Abstract
Nearly all RNA viruses produce double-stranded RNA (dsRNA) during their replication cycles—an important pathogen-associated molecular pattern recognized by the RNA interference (RNAi) pathway in invertebrates and plants. Nodamura virus (NoV) encodes a suppressor of RNA silencing termed B2, which binds to dsRNA and prevents the initiation of RNAi as well as the loading of silencing complexes. Using the published crystal structure of NoV-B2, we performed a series of molecular dynamics (MD) simulations to determine the relative electrostatic and van der Waals contributions of various residues in binding dsRNA, identifying four novel potential interactors: R56, E48, P68 and R69. Additionally, steered MD was used to simulate the binding affinity of NoV-B2 sequences bearing substitutions at positions F49, R56 or R59 to dsRNA, with F49S and R56L/R59L substitutions found to have a significant negative impact on the ability of NoV-B2 to bind dsRNA. NoV RNA1 variants were tested for self-directed replication in both vertebrate (RNAi−) and invertebrate (RNAi+) cultured cells. Consistent with a role in dsRNA binding, NoV replication in F49C and F49S variant constructs was affected negatively only in RNAi+ cells. Thus, we used a combination of MD simulations and experimental mutagenesis to further characterize residues important for NoV-dsRNA interactions.
Keywords: Nodamura virus, B2, Suppressor of RNA silencing, Steered molecular dynamics
Introduction
RNA interference (RNAi) refers to an RNA-guided post-transcriptional regulatory process found in eukaryotic organisms ranging from plants to mammals. RNAi is triggered by the pathogen-associated molecular pattern double-stranded RNA (dsRNA), which is cleaved into 21-nt small interfering RNAs (siRNAs) by the endonuclease Dicer [1]. These siRNAs are loaded into the RNA-induced silencing complex (RISC), which catalyzes the sequence-specific silencing of homologous transcripts through the action of an Argonaute family member [2, 3]. In plants and invertebrates, this process is an important part of the antiviral response (reviewed in [4]).
In response to RNAi, some viruses have been shown to express suppressors of RNA silencing (SRSs) upon infecting their plant or insect hosts (reviewed in [5]). Individual SRSs have been shown to act by sequestering dsRNA molecules, preventing siRNA-loading into RISC or through direct binding of a component of the RNAi machinery [6, 7]. One of the best characterized SRSs is the B2 protein encoded by flock house virus (FHV; family Nodaviridae, genus Alphanodavirus). FHV-B2 has been shown to bind both siRNA duplexes and long dsRNAs [8–10]. Thus, B2 is thought to act by preventing the protein components of the RNAi pathway access to siRNAs and dsRNAs associated with FHV infection [8–10]. The B2 protein is required for sustained FHV replication in RNAi-competent cells, and when components of the RNAi pathway are depleted viral RNA accumulation increases [6, 11–13].
Unlike FHV, Nodamura virus (NoV; family Nodaviridae, genus Alphanodavirus) is a vertebrate pathogen [14], and as such is currently the only known arthropod-borne virus to encode an SRS. Similar to its distant relative FHV, NoV RNA segment 1 (RNA1) encodes a B2 open reading frame (ORF) which overlaps with that of the viral replicase. This ORF is translated from a subgenomic mRNA, termed RNA3, during viral RNA replication. Amino acid similarity between FHV-B2 and NoV-B2 is less than 30% [6], yet the function of NoV-B2 appears to be conserved. NoV-B2 has been shown to bind dsRNA and mutations that prevent translation of NoV-B2 prevent the accumulation of NoV RNAs in cells that mount an RNAi response [13, 15–17].
The crystal structure of FHV-B2 has been solved both individually and bound to dsRNA [8, 9]. These structures suggest key binding residues that interact with dsRNA. Experimentally, predicted dsRNA-binding residues C44 and R54 of FHV-B2 have been subject to mutational analysis. FHV-B2 containing C44S or C44A substitutions were described as having a 100-fold reduced binding affinity to dsRNA [8]. Similarly, a C44Y substitution in FHV-B2 prevented the accumulation of FHV RNA in S2 cells and eliminated the ability of this protein to shield Sindbis virus (family Togaviridae, genus Alphavirus) dsRNA from the RNAi pathway in Aedes aegypti mosquitoes [18]. Likewise, an R54Q substitution in FHV-B2 eliminated its ability to bind long (but not short) dsRNA, resulting in a 20-fold reduction in FHV viral RNA accumulation in RNAi-competent cells [10]. The crystal structure of NoV-B2 has also been solved and its structure was superimposed onto FHV-B2, revealing potential equivalent binding residues [19]. An R59Q substitution in NoV-B2 abolished its ability to bind both long and short dsRNA, and prevented in vitro or in vivo dicing of NoV RNA [13].
In order to determine the importance of other residues of NoV-B2 in binding dsRNA, we used a combination of molecular dynamics (MD) simulations [20] and experimental mutagenesis. We constructed models of NoV-B2 or variant NoV-B2 sequences containing one or two amino acid substitutions in complex with dsRNA. Detailed electrostatic and van der Waals (VDW) interactions between individual NoV-B2 residues and dsRNA were determined in order to identify specific residues that contributed most to the binding interaction. Steered MD was used to simulate the binding affinity of variant NoV-B2 sequences to dsRNA. A subset of these data were used to construct NoV RNA1 variants that were tested for self-directed replication in both vertebrate (RNAi−) and invertebrate (RNAi+) cultured cells. Consistent with the targeted residues being important for dsRNA binding, NoV replication in variant constructs was affected negatively only in RNAi+ cells, confirming our simulations. Herein, we demonstrate a molecular model for predicting the effects of amino acid substitutions that correlates with in vivo experimental results.
Materials and methods
Computational details
Structure and topology
Crystal structures of FHV-B2 in complex with dsRNA (PDB ID: 2AZ0) [8], and unbound NoV-B2 (PDB ID: 3G80) [19] were obtained from the Protein Data Bank [21]. Although overall sequence similarity is less than 30 % [6], sequence identity for the RNA-binding region is 38.1 % (residues 30–71 in FHV-B2; 35–76 in NoV-B2). The backbone atoms of the conserved residues were used to align the NoV-B2 protein to FHV-B2. The resulting NoV-B2 coordinates were combined with the original coordinates of the dsRNA (18 nucleic acids in each chain) from 2AZ0 to make a complete starting structure for our simulations. In addition to the wild type (WT) NoV-B2 simulation, parallel systems containing NoV-B2 with one or two amino acid substitutions were also simulated. Mutations were performed on equivalent residues from both chains in the homodimer. The GROMACS 4.0.7 [22] suite of programs was used for all simulations in conjunction with the ffamber03 all-atom force field [23]. Each protein-dsRNA complex was solvated with the TIP3P water model [24] and sufficient ions were added to neutralize the charge of the system and crudely replicate in vivo conditions. Each system was minimized using a steepest descent integrator until the maximum force on any atom was less than 1,000 kJ mol−1.
Traditional MD simulations
During all simulations, explicit electrostatic interactions were restricted to 0.8 nm and VDW interactions to 1.4 nm. The particle mesh Ewald (PME) method [25, 26] was used to approximate long-range interactions. In addition, a linear constraint solver [22] was used to constrain all bond lengths, allowing for a 2 fs timestep. Following minimization, 200 ps of constant volume-temperature (NVT) MD simulations were performed at 301 K using the Berendsen thermostat [27] and with weak positional restraints on the protein and dsRNA backbone atoms. The systems were further equilibrated under constant pressure-temperature (NPT) conditions for 200 ps at 1 bar of pressure and 301 K. During NPT simulations, the Nosé-Hoover thermostat [28, 29] and Parrinello-Rahman barostat [30, 31] were employed. After lifting the position restraints on the protein backbone atoms, 60 ns of production MD was performed on all variant NoV-B2 systems, and 120 ns of production MD was performed on the NoV-B2 WT system. Equilibration for all systems was assessed on the basis of protein backbone, protein sidechain, and RNA root-mean-square deviation (RMSD) plots (see Supplementary Material, Fig. S1).
Steered MD simulations
Constant-velocity steered MD simulations were performed on the WT and each variant NoV-B2 sequence in quintuplicate. Full velocity and coordinate snapshots taken at 200 ps intervals at the end of the production MD served as the starting points for steered MD simulations. Position restraints were maintained on the dsRNA backbone atoms. The center-of-mass of all protein atoms were tethered to a steered MD reference point via a spring with a spring constant of 1,000 kJ mol−1 nm−2 (equivalent to 1,661 pN nm−1). The steered MD reference point was guided along the normal of the protein-dsRNA interface at a constant rate of 0.01 nm ps−1 for all systems. The present pull rate, spring constant, and tether groups were chosen following a series of ad hoc experiments (seven different pull rates from 0.001 to 0.010 nm ps−1; three spring constants from 500 to 10,000 kJ mol−1 nm−2; four tether groups including all protein heavy atoms, protein backbone atoms, select protein residues, and dsRNA backbone atoms) to find the combination for this specific system that yielded the smoothest force profile peaks without deforming the protein structure. All simulations were performed in parallel on Virginia Tech’s SystemX supercomputer (http://www.arc.vt.eu).
Experimental details
Cells and viruses
Baby hamster kidney (BHK- 21) and Drosophila (S2) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). BHK-21 cells were maintained in DMEM supplemented with penicillin, streptomycin, l-glutamine, and 10 % fetal bovine serum at 37 °C; S2 cells were maintained in Drosophila medium supplemented with penicillin, streptomycin, l-glutamine, and 10%fetal bovine serum at 28 °C. The pNoV RNA1 plasmid was synthesized de novo according to previous work [16, 32]. Untranslatable ΔB2 and all F49 substitution constructs were generated using the QuikChange® II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
Transfections
NoV RNA1 was in vitro transcribed using T7 RNA polymerase and the resulting RNA purified using the MEGAclearTM kit (Ambion, Austin, TX). One microgram of RNA was electroporated in 400 µL BHK-21 (460 V, 725Ω, 75 µF) or S2 (715 V, 1,575Ω, 50 µF) cells at a concentration of 1×107 cells mL−1 using a BTX ECM 630 (Genetronics, San Diego, CA). Electroporated cells were divided between three wells of a six well plate. Cells were harvested and RNA was extracted using TRI Reagent® (Molecular Research Center, Cincinnati, OH) at 1, 2, and 3 days post-transfection.
Northern blots
RNA was analyzed by Northern blot using standard procedures. Probes were generated with the Megaprime™ DNA Labeling System (Amersham, Little Chalfont, UK) from a fragment spanning the NotI and PflmI sites of pNoV RNA1. Detection was achieved using a Storm 840 phosphorimager (GE Healthcare, Piscataway, NJ). Images were analyzed with ImageQuant software (GE Healthcare).
Results
Energy profiles for NoV B2–dsRNA interaction
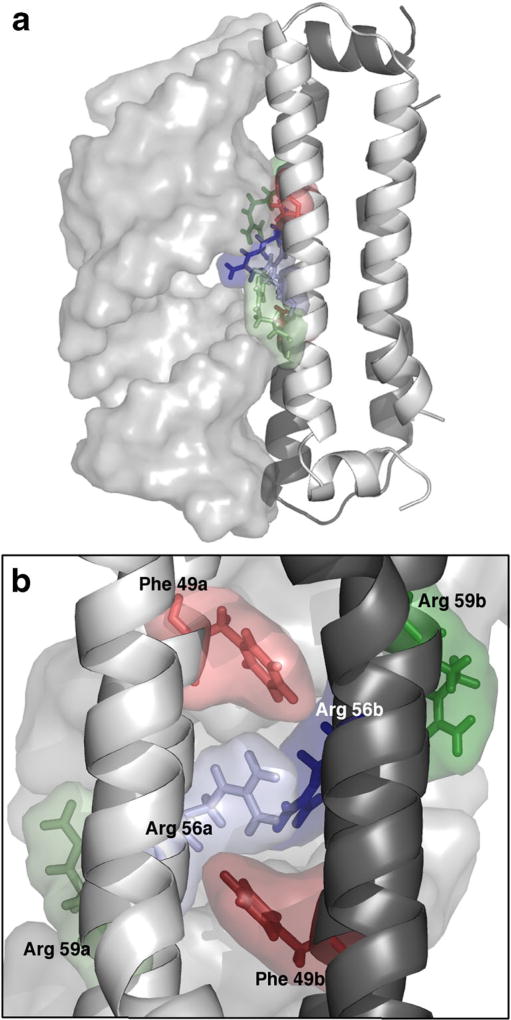
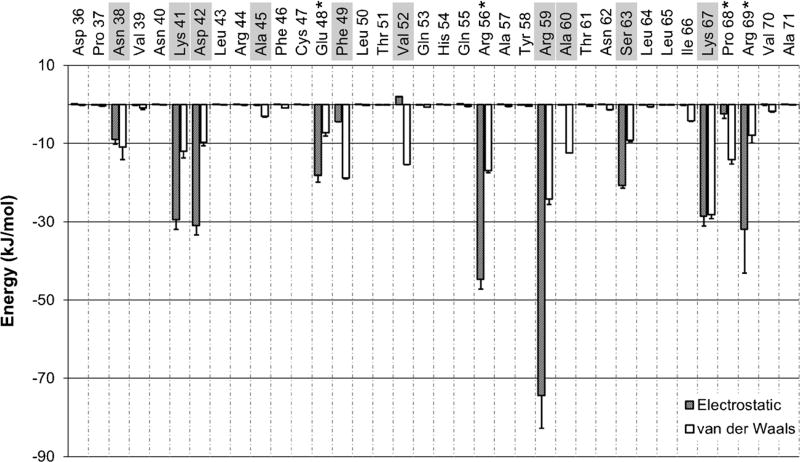
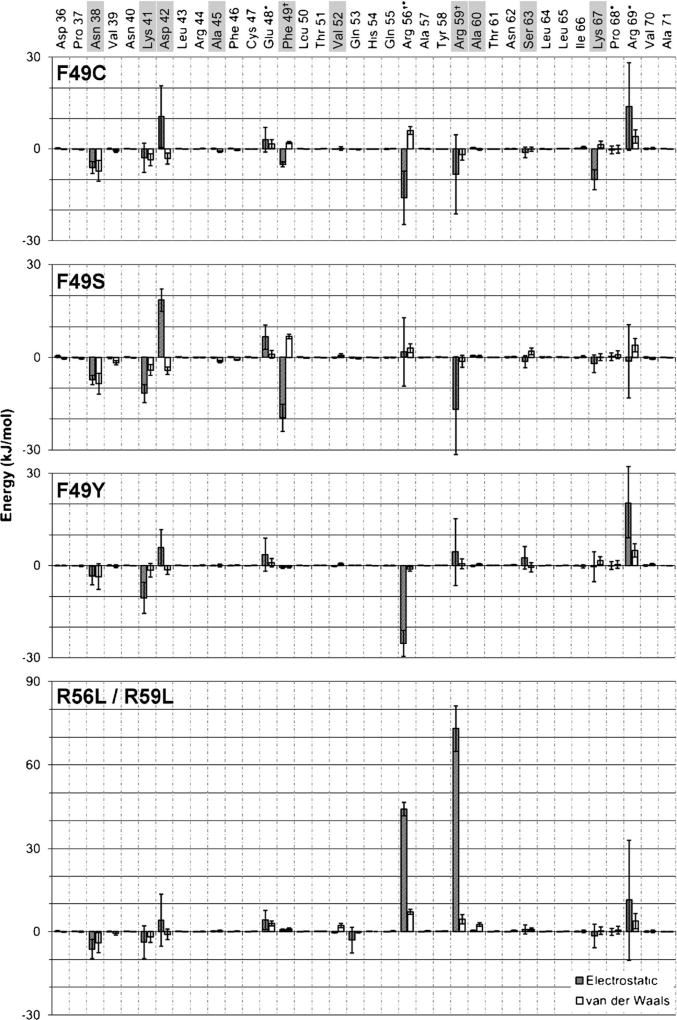
As NoV-B2 has not been crystallized in complex with dsRNA, we constructed a model by superimposing the crystal structure of NoV-B2 over the coordinates of the FHV-B2/dsRNA structure. The superimposed structures had a backbone RMSD of 1.29 Å for the RNA-binding region. The FHV-B2 atoms were deleted, leaving the NoV-B2 atoms in complex with dsRNA (Fig. 1a). We simulated NoV-B2 in complex with dsRNA for 120 ns. Excluding the first 20 ns of production MD, we measured the average energy of interaction between each NoV-B2 residue and the dsRNA molecule, broken down into individual electrostatic and VDW components (Fig. 2). As NoV-B2 binds dsRNA as a homodimer, the calculated energies were averaged between equivalent residues on each chain. Peaks in the energy profile appeared every 3–4 residues, a characteristic that is reflective of the helical conformation of NoV-B2 (Figs. 1, 2). Residue R59 (Fig. 1b) exhibited the greatest average potential energy of interaction, at −99±8 kJ mol−1, consistent with the critical role of this residue in binding dsRNA [13]. Nine additional residues were predicted to play an important role in dsRNA binding through previous work using static modeling (N38, K41, D42, A45, F49, V52, A60, S63, and K67) [19]. Our dynamic simulations support a role for eight of these nine, excluding only A45 (Fig. 2). Our simulations also predict a role for R56, R69, E48, and P68 in binding dsRNA, residues not identified using the crystal structure alone [19]. In particular, newly implicated residues R56 and R69 had the 2nd and 6th highest energy of dsRNA binding, respectively. Our observations indicate that R56, which falls in the major groove of dsRNA, formed a strong and consistent electrostatic interaction with a single phosphate group in the dsRNA backbone. The residue R69 also formed an electrostatic interaction with a phosphate group, but its distal location allowed the residue to explore more conformations that were directed toward the solvent. The acidic group of E48 formed a stable hydrogen bond with a 2′-hydroxyl group of dsRNA, and the P68 residue formed a stable VDW type contact with a 5′-carbon of dsRNA. Residues N-terminal to D36 or C-terminal to A71 were distant enough from the dsRNA during the simulations that all interactions in this region were negligible.
Fig. 1.
Model of Nodamura virus (NoV)-B2 protein in complex with dsRNA. a The B2 homodimer (right) with chainA(light gray) and chain B (dark gray, partly visible) indicated. The dsRNA is shown as a transparent surface rendering (left). b Alternate view of important NoV-B2/dsRNA-interacting residues. R56 (blue) and R59 (green) on each chain lay in dsRNA major groove. F49 (red) residues are adjacent to dsRNA backbone
Fig. 2.
Energy profile for NoV-B2 residues 36–71. Electrostatic contributions to dsRNA binding are shown as shaded bars, van der Waals (VDW) contributions are shown as white bars. Residues that were predicted to be important in dsRNA binding by static modeling [19] are highlighted. B2 residues predicted to be important for dsRNA binding by our dynamic simulations but not by previous static modeling are indicated (asterisk). Error bars Block-average standard error estimate using five blocks
Next, we sought to test the sensitivity of our model by determining how a residue predicted to be a relatively minor binding contributor impacted the overall interaction of NoV-B2 with dsRNA. The choice of residues and the number of possible NoV-B2 substitutions was limited by the presence of the open reading frame coding for proteins A and B1, which overlaps with that of B2 (Fig. 3a). In order to eventually assess the effects of each substitution on RNA1 replication experimentally, it was important that the variant sequences used in our expanded simulations contained alterations only in the B2 protein sequence—not in protein A or B1. Of the 13 residues predicted to participate in binding dsRNA, we selected a panel of three sequences where the F49 residue was altered: F49C, F49S, F49Y (Fig. 3b); and one theoretical construct containing an R56L/R59L double substitution. The F49 site was chosen because it was predicted to be a minor interactor (9th out of 14, Fig. 2), the overlap with protein A/B1 allowed three distinct biochemical substitutions (C, S, Y), and the homologous residue in FHV-B2 (C44) had been shown previously to be important in both binding dsRNA [8] and protecting replicating viral genomes from RNAi [18]. The R56L/R59L double mutant was included as a positive control as these were the strongest predicted interactors (1st and 2nd out of 14, Fig. 2), and an R59Q substitution had already been shown to abolish dsRNA binding [13].
Fig. 3.
NoV B2-F49 mutants are silent with respect to protein A/B1 amino acid sequence. a NoV RNA1 is shown with the subgenomic promoter depicted by the horizontal black arrow. During replication, RNA3 is synthesized and codes for proteins B1 (ORF1, white box) and B2 (ORF2, gray box). b F49 point mutations made in the NoV RNA1 sequence (nt 2881–2896 are shown). Altered nucleotides are in lower case and the affected residues are in bold
In the simulations of WT B2, F49 formed contacts with the dsRNA in a very consistent and predictable way. The sidechain of F49 was situated at the protein dimer interface, laying adjacent to, rather than coincident with, the dsRNA backbone. During the last 100 ns of simulation, a hydrogen atom from each F49 β-carbon formed a hydrogen bond with the 2′-oxygen of ribose (Fig. 4), accounting for the electrostatic interaction observed at this residue (Fig. 2). The large VDW contribution arose from the close and consistent contact of the F49 δ- and ε-carbons with the ribose carbons at the 4′ and 5′ positions. We did not observe any rotation of either F49 aromatic ring around the β-/γ-carbon bond during the simulation. When changed to a tyrosine (F49Y), the nature of the VDW interactions and the hydrogen bond between the β-carbon of Y49 and 2′-oxygen of ribose remained unchanged. The newly introduced hydroxyl group did not interact with the dsRNA; rather, it formed electrostatic interactions with Q53 of the same protein chain, in effect pulling the Y49 sidechain away from the dsRNA, resulting in some inconsistency in the interaction between Y49 and the dsRNA. This was observed as a greater fluctuation in the sidechain position when compared to WT (Fig. 4). For the F49C substitution, the residue lost its VDW contacts but formed an electrostatic interaction with the 3′-oxygen of dsRNA. The sidechain position, however, was still relatively stable during the course of the simulations (Fig. 4). Finally, with the serine substitution (F49S), the sidechain hydroxyl group intermittently formed a hydrogen bond with the 3′-oxygen of ribose, or with the 2′-oxygen of ribose, displacing the hydrogen bond between the β-carbon hydrogen and the 2′-oxygen. Unlike the F49C mutant, the serine residue in F49S rotated quite freely, and the hydrogen bond that it did form was short-lived. In addition to changes in interaction at the target residue, our simulations predicted various additional gains and losses among other residues of the NoV-B2 dsRNA-binding region in the presence of F49 substitutions. All three substitutions were predicted to result in a decrease in electrostatic interaction at positionD42 (positive values indicate a loss in potential energy of interaction, negative values indicate a gain of potential energy of interaction), and F49C and F49Y displayed large decreases in interaction at position R69 (Fig. 5a). Interactions at residues N38 and K41 were strengthened in all three substitutions, with interactions at positions R56, R59 and K67 affected in a subset of mutants (Fig. 5a).
Fig. 4.
Ensemble conformations of wild type (WT) and substituted residues at position 49. The F49 residue forms stable contacts with ribose atoms, whereas the F49Y substitution exhibits higher fluctuation. F49C substitution forms stable, but fewer contacts than does the WT F49, and F49S substitution fluctuates greatly. Protein backbone is shown as gray cartoon, dsRNA backbone is shown as orange cartoon, stick structures represent starting conformation of residues following equilibration (white H, gray C, blue N, red O), and dark gray lines represent conformations taken in 3-ns snapshots (WT) or 1.5-ns snapshots (all others)
Fig. 5.
Energy difference profiles for NoV-B2 F49 or R56/R59 substitutions. Shaded bars Electrostatic differences in dsRNA binding, white bars VDW differences. Positive values indicate a loss in binding energy (weaker interaction); negative values indicate a gain in binding energy (stronger interaction). dsRNA-interacting residues predicted by Körber et al. [19] are highlighted. B2 residues predicted to be important for dsRNA binding by our dynamic simulations but not by previous static modeling are indicated (asterisk). Black crosses indicate the substituted residue(s) at position F49 (top three panels) or R56/R59 (bottom panel). Error bars Block-average standard error estimate using five blocks
During the simulations of WT B2, the R56 and R59 residues of each chain formed many contacts in the major groove of dsRNA. Most notably, they formed highly charged electrostatic interactions, especially with the phosphate groups of the dsRNA backbone. These interactions were lost entirely when the sidechains were mutated to the non-polar leucine residues. Across the remainder of the binding surface, only residues R69 (loss) and N38 (gain) experienced a change in potential of interaction of at least 10 kJ mol−1 (Fig. 5b).
Steered MD simulations of NoV B2–dsRNA complex
The global changes in dsRNA interaction energies observed for F49 substitutions suggested that these changes may positively or negatively affect the ability of NoV-B2 to bind dsRNA. To predict the effects of each mutation on dsRNA binding, we performed a series of steered MD simulations. We hypothesized that the movement of the center of mass of the NoV-B2 protein in the direction away from the dsRNA would occur sooner or with less force for the variant proteins than for the WT, thus relieving the tension on the spring more quickly. Physiologically, this is comparable to a decreased affinity of the altered proteins for binding dsRNA.
We took five full-coordinate, full-velocity snapshots of each system in 200 ps intervals (as described in Materials and methods) at the conclusion of the traditional MD simulations to be used as the starting configurations for the steered MD simulations. Proteins bearing F49 or R56/R59 substitutions behaved similarly to WT during the first 100–150 ps of the pulled simulations, as the tension on the spring grew (Fig. 6a). During the last 100–150 ps, the force curves of each NoV-B2 variant typically re-converged with the WT protein curves, as it required the same amount of force to pull each protein through bulk solvent (Fig. 6a). During the middle of the simulations, however, especially between 150 and 300 ps, we observed a decrease in the peak of the force curves for each variant NoV-B2 sequence when compared to the WT (Fig. 6a). The average peak force required to pull NoV-B2 from the dsRNA was slightly lower for both F49Yand F49C substitutions, although this difference was not statistically significant (Fig. 6b). However, both the F49S and R56L/R59L sequences experienced a significant decrease in the peak force required to pull NoV-B2 from the dsRNA when compared to the WT protein (Fig. 6b). Based strictly on the means obtained, we predicted a rank order of B2-dsRNA binding strength of: WT > F49Y > F49C > F49S > R56L/R59L.
Fig. 6.
Force profiles for steered molecular dynamics (MD) simulations. a Force profiles obtained from five replicate simulations of pulling NoV-B2 protein (WT) away from dsRNA are shown in black on each plot for comparison. Blue (F49Y), red (F49C), green (F49S), and orange (R59L/R59L) curves indicate the force profiles obtained from five replicate simulations of pulling each variant B2 sequence. Inserts represent a magnified view of the peak force region. b Average peak force on the spring across replicates and c average time required to reach peak force for WT and variant B2 protein sequences. Error bars Standard deviation from the mean; asterisks statistical significance following one-way ANOVA and Dunnett’s multiple comparison test (* P<0.05; *** P<0.001)
We also analyzed the time to peak force for the WT NoVB2 sequence and each mutant (Fig. 6c). Though the average time to peak force was lower for all variant NoV-B2 sequences as compared to WT, none were statistically significant. Again using the calculated means, these data suggested slightly different rank order of NoV-B2-dsRNA binding strength: WT > F49C > F49Y > F49S > R56L/R59L.
Effect of F49 substitutions on the replication of NoV RNA1 in the presence or absence of RNAi
In order to determine the biological relevance of our simulations, we sought to obtain experimental evidence concerning the effect of each F49 substitution on the ability of NoV RNA1 to self-replicate in the presence or absence of a functional RNAi pathway. Previous work has established that NoV-B2 is critical to protecting the replicating viral RNA genome in the presence of an active RNAi response, whereas in the absence of RNAi NoV-B2 is expendable [13, 15–17]. Similar work with FHV-B2 has shown that the protective effect of B2 expression is dependent on its ability to bind dsRNA [8, 10]. In vitro synthesized RNA corresponding to WT NoV RNA1, or to RNA1 bearing F49Y, F49C or F49S substitutions was electro-transfected into BHK-21 or S2 cells (Fig. 7a). BHK-21 cells are not known to mount an RNAi response, and B2 does not appear to be required for NoV RNA1 replication in BHK-21 cells [15]. As expected, all of the NoV RNA1 F49 mutants were able to replicate to levels equivalent to wild-type NoV RNA1 in the absence of RNAi (Fig. 7). In contrast, Drosophila S2 cells mount a strong RNAi response triggered by the dsRNA generated through the self-replication of RNA1. The F49Y substitution did not appear to affect RNA1 replication in S2 cells, while the F49C substitution strongly reduced and the F49S mutation completely prevented RNA1 accumulation. This parallels the rank order predicted by our steered MD simulations using the peak force required to separate NoV-B2 from dsRNA. We conclude that a reduced ability of NoV-B2 to bind to, and thus protect the viral dsRNA, allows the RNAi machinery greater opportunity to target viral replicative intermediates and to form mature NoV-targeting RISC complexes.
Fig. 7.
NoV-B2 F49 substitutions reduce RNA1 replication in cells that mount an RNA interference (RNAi) response. a Northern blot analysis of NoV RNA1 and RNA3 in BHK-21 (top) and S2 (bottom) cells. RNA was extracted at days 1, 2, and 3 post-transfection; or from untransfected cells (−) as indicated. rRNA indicates equivalent RNA loading. b Average relative RNA levels of three biological replicates determined using a phosphorimager. All samples were normalized to the RNA1 band of the wild-type day 1 sample. Error bars Standard deviation from the mean; asterisks indicate statistical significance following one-way ANOVA and Dunnett’s multiple comparison test (* P<0.05; ** P<0.01). For S2 cells, F49Y samples were excluded during statistical analysis due to unequal variance compared with the rest of the samples
Three biological replicates of each NoV RNA1 accumulation experiment were performed, followed by densitometry analysis of both RNA1 and RNA3 band intensities (Fig. 7b). Statistical analysis confirmed that there was no difference in RNA1 or RNA3 levels between WT and F49 substitution constructs in BHK-21 cells (Fig. 7b). In S2 cells, accumulation of both RNA1 and RNA3 was significantly lower in both F49C (day 3 only) and F49S (all time points) (Fig. 7b). Thus our experimental data establishes a ranked order of: WT = F49Y > F49C > F49S, closely consistent with our MD simulations. These data also suggest that the absolute peak force required to separate NoV-B2 from the dsRNA molecule during steered MD simulations has more predictive power than the data concerning the time to peak force. This information will be valuable to future experiments where additional NoV-B2 residues predicted to participate in binding dsRNA are simulated, or as similar steered MD simulations are applied to other SRS-dsRNA interactions.
Discussion
We performed a series of MD simulations in an attempt to better understand the interactions between the NoV-B2 protein suppressor of RNA silencing and its dsRNA target. Our simulations have provided insight into the relative contributions of various residues in binding dsRNA in terms of both electrostatic and VDW interactions, and increased the number of amino acids predicted to play a role in dsRNA binding from 10 to 14 [19]. Simulations of single or double amino acid substitutions inNoV-B2 suggested that changes at residue F49 have an impact on predicted binding contributions across the entire surface of the dsRNA-binding site, while R56L/R59L changes had a more local effect. Steered MD simulations suggested that these changes in binding energies would negatively impact the ability of NoV-B2 to bind dsRNA. Indeed, we were able to confirm experimentally that NoV-B2 proteins carrying F49C or F49S substitutions were no longer able to protect the NoV genome from the RNAi pathway. It is important to emphasize that, in this manuscript, we focused on one type of SMD approach, but many other potentially useful experiments (including constant force SMD, umbrella sampling, etc.) and analyses (including examination of force vs distance, free energy calculations, etc.) are available to researchers.
Previously, Xia et al. [33] used steered MD techniques to simulate the separation of the p19 SRS from dsRNA and were able to distinguish weakly binding mutants from the more tightly binding wild-type protein. We used a variant of their technique to create a rank-order of dsRNA-binding affinity for NoV-B2 and each substitution mutant. Our computational model of NoV-B2 was similar to predictions made by Körber et al. [19], who identified putative dsRNA-binding residues through aligning the crystal structure of NoV-B2 with that of FHV-B2, which, in turn, had been crystallized in complex with dsRNA. Nine of the ten residues identified by Körber et al. were predicted to participate in dsRNA interactions in our study. However, our study predicted four novel interacting residues: E48, R56, P68 and R69; and called into question a role for A45. In fact, a ranking of the predicted binding energies places R56 second and R69 sixth in overall binding contribution. Also, all four newly identified residues had predicted binding energies at or greater than that of residue F49, which we found to be critical in this study, reinforcing the importance of these contacts. Overall, analogous to protein–protein interfaces, we would emphasize that it is not only the sum of all interactions that infer specificity in this system, but it is also the magnitude, distribution, and nature of those interactions [34].
FHV-B2 and NoV-B2 have both been shown to bind dsRNA [8, 9, 13, 17], and FHV and NoV expression systems that fail to express B2 have been used widely to study the role of the RNAi machinery in antiviral immunity [6, 11, 12, 15, 16, 35]. However, these studies presuppose that binding of dsRNA (either pre- or post-Dicer cleavage) is the only function attributable to FHV-B2 or NoV-B2. Ball [32] reported that loss of FHV-B2 expression reduces the efficiency of RNA1 replication and progeny production in BHK-21 cells, indicating a possible second, supplementary role for this protein. In contrast, Johnson et al. [15] did not observe a negative effect on NoV RNA1 replication in BHK-21 cells in the absence of B2. Our data (Fig. 6; ΔB2 samples) are consistent with those of Ball [32], and support the hypothesis that NoV-B2 may have additional (though not essential) roles beyond binding dsRNA. Indeed, Aliyari et al. [13] showed that FHV-B2 forms a stable complex with the viral replicase independent of dsRNA. A role for FHV-B2 in directly binding to Dcr-2 has also been suggested [36, 37]. Given this uncertainty, the use of NoV or FHV RNA1 constructs that express a B2 protein deficient only in dsRNA-binding may be more suitable to certain research questions.
Summary
Crystal structures are becoming more abundant for a variety of proteins. Traditionally, to interrogate critical interactions, multiple mutant forms are generated based on crystallographic or phylogenetic data. Subsequent biochemical characterization can then determine how each specific mutation affects the suspected function of the protein. Our data attest to the limitations of static crystal structures when predicting biophysical properties of a dynamic interaction. The relatively straightforward and concise simulations detailed in this report have provided valuable insight regarding critical binding residues for this system, which were confirmed by experiment. Being able to not only make better predictions, but also test them in silico will save valuable time and resources.
Supplementary Material
Acknowledgments
This work was supported by of the National Institutes of Health, National Institute of Allergies and Infectious Diseases (grants AI072038 to Z.N.A. and AI077726 to K.M.M.), as well as by the Fralin Life Science Institute at Virginia Tech. The authors thank the administrators of the Advanced Research Computing facility at Virginia Tech for computing hours.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00894-014-2092-0) contains supplementary material, which is available to authorized users.
Contributor Information
William J. Allen, Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061-0308, USA
Michael R. Wiley, Department of Entomology, and Fralin Life Science Institute, Virginia Tech, Blacksburg, VA 24061-0346, USA
Kevin M. Myles, Department of Entomology, and Fralin Life Science Institute, Virginia Tech, Blacksburg, VA 24061-0346, USA
Zach N. Adelman, Department of Entomology, and Fralin Life Science Institute, Virginia Tech, Blacksburg, VA 24061-0346, USA
David R. Bevan, Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061-0308, USA
References
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 4.Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 5.Wu Q, Wang X, Ding S-W. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe. 2010;8:12–15. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler J-L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 7.Marques JT, Carthew RW. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:359–364. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 9.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–1154. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X-H, Aliyari R, Li W-X, Li H-W, Kim K, Carthew R, Atkinson P, Ding W. RNA interference directs innate immunity against viruses in adult drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 13.Aliyari R, Wu Q, Li H-W, Wang X-H, Li F, Green LD, Han CS, Li W-X, Ding S-W. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ball LA, Amann JM, Garrett BK. Replication of nodamura virus after transfection of viral RNA into mammalian cells in culture. J Virol. 1992;66:2326–2334. doi: 10.1128/jvi.66.4.2326-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson KL, Price BD, Eckerle LD, Ball LA. Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J Virol. 2004;78:6698–6704. doi: 10.1128/JVI.78.12.6698-6704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W-X, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EWA, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding S-W. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Körber S, Shaik Syed Ali P, Chen JCH. Structure of the RNA-binding domain of Nodamura virus protein B2, a suppressor of RNA interference. Biochemistry. 2009;48:2307–2309. doi: 10.1021/bi900126s. [DOI] [PubMed] [Google Scholar]
- 20.Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 23.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 25.Darden T, York D, Pedersen LG. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 26.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 27.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 29.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 30.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 31.Nosé S, Klein ML. Constant pressure molecular dynamics for molecular systems. Mol Phys. 1983;50:1055–1076. [Google Scholar]
- 32.Ball LA. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Z, Zhu Z, Zhu J, Zhou R. Recognition mechanism of siRNA by viral p19 suppressor of RNA silencing: a molecular dynamics study. Biophys J. 2009;96:1761–1769. doi: 10.1016/j.bpj.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 35.Berry B, Deddouche S, Kirschner D, Imler J-L, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS ONE. 2009;4:e5866. doi: 10.1371/journal.pone.0005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh G, Popli S, Hari Y, Malhotra P, Mukherjee S, Bhatnagar RK. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 2009;23:1845–1857. doi: 10.1096/fj.08-125120. [DOI] [PubMed] [Google Scholar]
- 37.Singh G, Korde R, Malhotra P, Mukherjee S, Bhatnagar RK. Systematic deletion and site-directed mutagenesis of FHVB2 establish the role of C-terminal amino acid residues in RNAi suppression. Biochem Biophys Res Commun. 2010;398:290–295. doi: 10.1016/j.bbrc.2010.06.083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.