120–270 μM. The experiments were recorded at 298 K, using AV III 600 MHz Cryo or AV III 500 MHz Cryo Bruker spectrometers. The processing and analysis of the NMR data were performed using NMRPipe/Draw [43], and CCPN Analysis [44]. 1H, 15N chemical shifts and intensity of backbone amide signals for residues that neighbor the fluorophore attachment site were monitored to confirm complete labeling of the protein and rule out the presence of a mixture with fractional dye labeling during the measurements (Figure 2, marked residues on the spectrum).
Supplementary Material
Acknowledgments
We thank W. Schrimpf, A. Barth, and W. Kügel for providing the burst analysis software. We gratefully acknowledge the financial support of the Deutsche Forschungsgemeinschaft through the SFB1035 (to D.C.L., Project A11 and to M.S., Project B03), and GRK1721 (to D.C.L. and M.S.). D.C.L. acknowledges support from the Ludwig-Maximilians-Universität through the Center for NanoScience (CeNS) and the BioImaging Network (BIN). L.W. acknowledges a Longterm EMBO postdoctoral fellowship (ALTF 1520-2011) and National Institutes of Health Grants P20G M103408 andP20GM109095.
References
- 1.Marks KM, Nolan GP. Nat Meth. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 2.Owenius R, Osterlund M, Lindgren M, Svensson M, Olsen OH, Persson E, Freskgård PO, Carlsson U. Biophys J. 1999;77:2237–2250. doi: 10.1016/S0006-3495(99)77064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otting G. Journal of Biomolecular NMR. 2008;42:1–9. doi: 10.1007/s10858-008-9256-0. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Castañeda F, Haberz P, Leonov A, Griesinger C. Magnetic Resonance in Chemistry. 2006;44:S10–S16. doi: 10.1002/mrc.1811. [DOI] [PubMed] [Google Scholar]
- 5.Barthelmes K, Reynolds AM, Peisach E, Jonker HRA, DeNunzio NJ, Allen KN, Imperiali B, Schwalbe H. Journal of the American Chemical Society. 2011;133:808–819. doi: 10.1021/ja104983t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göbl C, Madl T, Simon B, Sattler M. Progress in Nuclear Magnetic Resonance Spectroscopy. 2014;80:26–63. doi: 10.1016/j.pnmrs.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Engels JW, Grünewald C, Wicke L. In: Chemical Biology of Nucleic Acids. Erdmann VA, Markiewicz WT, Barciszewski J, editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 385–407. [Google Scholar]
- 8.Erlenbach N, Endeward B, Schöps P, Gophane DB, Sigurdsson ST, Prisner TF. Phys Chem Chem Phys. 2016;18:16196–16201. doi: 10.1039/c6cp02475k. [DOI] [PubMed] [Google Scholar]
- 9.Gophane DB, Endeward B, Prisner TF, Sigurdsson ST. Chemistry - A European Journal. 2014;20:15913–15919. doi: 10.1002/chem.201403726. [DOI] [PubMed] [Google Scholar]
- 10.Jagtap AP, Krstic I, Kunjir NC, Hänsel R, Prisner TF, Sigurdsson ST. Free Radical Research. 2015;49:78–85. doi: 10.3109/10715762.2014.979409. [DOI] [PubMed] [Google Scholar]
- 11.Brumbaugh J, Schleifenbaum A, Gasch A, Sattler M, Schultz C. Journal of the American Chemical Society. 2006;128:24–25. doi: 10.1021/ja0562200. [DOI] [PubMed] [Google Scholar]
- 12.Dixit R, Cyr R, Gilroy S. The Plant Journal. 2006;45:599–615. doi: 10.1111/j.1365-313X.2006.02658.x. [DOI] [PubMed] [Google Scholar]
- 13.Shaner NC, Steinbach PA, Tsien RY. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 14.Chung HS, Louis JM, Eaton WA. Biophys J. 2010;98:696–706. doi: 10.1016/j.bpj.2009.12.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prisner TF, Marko A, Sigurdsson ST. Journal of Magnetic Resonance. 2015;252:187–198. doi: 10.1016/j.jmr.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Zanetti-Domingues LC, Tynan CJ, Rolfe DJ, Clarke DT, Martin-Fernandez M. PLoS ONE. 2013;8:e74200. doi: 10.1371/journal.pone.0074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes LD, Rawle RJ, Boxer SG. PLoS ONE. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dallmann A, Dehmel L, Peters T, Mügge C, Griesinger C, Tuma J, Ernsting NP. Angewandte Chemie International Edition. 2010;49:5989–5992. doi: 10.1002/anie.201001312. [DOI] [PubMed] [Google Scholar]
- 19.Brumbaugh J, Schleifenbaum A, Stier G, Sattler M, Schultz C. Nature Protocols. 2006;1:1044–1055. doi: 10.1038/nprot.2006.177. [DOI] [PubMed] [Google Scholar]
- 20.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcárcel J, Sattler M. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Warner LR, Sanchez C, Gabel F, Madl T, Mackereth CD, Sattler M, Blackledge M. Journal of the American Chemical Society. 2014;136:7068–7076. doi: 10.1021/ja502030n. [DOI] [PubMed] [Google Scholar]
- 22.Voith von Voithenberg L, Sánchez-Rico C, Kang H-S, Madl T, Zanier K, Barth A, Warner LR, Sattler M, Lamb DC. Proceedings of the National Academy of Sciences. 2016;113:E7169–E7175. doi: 10.1073/pnas.1605873113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stennett EMS, Ciuba MA, Levitus M. Chemical Society Reviews. 2014;43:1057. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 24.Kolmakov K, Belov V, Bierwagen J, Ringemann C, Müller V, Eggeling C, Hell S. Chemistry -A European Journal. 2010;16:158–166. doi: 10.1002/chem.200902309. [DOI] [PubMed] [Google Scholar]
- 25.Kudryavtsev V, Sikor M, Kalinin S, Mokranjac D, Seidel CAM, Lamb DC. Chem Phys Chem. 2012;13:1060–1078. doi: 10.1002/cphc.201100822. [DOI] [PubMed] [Google Scholar]
- 26.Widengren J, Schwille P. The Journal of Physical Chemistry A. 2000;104:6416–6428. [Google Scholar]
- 27.Tomov TE, Tsukanov R, Masoud R, Liber M, Plavner N, Nir E. Biophysical Journal. 2012;102:1163–1173. doi: 10.1016/j.bpj.2011.11.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sattler M, Schleucher J, Griesinger C. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34:93–158. [Google Scholar]
- 29.Hartmann A, Krainer G, Schlierf M. Molecules. 2014;19:13735–13754. doi: 10.3390/molecules190913735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcárcel J, Sattler M. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 31.Simon B, Madl T, Mackereth CD, Nilges M, Sattler M. Angewandte Chemie International Edition. 2010;49:1967–1970. doi: 10.1002/anie.200906147. [DOI] [PubMed] [Google Scholar]
- 32.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Tóth K, Langowski J, Seidel CA. Proceedings of the National Academy of Sciences. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buschmann V, Weston KD, Sauer M. Bioconjug Chem. 2003;14:195–204. doi: 10.1021/bc025600x. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa M, Kosaka N, Longmire MR, Urano Y, Choyke PL, Kobayashi H. Molecular Pharmaceutics. 2009;6:386–395. doi: 10.1021/mp800115t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosch PJ, Corrêa IR, Sonntag MH, Ibach J, Brunsveld L, Kanger JS, Subramaniam V. Biophysical Journal. 2014;107:803–814. doi: 10.1016/j.bpj.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris SR, Núñez MF, Verhey KJ. Biophysical Journal. 2015;108:1133–1143. doi: 10.1016/j.bpj.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brabetz W, Weber C. WO2010063732 A1 Novel Combination of Fluorescent Dyes for the Detection of Nucleic Acids. n.d
- 38.Cooper M, Ebner A, Briggs M, Burrows M, Gardner N, Richardson R, West R. J Fluoresc. 2004;14:145–150. doi: 10.1023/b:jofl.0000016286.62641.59. [DOI] [PubMed] [Google Scholar]
- 39.Wurm CA, Kolmakov K, Göttfert F, Ta H, Bossi M, Schill H, Berning S, Jakobs S, Donnert G, Belov VN, et al. Optical Nanoscopy. 2012;1:1–7. [Google Scholar]
- 40.Altman RB, Terry DS, Zhou Z, Zheng Q, Geggier P, Kolster RA, Zhao Y, Javitch JA, Warren JD, Blanchard SC. Nature Methods. 2011;9:68–71. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altman RB, Zheng Q, Zhou Z, Terry DS, Warren JD, Blanchard SC. Nature methods. 2012;9:428–429. doi: 10.1038/nmeth.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinnefeld P, Cordes T. Nat Methods. 2012;9:426–427. doi: 10.1038/nmeth.1977. author reply 427–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 45.Sikor M, Mapa K, Voith von Voithenberg L, Mokranjac D, Lamb DC. The EMBO Journal. 2013;32:1639–1649. doi: 10.1038/emboj.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. Nature Structural & Molecular Biology. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.