Abstract
Thrombin is a serine protease that plays a crucial role in hemostasis, fibrinolysis, cell proliferation, and migration. Thrombin binding aptamer (TBA) is able to inhibit the activity of thrombin molecule via binding to its exosite I. This 15-nt DNA oligonucleotide forms an intramolecular, antiparallel G-quadruplex structure with a chair-like conformation. In this paper, we report on our investigations on the influence of certain modified nucleotide residues on thermodynamic stability, folding topology, and biological properties of TBA variants. In particular, the effect of single incorporation of a novel 4-thiouracil derivative of unlocked nucleic acid (UNA), as well as single incorporation of 4-thiouridine and all four canonical UNAs, was evaluated. The studies presented herein have shown that 4-thiouridine in RNA and UNA series, as well as all four canonical UNAs, can efficiently modulate G-quadruplex thermodynamic and biological stability, and that the effect is strongly position dependent. Interestingly, TBA variants containing the modified nucleotide residues are characterized by unchanged folding topology. Thrombin time assay revealed that incorporation of certain UNA residues may improve G-quadruplex anticoagulant properties. Noteworthy, some TBA variants, characterized by decreased ability to inhibit thrombin activity, possess significant antiproliferative properties reducing the viability of the HeLa cell line even by 95% at 10 μM concentration.
Keywords: thrombin binding aptamer, G-quadruplexes, thrombin, antiproliferative agents, anticoagulants
Graphical Abstract
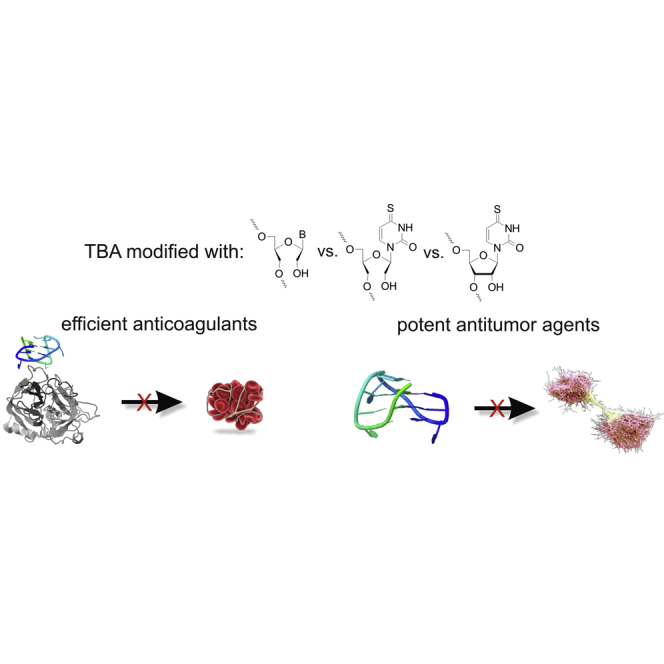
Introduction
In recent years there has been growing interest in discovery and application of therapeutic oligonucleotides. They constitute a wide group of compounds consisting of single- or double-stranded DNA or RNA constructs. According to structure and mechanism of action, therapeutic oligonucleotides could be divided into a few classes including antisense oligonucleotides, small interfering RNAs, ribozymes, DNAzymes, decoys, CpG oligonucleotides, and aptamers.1
Aptamers are short, single-stranded oligonucleotides whose sequences determine their folding into secondary structure.2, 3 Accordingly, they are able to bind with high affinity and selectivity to various targets including proteins,4, 5 peptides,5 small chemical compounds,4, 6 and metal ions,7 and thereby to initiate a biological response. Due to those interesting properties they are termed chemical antibodies. The main method to evolve a new aptamer is in vitro selection via a process termed systematic evolution of ligands by exponential enrichment (SELEX).3, 8
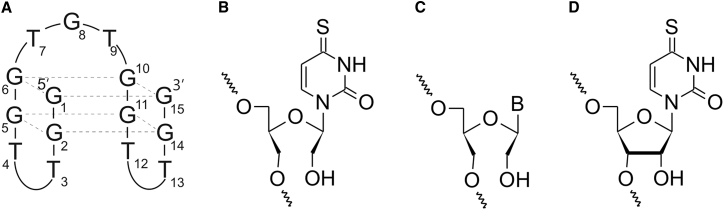
One of the first aptamers discovered was thrombin binding aptamer (TBA), selected by Louis Bock in 1992.9 It is a 15-nt DNA oligonucleotide, which forms an intramolecular, antiparallel G-quadruplex structure with a chair-like conformation (Figure 1A).10 The core of TBA consists of the two G-tetrads, each built up with four guanosine residues stabilized by Hoogsteen hydrogen bonds11, 12 and displaying syn or anti conformation of the glycosidic bonds.13, 14 G-tetrads within TBA are linked by three loops: two TT edge-wise loops and one TGT loop.15, 16 Structural studies indicate that T4 and T13 residues stabilize G-quadruplex structure due to stacking on the neighboring G-quartet and formation of T-T base pair via carbonyl and imino proton of each base. On the contrary, T3, T7, and T12 residues are not involved in intramolecular interactions and are pointed outside the G-quadruplex core. Based on crystallographic and nuclear magnetic resonance (NMR) studies it was determined that TBA interacts with thrombin anion exosite I via two TT loops.16, 17, 18 Comprehensive structural studies showed that T3 and T12 nucleobases form a pincer-like motif that grips the fibrinogen recognition site region.
Figure 1.
Schematic Representation of TBA and Modified Residues Applied in the Studies
Structure of (A) thrombin binding aptamer (TBA), (B) novel 4-thiouracil derivative of UNA, (C) UNA nucleotide monomer, and (D) 4-thiouridine.
The superior anticoagulant properties of TBA were noticed simultaneously with its discovery. However, the therapeutic dose was too high to successfully accomplish clinical trials.19 Fortunately, the promising properties of TBA such as reversibility of action, small size, and simplicity of chemical synthesis prompted scientists to make attempts to create new TBA variants that would provide significant therapeutic benefit, outweighing the side effects.12 Thus far, a wide variety of modifications were tested for the ability to improve anticoagulant properties of TBA, including 4-thio-2′-deoxyuridine,20 LNAs (locked nucleic acids),21 UNAs (unlocked nucleic acids),22, 23 2′-deoxy-isoguanosine,24 RNA25 and 2′-O-methyl-RNA nucleotides,25 methylphosphonate25 and phosphorothioate internucleoside linkages,25, 26 partial inversion of TBA polarity with a 5′-5′27, 28 and a 3ʹ-3ʹ internucleoside linkage,29 and changes of the loop size and sequence.30 Three of these modifications, i.e., 4-thio-2′-deoxyuridine, 2′-deoxy-isoguanosine, and unlocked nucleic acids, were found as favorable for TBA anticoagulant properties. In addition, the latest literature has demonstrated the possibility of changing the anticoagulant activity of TBA for antiproliferative activity via introduction of a dibenzyl linker.31
Herein, a novel 4-thiouracil derivative of UNA (UNA-s4U) has been synthesized for the first time and introduced into TBA. We have examined the influence of single and multiple introductions of UNA-s4U, as well as of regular UNA-A, UNA-C, UNA-G, UNA-U, and 4-thiouridine (RNA-s4U) on TBA folding topology (Figure 1). Furthermore, the potential changes in G-quadruplex thermodynamic stability induced by the modified nucleoside residues were investigated. Finally, we have analyzed the anticoagulant and antiproliferative potential of the novel, chemically modified, TBA variants.
Results and Discussion
It has been demonstrated that the replacement of T or dG within the TGT loop by other 2′-deoxynucleosides does not perturb G-quadruplex structure and can be favorable for thermodynamic stability of the TBA molecule.30 Moreover, previously published data concerning UNA-modified TBA variants described the consequences of substitution of dG by UNA-G and T by UNA-U.22 Herein we have extended these studies by the introduction of other types of UNA residues, i.e., UNA-A, UNA-C, or UNA-G instead of T and UNA-A, UNA-C, or UNA-U instead of dG. According to already published data, the thermodynamically and biologically most unfavorable positions for the introduction of UNA residues into TBA was any position of the G-quartet. Therefore, in this paper, UNA residues were introduced only at the positions of TBA loops: T3, T4, T7, G8, T9, T12, or T13.
It was reported that TBA containing four 4-thio-2′-deoxyuridine nucleotides at positions T3, T7, T9, and T13 possess improved anticoagulant activity.20 Increased biological activity was also described for TBA modified by UNA-U at position T7.22 Based on these findings, we designed TBA variants with single substitution of a novel UNA derivative, i.e., UNA-4-thiouracil, thus combining both structural components reported to be favorable for biological activity.18, 20 Novel TBA variants containing a single substitution of UNA-4-thiouracil or 4-thiouridine at all possible positions of the loops were synthesized and studied, and we also analyzed the TBA variants with four UNA-4-thiouracil or 4-thiouridine residues at positions T3, T7, T9, and T12 or T3, T7, T9, and T13.
CD Spectroscopy
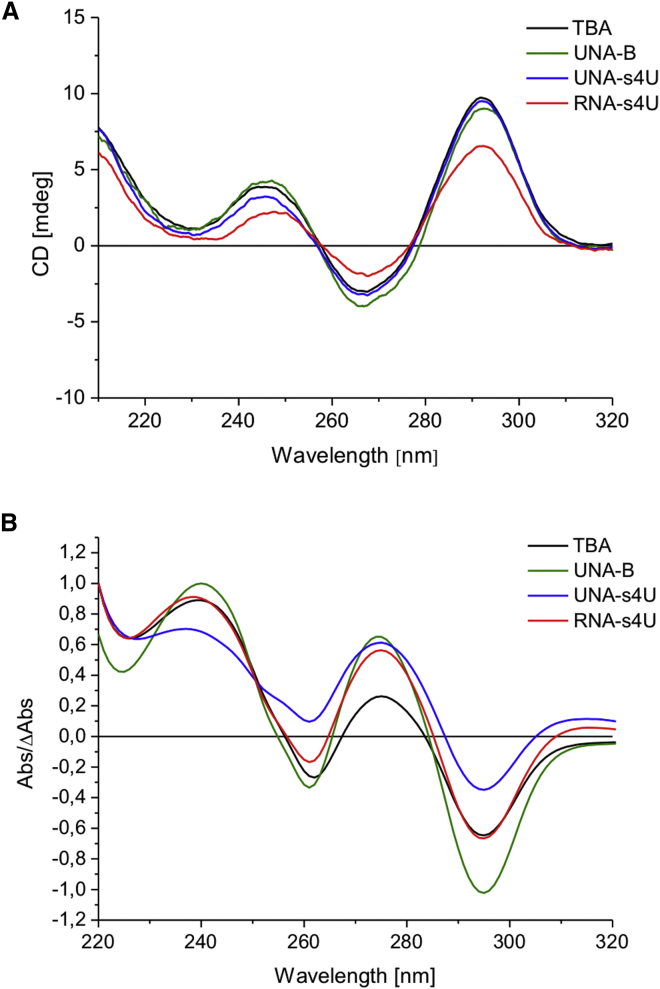
Circular dichroism (CD) spectroscopy is a technique that allows determination of probe conformation based on the differences in absorbance of right- or left-handed circularly polarized light.32 Nucleotide residues are optically active elements that generate CD signal. Different spatial orientation of the heterocyclic bases in certain nucleic acid secondary structures results in occurrence of characteristic CD spectra providing information about specific structural geometry or folding topology. G-quadruplexes may possess one of two folding topologies: parallel or antiparallel.33 The parallel folding topology occurs when all strands display the same orientation and is characterized by the presence of one positive CD band around 260 nm and one negative near 240 nm. When the antiparallel folding topology is observed, CD spectra are characterized by the presence of one positive band around 290 nm and one negative band near 260 nm.
The CD spectra obtained for TBA variants containing single or multiple substitution of RNA-s4U, UNA-s4U, UNA-A, UNA-C, UNA-G, and UNA-U display the typical profile for an antiparallel G-quadruplex structure. Two positive bands around 245 and 295 nm and one negative band near 265 nm were observed for all TBA variants (Figure 2A).10, 34 Based on this result, it can be concluded that introduction of modified nucleotides into TBA at the studied positions has no influence on the overall G-quadruplex folding topology, and all analyzed TBA variants form antiparallel G-quadruplex structure at physiological temperature.
Figure 2.
Spectral Analysis of G-quadruplex Folding Topology
Representative circular dichroism spectra (A) and thermal difference spectra (B) of unmodified TBA (black lines) and TBA variants modified with canonical UNA (green lines), UNA-s4U (blue lines), and RNA-s4U (red lines).
Thermal Difference Spectra
Thermal difference spectroscopy (TDS) constitutes a complementary technique to CD spectroscopy and enables evaluation of oligonucleotides structure in solution.35 Specific nucleic acids structures possess characteristic TDS patterns reflecting subtle alterations in stacking interactions.
The TDS obtained for TBA variants containing single or multiple substitution of RNA-s4U, UNA-s4U, UNA-A, UNA-C, UNA-G, and UNA-U is characterized by a similar pattern with two positive bands around 240 and 270 nm and two negative bands near 260 and 295 nm (Figure 2B). According to previously published data and to the TDS of unmodified TBA, the profile of the spectra is characteristic for an antiparallel G-quadruplex structure.22 Taking under consideration the above data, it can be concluded that the introduction of modified nucleotides, i.e., canonical UNAs, UNA-s4U, and RNA-s4U, into TBA has no influence on the overall G-quadruplex folding topology. The results are consistent with those derived from the CD spectra and thermodynamic studies.
Thermal Denaturation Measurements
Thermodynamic Analysis of TBA Variants Modified with UNA-A, UNA-C, UNA-G, or UNA-U
Thermodynamic analysis revealed that the most energetically favorable is replacement of thymidine at position T7 by UNA-A, UNA-C, or UNA-G (Table 1). The presence of UNA-A, UNA-C, and UNA-G at this position induced change of Gibbs free energy by −0.37, −0.59, and −0.53 kcal/mol, respectively. Previously published data indicate that the presence of UNA-U at position T7 caused stabilization by 0.50 kcal/mol.22 Our results are strictly comparable with the influence of UNA-U. It suggests that stabilization of TBA structure by UNAs at position T7 is rather due to increased flexibility of ribose moiety or the presence of additional 2′OH group, and that the type of heterocyclic base at this position is not crucial for structure stabilization. Analysis of the published NMR and X-ray structures of unmodified TBA supports this observation, because thymidine residue at position T7 is flipped out toward solution.36, 37, 38 If the residue does not interact with the rest of the G-quadruplex structure, the high tolerance as observed at this position toward changes of the heterocyclic part of the residue, i.e., different size of aromatic surface and various exocyclic groups, is not surprising.
Table 1.
Thermodynamic Parameters of G-Quadruplex Formation of TBA Variants Modified with UNA-A (AU), UNA-C (CU), UNA-G (GU), and UNA-U (UU)
| Position of Modification | Sequence (5ʹ-3ʹ) | Average of Curve Fits |
|||||
|---|---|---|---|---|---|---|---|
| −ΔH° (kcal/mol) | −ΔS° (eu) | ΔG°37 (kcal/mol) | TM (°C) | ΔΔG°37 (kcal/mol) | ΔTM (°C) | ||
| GGTTGGTGTGGTTGG | 41.2 ± 0.9 | 127.2 ± 2.7 | −1.74 ± 0.02 | 50.7 | 0 | 0 | |
| T3 | GGAUTGGTGTGGTTGG | 39.5 ± 0.8 | 122.7 ± 2.4 | −1.43 ± 0.04 | 48.7 | 0.31 | −2.0 |
| T3 | GGCUTGGTGTGGTTGG | 40.3 ± 0.4 | 124.3 ± 1.1 | −1.74 ± 0.01 | 51.0 | 0 | 0.3 |
| T3 | GGGUTGGTGTGGTTGG | 40.5 ± 0.9 | 125.1 ± 2.9 | −1.66 ± 0.05 | 50.2 | 0.08 | −0.5 |
| T4 | GGTAUGGTGTGGTTGG | 33.5 ± 0.3 | 107.4 ± 0.9 | −0.21 ± 0.02 | 39.0 | 1.53 | −11.7 |
| T4 | GGTCUGGTGTGGTTGG | 34.7 ± 1.0 | 110.9 ± 3.2 | −0.29 ± 0.04 | 39.6 | 1.45 | −11.1 |
| T7 | GGTTGGAUGTGGTTGG | 43.6 ± 1.0 | 133.9 ± 3.1 | −2.11 ± 0.05 | 52.8 | −0.37 | 2.1 |
| T7 | GGTTGGCUGTGGTTGG | 44.4 ± 1.4 | 135.7 ± 4.3 | −2.33 ± 0.06 | 54.2 | −0.59 | 3.5 |
| T7 | GGTTGGGUGTGGTTGG | 44.1 ± 1.2 | 134.8 ± 3.6 | −2.27 ± 0.10 | 53.9 | −0.53 | 3.2 |
| G8 | GGTTGGTAUTGGTTGG | 34.9 ± 1.8 | 112.7 ± 5.7 | 0.00 ± 0.01 | 37.0 | 1.74 | −13.7 |
| G8 | GGTTGGTCUTGGTTGG | 35.2 ± 1.3 | 114.4 ± 4.2 | 0.29 ± 0.03 | 34.5 | 2.03 | 16.2 |
| G8 | GGTTGGTUUTGGTTGG | 36.3 ± 0.5 | 117.0 ± 1.6 | 0.02 ± 0.02 | 36.8 | 1.76 | −13.9 |
| T9 | GGTTGGTGAUGGTTGG | 43.4 ± 1.1 | 136.1 ± 3.4 | −1.21 ± 0.05 | 45.9 | 0.53 | −4.8 |
| T9 | GGTTGGTGCUGGTTGG | 40.6 ± 2.8 | 129.5 ± 9.0 | −0.44 ± 0.07 | 40.4 | 1.30 | −10.3 |
| T9 | GGTTGGTGGUGGTTGG | 47.3 ± 1.7 | 146.1 ± 5.1 | −2.03 ± 0.15 | 50.9 | −0.29 | 0.2 |
| T12 | GGTTGGTGTGGAUTGG | 37.8 ± 0.7 | 117.3 ± 2.2 | −1.37 ± 0.02 | 48.7 | 0.37 | −2.0 |
| T12 | GGTTGGTGTGGCUTGG | 39.6 ± 0.6 | 122.3 ± 1.8 | −1.63 ± 0.03 | 50.3 | 0.11 | −0.4 |
| T12 | GGTTGGTGTGGGUTGG | 39.9 ± 1.3 | 123.3 ± 4.0 | −1.64 ± 0.08 | 50.3 | 0.1 | −0.4 |
| T13 | GGTTGGTGTGGTAUGG | 34.8 ± 0.2 | 111.7 ± 0.7 | −0.19 ± 0.02 | 38.7 | 1.55 | −12.0 |
| T13 | GGTTGGTGTGGTCUGG | 34.3 ± 1.0 | 109.9 ± 3.0 | −0.16 ± 0.03 | 38.4 | 1.58 | −12.3 |
| T13 | GGTTGGTGTGGTGUGG | 35.3 ± 0.7 | 113.4 ± 2.1 | −0.16 ± 0.05 | 38.4 | 1.58 | −12.3 |
Buffer: 100 mM KCl, 20 mM sodium cacodylate, 0.5 mM EDTA(Na)2 (pH 7.0).
On the contrary, significant dependence between TBA thermodynamic stability and the type of heterocyclic base is observed at position T9 (Table 1). The presence of UNA-G caused increase of thermodynamic stability by 0.29 kcal/mol (ΔΔG°37 = −0.29 kcal/mol), whereas introduction of UNA-A or UNA-C induced destabilization by 0.53 and 1.30 kcal/mol (ΔΔG°37 = +0.53 and +1.30 kcal/mol), respectively. Similarly, also UNA-U at position T9 decreased G-quadruplex thermodynamic stability (increased ΔG°37) by 1.08 kcal/mol.22 These data indicate that UNA pyrimidines placed at position T9 are more unfavorable for G-quadruplex formation than UNA purines. According to TBA structure, thymidine residue at position T9 stack over the G-quartet, and apparently the presence of different heterocyclic bases at this position may interrupt this type of interaction.36, 37 Additionally, a more flexible ribose moiety might hinder optimal orientation for the interfaces between the G-tetrad and the aromatic surfaces of the nucleobase. Nevertheless, the increase of TBA thermodynamic stability by UNA-G at position T9 might be due to some additional, favorable interactions between the guanine moiety and the G-quartet.
The highest destabilization was observed for TBA variants modified with UNAs at position G8 (Table 1). The replacement of dG at this position by UNA-A, UNA-C, or UNA-U induced a decrease in thermodynamic stability of the resulting G-quadruplexes by 1.74, 2.03, and 1.76 kcal/mol, respectively. The effects are definitely more unfavorable relative to introduction of UNA-G at position G8 (ΔΔG°37 = 1.08 kcal/mol).22 The guanosine residue at position G8 forms hydrogen bonds with the G-tetrad within TBA.39 Therefore, optimal orientation of the residue to interact with the G-quartet is likely hindered by the increased flexibility of UNA, and as a consequence destabilization is observed. Moreover, higher destabilization induced by UNA-A, UNA-C, or UNA-U in reference to UNA-G is probably due to the lack or spatial shift of exocyclic groups that would be able to form stable, effective hydrogen bonds with the G-tetrad.
Introduction of UNA-A or UNA-C at position T4 and UNA-A, UNA-C, or UNA-G at position T13 decreased the thermodynamic stability of TBA by approximately 1.5 kcal/mol (Table 1). Both thymidine residues in unmodified TBA are involved in favorable hydrogen bonding and stacking interactions with the G-quadruplex core, and thus increase of sugar flexibility and change to other types of nucleobase cause clear destabilization.18, 40 The results stay in accordance with previously published data regarding the substitution of thymidine by UNA-U, which destabilized the TBA structure by 0.8 and 1.4 kcal/mol at positions T4 and T13, respectively.22
Minor destabilization of the G-quadruplex structure was observed when UNAs were placed at position T3 or T12 (Table 1). A UNA-A residue decreased the thermodynamic stability by 0.31–0.37 kcal/mol, whereas UNA-C and UNA-G caused only negligible destabilization up to 0.10 kcal/mol. High tolerance to chemical modification of those two positions is probably due to the lack of interactions between T3 and T12 residues with the rest of the TBA molecule.
Thermodynamic Analysis of TBA Variants Modified with UNA-4-Thiouracil and 4-Thiouridine
Thermodynamic analysis showed that the most energetically favorable was introduction of 4-thiouridine at position T12 (ΔΔG°37 = −0.53 kcal/mol; Table 2). On the contrary, the presence of UNA-4-thiouracil at the same position did not influence the thermodynamic stability of TBA. Similarly, also the substitution of thymidine by UNA-4-thiouracil at position T3 was neutral for thermodynamic stability of TBA, but the presence of the RNA counterpart caused stabilization by 0.31 kcal/mol in reference to unmodified TBA. Previously reported data demonstrated that the presence of UNA-U at position T3 or T12 increased the thermodynamic stability of TBA by 0.23 and 0.15 kcal/mol, respectively.22 Less favorable influence of UNA-s4U in reference to UNA-U is therefore presumably due to the presence of the chemically modified nucleobase.
Table 2.
Thermodynamic Parameters of G-Quadruplex Formation of TBA Variants Modified with RNA-s4U (s4UR) and UNA-s4U (s4UU)
| Position of Modification | Sequence (5ʹ-3ʹ) | Average of Curve Fits |
|||||
|---|---|---|---|---|---|---|---|
| −ΔH° (kcal/mol) | −ΔS° (eu) | ΔG°37 (kcal/mol) | TM (°C) | ΔΔG°37 (kcal/mol) | ΔTM (°C) | ||
| GGTTGGTGTGGTTGG | 41.2 ± 0.9 | 127.2 ± 2.7 | −1.74 ± 0.02 | 50.7 | 0 | 0 | |
| T3 | GGs4URTGGTGTGGTTGG | 41.7 ± 2.6 | 127.8 ± 7.8 | −2.05 ± 0.16 | 52.7 | −0.31 | 2.0 |
| T3 | GGs4UUTGGTGTGGTTGG | 40.3 ± 1.1 | 124.3 ± 3.5 | −1.74 ± 0.04 | 51.0 | 0 | 0.3 |
| T4 | GGTs4URGGTGTGGTTGG | 40.4 ± 3.3 | 127.8 ± 7.5 | −0.74 ± 0.05 | 42.8 | 1.00 | −7.9 |
| T4 | GGTs4UUGGTGTGGTTGG | 45.6 ± 8.6 | 144.4 ± 27.7 | −0.80 ± 0.10 | 42.5 | 0.94 | −8.2 |
| T7 | GGTTGGs4URGTGGTTGG | 44.0 ± 1.1 | 134.8 ± 3.4 | −2.22 ± 0.05 | 53.5 | −0.48 | 2.8 |
| T7 | GGTTGGs4UUGTGGTTGG | 40.2 ± 0.6 | 122.8 ± 1.9 | −2.08 ± 0.03 | 53.9 | −0.34 | 3.2 |
| G8 | GGTTGGTs4URTGGTTGG | 39.6 ± 2.3 | 123.5 ± 7.1 | −1.27 ± 0.07 | 47.3 | 0.47 | −3.4 |
| G8 | GGTTGGTs4UUTGGTTGG | 37.3 ± 0.7 | 120.2 ± 2.1 | −0.04 ± 0.02 | 37.3 | 1.70 | −13.4 |
| T9 | GGTTGGTGs4URGGTTGG | 43.2 ± 3.2 | 132.5 ± 10.0 | −2.16 ± 0.09 | 53.3 | −0.42 | 2.6 |
| T9 | GGTTGGTGs4UUGGTTGG | 41.6 ± 2.9 | 131.6 ± 9.1 | −0.79 ± 0.08 | 43.0 | 0.95 | −7.7 |
| T12 | GGTTGGTGTGGs4URTGG | 44.2 ± 1.3 | 135.1 ± 4.2 | −2.27 ± 0.05 | 53.8 | −0.53 | 3.1 |
| T12 | GGTTGGTGTGGs4UUTGG | 40.6 ± 0.6 | 125.2 ± 2.0 | −1.74 ± 0.03 | 50.9 | 0 | 0.2 |
| T13 | GGTTGGTGTGGTs4URGG | 35.9 ± 0.8 | 114.0 ± 2.7 | −0.56 ± 0.04 | 41.9 | 1.18 | −9.5 |
| T13 | GGTTGGTGTGGTs4UUGG | 37.3 ± 1.1 | 118.2 ± 3.5 | −0.66 ± 0.02 | 42.6 | 1.08 | −8.1 |
| T3, T7, T9, T12 | GGs4URTGGs4URGs4URGGs4URTGG | 46.6 ± 1.2 | 139.5 ± 3.5 | −3.30 ± 0.08 | 60.6 | −1.56 | 9.9 |
| T3, T7, T9, T12 | GGs4UUTGGs4UUGs4UUGGs4UUTGG | 37.8 ± 2.8 | 118.0 ± 8.9 | −1.23 ± 0.08 | 47.4 | 0.51 | −3.3 |
| T3, T7, T9, T13 | GGs4URTGGs4URGs4URGGTs4URGG | 37.9 ± 1.1 | 117.6 ± 3.4 | −1.39 ± 0.05 | 48.8 | 0.35 | −1.9 |
| T3, T7, T9, T13 | GGs4UUTGGs4UUGs4UUGGTs4UUGG | 26.2 ± 3.6 | 84.0 ± 11.5 | −0.16 ± 0.08 | 38.9 | 1.58 | −11.8 |
Buffer: 100 mM KCl, 20 mM sodium cacodylate, 0.5 mM EDTA(Na)2 (pH 7.0).
The presence of 4-thiouridine at position T7 increases the thermodynamic stability of TBA by 0.48 kcal/mol, whereas UNA-4-thiouracil stabilizes by 0.34 kcal/mol (Table 2). The only minor difference observed for the two modifications at position T7 suggests that the flexibility of the ribose moiety at this position is not governing the thermodynamic stability. Again, this is probably due to the structural orientation of nucleobase at position T7, which is headed toward the solution without interacting with the rest of the G-quadruplex molecule.38 Interestingly, 4-thiouridine introduced at position T9 likewise stabilizes TBA (ΔΔG°37 = −0.42), but the effect of UNA-4-thiouracil at position T9 is opposite and substantial destabilization is observed (ΔΔG°37 = 0.95 kcal/mol). The difference of ΔΔG°37 between UNA-s4U and RNA-s4U placed at position T9 might be because of increased flexibility of UNA, which hinders optimal orientation of the nucleobase for stacking interactions with the G-tetrad. A similar effect was observed previously for UNA-U at position T9 (ΔΔG°37 = 1.08 kcal/mol).22
Introduction of 4-thiouridine or UNA-4-thiouracil at position T4 or T13 decreased the thermodynamic stability of TBA. The presence of the above modifications caused destabilization by 1.00 and 0.94 kcal/mol for RNA-s4U and UNA-s4U at position T4, and 1.18 and 1.08 kcal/mol for RNA-s4U and UNA-s4U at position T13, respectively (Table 2). Comparable thermodynamic effects observed for UNA-s4U and RNA-s4U at the above positions suggest that the change of the thermodynamic stability of TBA strongly depends on the type of nucleobase (less electronegative sulfur atom in reference to oxygen), not the type of sugar moiety. The observed decrease of thermodynamic stability is probably due to the disturbance of stacking and/or hydrogen bonding between the nucleobase at position T4 or T13 and the remaining part of the G-quadruplex.
The most unfavorable change of the thermodynamic stability of TBA was induced by the presence of UNA-4-thiouracil at position G8 (ΔΔG°37 = 1.70 kcal/mol; Table 2). Previously discussed data showed that UNA-U at the same position caused similar destabilization (ΔΔG°37 = 1.76 kcal/mol). This suggests that decrease of TBA thermodynamic stability observed for UNA-modified variants at position G8 is rather an effect of substitution of purine by pyrimidine moiety and that further modification of base (O→S) does not contribute additionally to the destabilization. On the contrary, the presence of 4-thiouridine at position G8 decreases thermodynamic stability of TBA by only 0.47 kcal/mol. Thus, we can conclude that not only substitution of purine by pyrimidine, but also sugar flexibility at position G8 is pivotal for the thermodynamic stability of TBA.
As expected, the thermodynamic effects observed for single modifications within TBA were additive, i.e., the theoretical ΔG°37 values of multi-modified TBA variants were approximately the sum of the experimental ΔG°37 value of unmodified TBA and the thermodynamic effects (ΔΔG°37) caused by single modification at particular positions. The additivity for the majority of studied TBA variants was in the range of 83% to 94% (data not shown). Simultaneous introduction of four 4-thiouridines at positions T3, T7, T9, and T12 into TBA caused an increase of TBA thermodynamic stability by 1.56 kcal/mol (Table 2). On the contrary, the presence of UNA counterparts at the same positions caused destabilization of the G-quadruplex structure by 0.51 kcal/mol. The presence of 4-thiouridine or UNA-4-thiouracil at positions T3, T7, T9, and T13 always caused destabilization (ΔΔG°37 = 0.35 and 1.58 kcal/mol for RNA-s4U and UNA-s4U, respectively).
In general, the thermodynamic analysis discussed above showed that the presence of 4-thiouridine is usually more favorable for thermodynamic stability of TBA than UNA-4-thiouracil. Nevertheless, the majority of modified TBA variants described in this paper possess negative ΔG°37 value, indicating that at physiological temperature (37°C) the predominant species would be a folded G-quadruplex structure. Accordingly, analysis of Tm (melting temperature) versus concentration independence indicates that the modified TBA variants form intramolecular G-quadruplexes. Surprisingly, one TBA molecule modified with UNA-G at position T4 showed intermolecular G-quadruplex folding (Supplemental Information; Table S1). Nevertheless, at this moment it is difficult to explain the exact nature and origin of folding molecularity change of this particular TBA variant.
Thrombin Time Assay
The influence of single or multiple introduction into TBA of 4-thiouridine in RNA and UNA series or canonical UNAs on the anticoagulant properties was evaluated by the thrombin time assay. It is a standard method that allows determination of the time needed to form fibrin clot after addition of exogenous thrombin to blood plasma. The anticoagulant effect (AE) used herein to compare the anticoagulant properties of TBA variants was calculated by subtraction of the clotting time for plasma with exogenous thrombin and oligonucleotides from the clotting time for plasma with exogenous thrombin only.
It was previously reported that the presence of a single UNA residue at certain positions of TBA has a beneficial influence on its anticoagulant properties.22, 23 The data obtained herein revealed that the introduction of UNA residues has a different influence on the anticoagulant activity of TBA depending on their exact positioning. Modification of position T7 was the most beneficial for maintaining TBA anticoagulant activity. TBA variant modified with UNA-C at T7 possesses the most favorable, comparable with TBA, anticoagulant properties (AE = 16.3 s; Table 3). On the contrary, the introduction of UNA-A or UNA-G at this position causes a decrease of anticoagulant activity (AE was 15.3 and 10.2 s, respectively; Table 3). According to recent literature, the thymidine residue at T7 position is not involved in thrombin binding or aptamer stabilizing interactions, reflecting the high tolerance at this position toward substitution with UNA residues.38 Introduction of UNA into the aptamer loops caused distinct decrease in TBA anticoagulant properties. Thus, the TBA variants modified with UNA residues at positions T3, G8, T9, or T12 display significantly reduced ability to inhibit thrombin activity (the AE was in the range between 3.2 and 8.0 s; Table 3). The lowest value of anticoagulant effect was observed for the TBA variant modified with UNA-A, UNA-C, or UNA-G residue at position T4 or T13 (the AE was in range −0.1 to 0.5 s; Table 3). This can be explained by the thrombin-TBA interaction model that assumes that the two TT loops act as a pincer-like system that embraces the protruding region of thrombin exosite I.18 In this mode of action, T4 and T13 nucleotides play the crucial role in hydrogen bonds formation with thrombin amino acid residues. What is more, the T4 and T13 residues contribute to the TBA stability by π-stacking interactions with the guanine tetrads. Thus, the increase in flexibility of this TBA fragment via introduction of UNA residues results in disruption of intramolecular and intermolecular interactions. The results obtained are consistent with previously published data showing that the introduction of UNA-U at positions T4 or T13 significantly decreases the anticoagulant properties of TBA.22
Table 3.
The Anticoagulant Properties of TBA and TBA Variants Modified with Canonical UNAs (AU, CU, GU, UU), UNA-s4U (s4UU), and RNA-s4U (s4UR)
| Position of Modification | Sequence (5ʹ-3ʹ) | AE (s) | Position of Modification | Sequence (5ʹ-3ʹ) | AE (s) |
|---|---|---|---|---|---|
| GGTTGGTGTGGTTGG | 17.3 | T13 | GGTTGGTGTGGTCUGG | 0.6 | |
| T3 | GGAUTGGTGTGGTTGG | 8.0 | T13 | GGTTGGTGTGGTGUGG | 0.5 |
| T3 | GGCUTGGTGTGGTTGG | 4.7 | T3 | GGs4URTGGTGTGGTTGG | 4.0 |
| T3 | GGGUTGGTGTGGTTGG | 6.0 | T3 | GGs4UUTGGTGTGGTTGG | 8.7 |
| T4 | GGTAUGGTGTGGTTGG | 0.0 | T4 | GGTs4URGGTGTGGTTGG | 1.0 |
| T4 | GGTCUGGTGTGGTTGG | −0.1 | T4 | GGTs4UUGGTGTGGTTGG | 0.4 |
| T4 | GGTGUGGTGTGGTTGG | 0.2 | T7 | GGTTGGs4URGTGGTTGG | 4.8 |
| T7 | GGTTGGAUGTGGTTGG | 15.3 | T7 | GGTTGGs4UUGTGGTTGG | 15.5 |
| T7 | GGTTGGCUGTGGTTGG | 16.3 | G8 | GGTTGGTs4URTGGTTGG | 5.7 |
| T7 | GGTTGGGUGTGGTTGG | 10.2 | G8 | GGTTGGTs4UUTGGTTGG | 3.7 |
| G8 | GGTTGGTAUTGGTTGG | 4.2 | T9 | GGTTGGTGs4URGGTTGG | 1.3 |
| G8 | GGTTGGTCUTGGTTGG | 3.2 | T9 | GGTTGGTGs4UUGGTTGG | 6.9 |
| G8 | GGTTGGTUUTGGTTGG | 4.6 | T12 | GGTTGGTGTGGs4URTGG | 5.0 |
| T9 | GGTTGGTGAUGGTTGG | 5.5 | T12 | GGTTGGTGTGGs4UUTGG | 8.3 |
| T9 | GGTTGGTGCUGGTTGG | 6.9 | T13 | GGTTGGTGTGGTs4URGG | 2.4 |
| T9 | GGTTGGTGGUGGTTGG | 5.5 | T13 | GGTTGGTGTGGTs4UUGG | 0.1 |
| T12 | GGTTGGTGTGGAUTGG | 6.9 | T3, T7, T9, T12 | GGs4URTGGs4URGs4URGGs4URTGG | 0.8 |
| T12 | GGTTGGTGTGGCUTGG | 5.1 | T3, T7, T9, T12 | GGs4UUTGGs4UUGs4UUGGs4UUTGG | 0.5 |
| T12 | GGTTGGTGTGGGUTGG | 6.3 | T3, T7, T9, T13 | GGs4URTGGs4URGs4URGGTs4URGG | −0.3 |
| T13 | GGTTGGTGTGGTAUGG | 0.2 | T3, T7, T9, T13 | GGs4UUTGGs4UUGs4UUGGTs4UUGG | 0.6 |
In the past decade 4-thio-2′-deoxyuridine has received much attention because of its capability of improving oligonucleotide binding affinity to target protein,41, 42 and a beneficial influence of 4-thio-2′-deoxyuridine on TBA anticoagulant properties has been reported.20 In light of the above we decided to evaluate the influence of single or multiple introduction of 4-thiouridine and UNA-4-thiouracil on TBA anticoagulant properties. The TBA variant modified with UNA-s4U residue at position T7 displays the most favorable anticoagulant effect (the AE was 15.5 s; Table 3), almost being comparable with TBA. The introduction of UNA-s4U and RNA-s4U residues at other TBA positions reduced its ability to inhibit thrombin activity. Further analysis revealed that the anticoagulant effect caused by single substitution of UNA-s4U is more favorable than that induced by RNA-s4U. The effects for both groups of modifications were in the range between 6.9–15.5 s (UNA-s4U; Table 3) and 1.3–5.0 s (RNA-s4U; Table 3). The observed differences can be a result of increased flexibility of loop fragments caused by the presence of the UNA moieties. Interestingly, the above trend was opposite when RNA-s4U or UNA-s4U residues were introduced at positions T4, G8, or T13. The T4 and T13 residues are involved in the interactions with the thrombin molecule,18 and their proper arrangement to interact with the protein active site is therefore crucial. Presumably, the introduction of UNA-s4U at positions T4 or T13 causes local, unfavorable structure relaxation, which is manifested in the decrease of TBA anticoagulant properties. TBA variants possessing multiple substitution of RNA-s4U or UNA-s4U at positions T3, T7, T9, and T12 or T13 were characterized by having in general lowest AE values (Table 3).
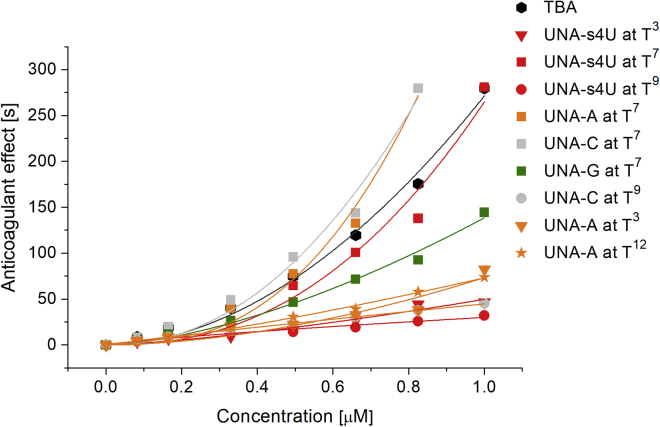
The data analysis of thrombin time assay conducted with different solutions of selected TBA variants (0.0825, 0.165, 0.330, 0.495, 0.660, 0.825, and 1 μM) revealed that the observed anticoagulant effect is strictly dependent on the concentration of oligomers. It was possible to improve inhibitory properties of TBA variants modified with UNA-A and UNA-C at position T7 via increasing their concentration (Figure 3; Supplemental Information; Tables S2 and S3). The thrombin time values achieved for those oligomers were 1.5-fold higher than for unmodified TBA (AE = 279.7 s for UNA-A and UNA-C at T7, and 175.5 s for unmodified TBA). Improvement of TBA anticoagulant properties was also observed when UNA-s4U and UNA-G were introduced at the position T7. However, in this case, the AE value was always below the AE value determined for TBA. The remaining TBA variants were characterized only by a slight improvement of anticoagulant properties provoked by the increase of oligonucleotide concentration.
Figure 3.
Concentration Dependence of Anticoagulant Effect of Unmodified TBA and TBA Variants
Analyzed TBA variants possessed: UNA-s4U at T3 (red triangles), UNA-s4U at T7 (red squares), UNA-s4U at T9 (red circles), UNA-A at T7 (orange squares), UNA-C at T7 (gray squares), UNA-G at T7 (green squares), UNA-C at T9 (gray circles), UNA-A at T3 (orange triangles), and UNA-A at T12 (orange stars).
The results indicate a particularly favorable influence of introducing nucleotide modification at position T7 on TBA anticoagulant properties. Furthermore, the observed anticoagulant effect of the analyzed aptamers is dependent on their concentration. It is possible to increase the inhibitory potential of these oligonucleotides by raising its concentration during injection to the plasma. All analyzed variants possess anticoagulant properties, but only two of them are characterized by improved AE value in reference to the unmodified TBA.
Amidolytic Assay
Three main functional domains can be distinguished in the thrombin structure: exosite I, exosite II, and active site.43, 44, 45 The exosite I has been indicated as playing a key role for binding to fibrinogen, protease-activated receptors,44 and activity modulators such as TBA.20 In order to verify whether the introduction of modified nucleotides into TBA has an influence on aptamer binding to this region, the amidolytic assay was applied. It is a simple method to determine the availability of the thrombin catalytic center by the addition of the chromogenic substrate H-D-Phe-Pip-Arg-pNA and analysis of the spectrophotometric change in the sample color intensity. H-D-Phe-Pip-Arg-pNA is a short peptide, able to bind to the thrombin active site even though other functional domains are occupied by enzyme substrates or its inhibitors. Based on the appearance of yellow color, as a result of peptide bond hydrolysis within the H-D-Phe-Pip-Arg-pNA substrate and generation of p-nitroaniline, it can be concluded that the thrombin catalytic center is available. The thrombin ability to hydrolyze short peptide substrate in the presence of tested TBA variants was determined by calculation of the percentage of its activity maintenance (At). The absorbance for the thrombin solution at 405 nm was assumed as 100% of At parameter value.
The highest level of thrombin activity was observed for TBA variants containing UNA-C, UNA-G, UNA-s4U at position T7, UNA-C at position T9, UNA-A at position T12, and unmodified TBA (Supplemental Information; Figure S1). The At value was above 100% for all mentioned TBA variants. A slight decrease in thrombin At levels (At < 100%) was observed when solutions contained TBA variants modified with UNA-A at positions T3 or T7 and UNA-s4U at positions T3 or T12. Statistical analysis by the p-Student’s test was performed and the p-parameter for all observed differences in thrombin At level in the presence of analyzed TBA variants determined (data not shown). In all of the above cases, the p-parameter value was greater than 0.05, and it was therefore concluded that the resulting variation in thrombin activity levels in the presence of the studied TBA variants was not statistically significant. Hence it can be stated that neither of the analyzed aptamers affect the ability of thrombin to hydrolyze peptide substrate H-D-Phe-Pip-Arg-pNA, and that the tested TBA variants exert their inhibitory effect by binding to exosite I of thrombin molecule.
Surface Plasmon Resonance
In this paper, the dissociation constant (KD value) of thrombin-TBA complex was determined using real-time surface plasmon resonance spectroscopy. Surface plasmon resonance is an optical technique that allows to determine the strength of interactions between two molecules: one suspended in solution and the other immobilized on a metallic surface, all based on the registration of the change in refractive index.46
It was previously reported that a favorable KD value is predominantly obtained when introduction of modified nucleotides has no influence on G-quadruplex structure and is localized in one of three TBA loops.12 Herein, surface plasmon resonance measurements were conducted for the group of TBA variants characterized by the most beneficial thermodynamic parameters and anticoagulant properties.
The most favorable KD values were obtained for TBA variants modified with canonical UNAs and UNA-s4U residues at position T7 (the KD values were 4.25, 4.39, 4.75, and 5.00 nM for UNA-G, UNA-C, UNA-A, and UNA-s4U, respectively; Table 4). The surface plasmon resonance data remain in a close correlation with thrombin time assay results, what evidences for pronounced tolerance of position T7 toward substitution with UNA residues. The similar observation was made earlier upon analysis of the influence of single introduction of UNA-U or UNA-G into TBA.22 Higher KD values were measured for TBA variants containing canonical UNAs and UNA-s4U residues at positions T3 or T9 (the KD values were 5.99, 6.67, and 8.77 nM for UNA-A at T3, UNA-s4U at T3, and UNA-C at T9, respectively; Table 4). The decrease in the strength of thrombin-TBA interactions was also observed when UNA-A and UNA-s4U residues were introduced into position T12 (the KD values were 10.91 and 15.99 nM for UNA-A and UNA-s4U, respectively; Table 4).
Table 4.
KD and Biostability (T1/2) of TBA and TBA Variants Modified with Canonical UNAs (AU, CU, GU) and UNA-s4U (s4UU)
| Position of Modification | Sequence (5ʹ-3ʹ) | KD (nM) | T1/2 (s) |
|---|---|---|---|
| GGTTGGTGTGGTTGG | 4.23 | 59.3 | |
| T3 | GGAUTGGTGTGGTTGG | 5.99 | ND |
| T7 | GGTTGGAUGTGGTTGG | 4.75 | 78.9 |
| T7 | GGTTGGCUGTGGTTGG | 4.39 | 89.2 |
| T7 | GGTTGGGUGTGGTTGG | 4.25 | 89.2 |
| T9 | GGTTGGTGCUGGTTGG | 8.77 | ND |
| T12 | GGTTGGTGTGGAUTGG | 10.91 | ND |
| T3 | GGs4UUTGGTGTGGTTGG | 6.67 | ND |
| T7 | GGTTGGs4UUGTGGTTGG | 5.00 | 66.9 |
| T12 | GGTTGGTGTGGs4UUTGG | 15.99 | ND |
ND, not determined.
In general, all TBA variants containing UNA-s4U residues were characterized by higher KD values in comparison with variants modified with canonical UNA residues. The results were partially reflected in the AE values measured at 0.165 μM concentration (the higher AE, the lower KD). A noticeable disagreement was observed for two TBA variants containing UNA-G or UNA-s4U residues at positions T7 and T12, respectively. In both cases, the KD values could not be directly translated into the thrombin test results. This reflects the complexity of blood coagulation processes and indicates that the knowledge of the KD value alone is not sufficient for the estimation of the anticoagulant properties of TBA variants.
Antiproliferative Assay
In recent years there has been growing interest in the antiproliferative potential of guanosine-rich oligonucleotides. These molecules usually fold into G-quadruplex structures and are capable of binding to specific proteins involved in the cell cycle that may result in cell growth inhibition.47, 48 The L. Mayol research group31 has shown that the introduction of dibenzyl linker at specific TBA positions causes a decrease in the anticoagulant properties of this aptamer while increasing its antiproliferative potential.
Based on these reports, experiments on the inhibition of cervical cancer cell growth (HeLa) by selected TBA variants using the MTT test were performed. This method allows for evaluation of cell viability based on the reduction of the water-soluble, yellow tetrazole salt (MTT) into the insoluble dark blue formazan. The amount of reduced MTT is directly proportional to the number of living cells on the experimental plates. The initial selection of candidates from the total pool of TBA variants was conducted via MTT screening test carried out for the aptamers at two concentrations (0.1 and 5.0 μM; Supplemental Information; Table S4). The results of these experiments indicated that introduction of canonical UNA residues into TBA at positions T3, T4, T7, G8, T9, T12, or T13 caused only a slight decrease in HeLa cell proliferation (the mean cell viability was 89.5% for UNA-A, 86.5% for UNA-C, and 85.8% for UNA-G in 5.0 μM). TBA variants modified with UNA-s4U possessed more favorable inhibitory properties with a reduction of HeLa cell viability to around 75% at the highest concentration tested. The most pronounced antiproliferative activity was observed for aptamers bearing multiple RNA-s4U substitution at positions T3, T7, T9, and T12 (or T13). Interestingly, these TBA variants containing four RNA-s4U residues were able to reduce HeLa cell viability to values below 50%.
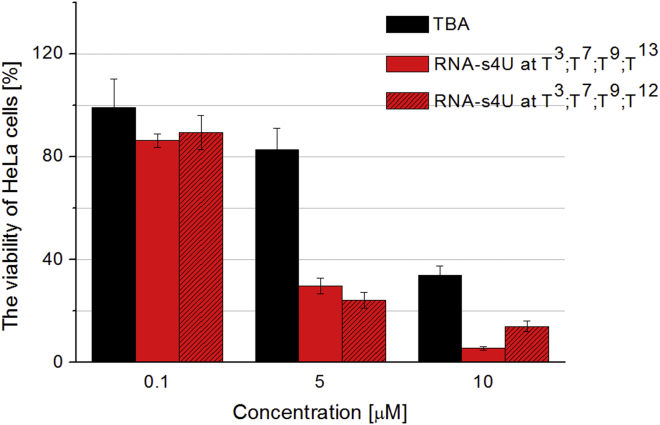
Based on the above data, the detailed analysis of antiproliferative properties was performed using 0.1, 5.0, and 10.0 μM concentration of unmodified TBA and the two potent TBA variants modified with RNA-s4U residues at positions T3, T7, T9, and T12 (or T13).
The data analysis revealed that the HeLa cell viability was reduced by 11% in the presence of 0.1 μM TBA variant modified with RNA-s4U residues at positions T3, T7, T9, and T12 (Figure 4; Supplemental Information; Table S5). The effect was gradually enhanced with an increase in oligomer concentration in the cell medium, reaching the 85% inhibition at 10 μM. A similar trend was observed for the aptamer modified with RNA-s4U residues at positions T3, T7, T9, and T13. Notably, the inhibitory effect for the RNA-s4U modified aptamer at 10 μM concentration was 95%. Interestingly, the unmodified TBA in 5 μM concentration was 3-fold less potent than the two analyzed TBA variants, and at 10.0 μM concentration TBA caused a reduction in HeLa cell viability by only 66%. These results highlight that switching the TBA activity from anticoagulant to antiproliferative is possible not only by replacement of a single loop nucleoside with a dibenzyl linker,31 but also via nucleobase and sugar moiety modification.
Figure 4.
The Antiproliferative Activity of Unmodified TBA (Black Bars) and TBA Variants Modified with RNA-s4U at T3, T7, T9, and T13 (Red Bars) or at T3, T7, T9, and T12 (Red Crosshatch Bars)
All data are presented as the mean ± SEM from three independent experiments (*p < 0.05, Student’s t test; n = 3).
Viability in Human Serum
The rapid elimination of aptamers from the bloodstream and their susceptibility on nucleases digestion are major factors limiting the biological effects of oligonucleotides in the human body.49 The biostability of oligonucleotides in human serum can be assessed by designation of their half-life (T1/2), defined as the time needed to reduce the amount of tested substance by half. It is known that the introduction of modified nucleotide residues into an aptamer can result in a favorable T1/2 value as shown for UNA-containing G-quadruplex structures.50
Herein, the T1/2 value for selected TBA variants characterized by the most beneficial anticoagulant or antiproliferative properties was determined via incubation in human serum at 37°C. The TBA variants modified with UNA-C or UNA-G at position T7 displayed the most favorable value of serum stability. In both cases, T1/2 was equal to 89.2 min in comparison with 59.3 min for unmodified TBA (Table 4). The T7 substitution by UNA-A also increased resistance of the aptamer to degradation; however, in this case, the T1/2 was equal to 78.9 min (Table 4). Only slight extension of biostability was observed for TBA variants containing UNA-s4U at position T7 (T1/2 was 66.9 min; Table 4). The achieved results are consistent with previous literature data, thus confirming that the introduction of UNA modifications can have a beneficial influence on in vitro stability of DNA G-quadruplex in serum.50
Interestingly, the determination of biostability of the two TBA variants modified with four RNA-s4U residues at positions T3, T7, T9, and T12 (or T13) was impossible due to the presence of additional products characterized by retarded electrophoretic mobility (Supplemental Information; Figure S2). Previously, it was described that the introduction of sulfur modification of nucleotide residues or the phosphodiester linkage of an oligonucleotide chain can result in increased affinity to proteins.20, 51, 52 Taking under consideration the above literature reports, the additional bands could originate from the formation of complexes between the TBA variants and serum proteins.
Conclusions
In this paper, a novel UNA derivative, i.e., UNA-4-thiouracil, has been synthesized and introduced into oligonucleotides for the first time. Furthermore, detailed analysis of the influence of single and multiple introduction of 4-thiouridine or UNA-4-thiouracil, as well as regular UNA-A, UNA-C, UNA-G, and UNA-U into TBA, on thermodynamic and biological properties was performed.
The most favorable thermodynamic parameters were achieved by the introduction of RNA-s4U or UNA-s4U at positions T3, T7, or T12 and UNA-A, UNA-C, and UNA-G residues at position T7 of the TBA aptamer. In addition, RNA-s4U substitution resulted in stronger stabilization of G-quadruplex structure in comparison with UNA-s4U substitution. Interestingly, the thermodynamic effects obtained by simultaneous introduction of the nucleobase-modified UNA or RNA nucleotides were additive. The structural studies confirmed that the TBA variants had preserved folding topology and retained the intramolecular, antiparallel G-quadruplex structure of TBA at 37°C. Thrombin time assay revealed that the oligonucleotides possess concentration-dependent anticoagulant properties. Notably, at higher concentration two variants modified with UNA-A or UNA-C at position T7 displayed more efficiently thrombin inhibition than unmodified TBA. The most favorable KD value and extended biostability in human serum were achieved for TBA variants possessing UNA-s4U or canonical UNA nucleotides at position T7. Taken together, these results suggest that T7 nucleotide substitution allows for maintenance of favorable thermodynamic stability and biological activity. Furthermore, the introduction of canonical UNA nucleotides or UNA-s4U into position T7 of TBA was beneficial not only for its thermodynamic parameters and anticoagulant properties, but also for improved TBA biostability in human serum. In general, it can be concluded that G-quadruplex structure is crucial for TBA anticoagulant properties. Nevertheless, the improvement of thermodynamic stability does not guarantee the increase in TBA inhibitory activity against thrombin. Interestingly, some TBA variants with poor anticoagulant properties were shown to be efficient antiproliferative agents. Herein, we demonstrate that two compounds containing multiple substitution of RNA-s4U at positions T3, T7, T9, and T12 (or T13) exhibited a potential to inhibit HeLa cell growth. In the future, our findings have the potential to serve as the basis for design and selection of compounds with the necessary improved biological properties for therapeutic applications.
Materials and Methods
Chemical Synthesis of 2′-O-Benzoyl-3′-O-(2-cyanoethoxy(diisopropylamino)phosphino)-5′-O-(4,4′-dimethoxytrityl)-2′,3′-seco-4-thiouridine
The synthesis of UNA-4-thiouridine derivative was performed according to the general published procedure for synthesis of UNA phosphoramidites53 with some modifications. Uridine was treated with TBDMSCl followed by NaIO4 and NaBH4. O5′-TBDMS-protected 2′,3′-secouridine was acetylated and then converted into 4-thiouridine derivative using Lawesson reagent. This acyclic derivative of 4-thiouridine was afterward treated with Et3N·3HF followed by DMTCl and Et3N/MeOH to give O5′-DMT-protected UNA-4-thiouracil. Selective benzoylation of O2′ was performed according to the previously published protocol.53 Treatment of the resulting compound with 2-cyanoethyl-N,N,N′,N′-tetraisopropylphosphordiamidite gave the final UNA-4-thiouracil building block suitable for oligonucleotide synthesis in an overall yield ca. 25%. Synthetic details are described in the Supplemental Information.
Chemical Synthesis of Oligonucleotides
All oligonucleotides were synthesized on an automated RNA/DNA synthesizer using standard phosphoramidite chemistry.54 The details of deprotection and purification procedures used for unmodified oligoribonucleotides or TBA variants containing nucleotide residues in the UNA series were described previously.53, 55 Deprotection of TBA variants modified with RNA-s4U or UNA-s4U was performed by treatment with methanol/aqueous ammonia solution (1:1 v/v) at room temperature for 48 hr. Purification was accomplished according to general procedures.56 The composition of all oligonucleotides was confirmed by MALDI-TOF mass spectrometry.
UV Melting Analysis
Oligonucleotides were dissolved in a buffer containing 100 mM potassium chloride, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA (pH 7.0). Single-stranded oligonucleotide concentrations were calculated based on the absorbance measured above 80°C,57 and the extinction coefficients were calculated using Oligo Calculator (http://www.ribotask.com). The samples were denatured at 95°C for 5 min and then cooled to room temperature overnight. The measurements were performed for nine different concentrations of the G-quadruplexes in the concentration range of 10−4 to 10−6 M. Absorbance versus temperature curves were obtained using the UV melting method at 295 nm in the temperature range of 4°C to 90°C with a heating rate of 0.2°C/min on a JASCO V-650 spectrophotometer equipped with a thermoprogrammer. Melting curves (Supplemental Information; Figure S3) were analyzed and the thermodynamic parameters determined by nonlinear curve fitting using the MeltWin 3.5 software. Melting temperatures calculated for a 10−4 M concentration of oligonucleotide are denoted by TM, and melting points for any other concentration of oligonucleotide are denoted by Tm.
Circular Dichroism Spectra
CD spectra were recorded on a JASCO J-815 spectropolarimeter. The oligonucleotides were dissolved in a buffer containing 100 mM potassium chloride, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA (pH 7.0) to achieve a sample concentration of 3.0 μM. All samples were denatured at 95°C for 5 min and then slowly cooled to room temperature overnight prior to data collection. The spectra were recorded in triplicate at 37°C in the 205 to 320 nm wavelength range. The buffer spectrum was subtracted from the sample spectra. The data analysis was performed using the Origin 8.0 software.
Thermal Difference Spectra
The TDS measurements were performed on a JASCO V-650 spectrophotometer equipped with a thermoprogrammer. The oligonucleotides were dissolved in a buffer containing 100 mM potassium chloride, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA (pH 7.0), to achieve a sample concentration of 0.1 μM. Absorbance spectra were recorded in triplicate at 4°C and 90°C in the 220 to 335 nm wavelength range. The scan speed was 1,000 nm/min with a 1-nm data interval. Thermal difference spectra were obtained by subtraction of the low-temperature absorbance spectra from the high-temperature absorbance spectra using the Origin 8.0 software. The differential spectra were normalized by dividing the data by its maximum value.35
Thrombin Time Assay
The thrombin time assay was performed using commercially available Dia-TT kit (DIAGON) and coagulometer K-3002 Optic. Each reaction mixture consisted of TBA variant (0.33 μM) dissolved in 100 μL of Dia-TT reagent. The reaction mixture was incubated at 37°C for 5 min and put into a sample well of coagulometer K-3002 Optic. Next, 100 μL of citrate plasma was added. Each TBA variant was analyzed using plasma samples derived from five healthy volunteers. The sample from each volunteer was measured twice, and the thrombin time values obtained from the double measurements did not exceed 6% (Supplemental Information; Table S6). The anticoagulant effect was obtained by subtraction of the time needed for plasma clotting in the presence of the aptamers from the time needed for clotting of plasma without aptamer. In addition, thrombin time for TBA variants with the best anticoagulant properties in seven different concentrations, 0.0825, 0.165, 0.330, 0.495, 0.660, 0.825, and 1 μM, was assigned using plasma samples derived from three healthy volunteers (Supplemental Information; Table S2). Each experiment was repeated twice and the result expressed as mean value.
Surface Plasmon Resonance
The value of KD for chosen TBA variants was measured using a BIAcore X100 system (GE Healthcare Life Science), human alpha-thrombin solution (Haematologic Technologies), commercially available Biotin CAPture KIT, and Sensor Chip SA (GE Healthcare Life Science). The 5′-biotinylated oligonucleotides were dissolved in a running buffer, 10 mM PBS (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, 0.05% Tween 20 [pH 7.4]), to achieve a sample concentration of 1 μM. The samples were denatured at 95°C for 6 min and then cooled to room temperature overnight. The oligonucleotides were immobilized on a chip surface via the streptavidin-biotin coupling method. The flow rate was set to 30 μL/min, and the unbound oligonucleotide was removed by the treatment with 50 mM aqueous NaOH. Next, serially diluted thrombin solution in the concentration range of 12.5 to 200 nM along with a BSA (Sigma-Aldrich) and sample with only running buffer were injected. The measurements were performed at 25°C. After each run, the system was regenerated by treatment with 50 mM aqueous NaOH. Data analysis was performed using the BIAcore X100 Evaluation software and a 1:1 binding mode.
Amidolytic Assay
Amidolytic assay was used to evaluate the ability of human alpha-thrombin (Haematologic Technologies) to hydrolyze chromogenic substrate H-D-phenylalanyl-L-arginine-p-nitroaniline (H-D-Phe-Pip-Arg-pNA, trade name: S-2238; CHROMOGENIX) in the presence of chosen TBA variants. The reaction mixture (150 μL), consisting of the chosen TBA variants (1 μM), thrombin (0.4 U/mL) and reaction buffer (0.1 M Tris [pH 8.2], 150 NaCl, 5 mM KCl, 0.1% BSA), was incubated at 37°C for 5 min. Next, 50 μL of 2 mM chromogenic substrate H-D-Phe-Pip-Arg-pNA was added. The amount of released p-nitroaniline was measured at 405 nm using a microplate reader xMark (Bio-Rad). Data analysis was performed using Microsoft Excel 2013 software. Each experiment was repeated in triplicate and the result expressed as mean value.
Viability of Oligonucleotides in Human Serum
The amount of 1 × 106 cpm of 32P-labeled oligonucleotides was dissolved in 1× PBS buffer containing 100 mM potassium chloride. The samples were denatured at 90°C for 6 min and then cooled to room temperature overnight. After this time, 200 μL of human serum from human male AB plasma (Sigma-Aldrich) was added, and the samples were incubated at 37°C. Aliquots of 5 μL were removed after 0, 10, 20, 40, 60, 120, 180, and 1,440 min of incubation and were then mixed with 5 μL of a 70% deionized formamide solution containing 50 mM EDTA and cooled on dry ice to quench the reaction. The samples were loaded on a 12% denaturing polyacrylamide gel prepared in 1× TBE buffer. PAGE in denaturing condition was performed in 1× TBE buffer at 20 W for 3 hr at room temperature. The resultant gel was imaged and quantified by storage phosphor technology using a Fuji PhosphorImager and MultiGauge Analysis Software.
Antiproliferative Assay
The antiproliferative properties of TBA variants were evaluated via MTT assay. The oligonucleotide solutions in concentrations of 1, 50, and 100 μM were prepared in 1× PBS buffer with 100 mM KCl. The samples were denatured at 95°C for 6 min and then cooled to room temperature overnight. The experiments were conducted on cervical carcinoma cells (HeLa cell line), which were seeded in a 96-well plates at a density of 250 cells/well in 100 μL of RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1× vitamin solution (Sigma-Aldrich). The plates were cultured at 37°C, 5% CO2, and a relative humidity of 100% for 24 hr. Next, the HeLa cells were exposed to varying concentrations of the chosen TBA variants (0.1, 5.0, 10.0 μM, final working volume: 200 μL) for 7 days. All appropriate control reactions were conducted. Afterward, the growth medium was removed and 1× MTT solution (Sigma-Aldrich) in RPMI 1640 media was added. The cells were incubated at 37°C in an atmosphere of 5% CO2 and a relative humidity of 100% for 2 hr. Next, the medium was removed and replaced with an aqueous mixture of 70% isopropanol and 40 mM HCl (100 μL/well) in order to dissolve the blue-purple crystals of formazan. The plates were shaken at 80 rpm for 30 min at room temperature. The amount of released formazan was measured at 595 nm using a microplate reader xMark (Bio-Rad). Data analysis was performed using Microsoft Excel 2013 software. Each experiment was repeated in triplicate and the result expressed as mean ± SD.
Author Contributions
W.K. performed all experiments except the initial MTT screening test and wrote the paper. J.L.-W. performed the initial MTT screening test and participated in publication editing. J.G. prepared the phosphoramidite of UNA-s4U. R.K. participated in oligonucleotide synthesis. J.W. supervised the surface plasmon resonance measurements and participated in publication editing. A.P. prepared the phosphoramidite of UNA-s4U, performed oligonucleotide synthesis, supervised all the experiments, and wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work has been supported by the National Science Center (UMO-2015/19/N/NZ5/02246 to W.K., UMO-2013/10/E/NZ1/00741 to A.P., and UMO-2016/21/D/NZ5/01906 to J.L.-W.) and the Polish Ministry of Science and Higher Education under the KNOW program.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, three figures, six tables, one scheme, and procedure of chemical synthesis and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.013.
Contributor Information
Weronika Kotkowiak, Email: kawecka@ibch.poznan.pl.
Anna Pasternak, Email: apa@ibch.poznan.pl.
Supplemental Information
References
- 1.Goodchild J. Therapeutic oligonucleotides. Methods Mol. Biol. 2011;764:1–15. doi: 10.1007/978-1-61779-188-8_1. [DOI] [PubMed] [Google Scholar]
- 2.Woodruff R.S., Sullenger B.A. Modulation of the coagulation cascade using aptamers. Arterioscler. Thromb. Vasc. Biol. 2015;35:2083–2091. doi: 10.1161/ATVBAHA.115.300131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santosh B., Yadava P.K. Nucleic acid aptamers: research tools in disease diagnostics and therapeutics. BioMed Res. Int. 2014;2014:540451. doi: 10.1155/2014/540451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamula C.L.A., Guthrie J.W., Zhang H., Li X.-F., Le X.C. Selection and analytical applications of aptamers. Trends Analyt. Chem. 2006;25:681–691. doi: 10.1016/j.trac.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agyei D., Acquah C., Tan K.X., Hii H.K., Rajendran S.R.C.K., Udenigwe C.C., Danquah M.K. Prospects in the use of aptamers for characterizing the structure and stability of bioactive proteins and peptides in food. Anal. Bioanal. Chem. 2018;410:297–306. doi: 10.1007/s00216-017-0599-9. [DOI] [PubMed] [Google Scholar]
- 6.Yang K.-A., Pei R., Stojanovic M.N. In vitro selection and amplification protocols for isolation of aptameric sensors for small molecules. Methods. 2016;106:58–65. doi: 10.1016/j.ymeth.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Cheng H., Wang J., Xu L., Chen H., Pei R. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II) Talanta. 2016;154:498–503. doi: 10.1016/j.talanta.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Darmostuk M., Rimpelova S., Gbelcova H., Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015;33:1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 10.Nagatoishi S., Tanaka Y., Tsumoto K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007;352:812–817. doi: 10.1016/j.bbrc.2006.11.088. [DOI] [PubMed] [Google Scholar]
- 11.Zhao D., Dong X., Jiang N., Zhang D., Liu C. Selective recognition of parallel and anti-parallel thrombin-binding aptamer G-quadruplexes by different fluorescent dyes. Nucleic Acids Res. 2014;42:11612–11621. doi: 10.1093/nar/gku833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avino A., Fabrega C., Tintore M., Eritja R. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012;18:2036–2047. doi: 10.2174/138161212799958387. [DOI] [PubMed] [Google Scholar]
- 13.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 15.Wang K.Y., Krawczyk S.H., Bischofberger N., Swaminathan S., Bolton P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 16.Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 17.Russo Krauss I., Merlino A., Giancola C., Randazzo A., Mazzarella L., Sica F. Thrombin-aptamer recognition: a revealed ambiguity. Nucleic Acids Res. 2011;39:7858–7867. doi: 10.1093/nar/gkr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pica A., Russo Krauss I., Merlino A., Nagatoishi S., Sugimoto N., Sica F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013;280:6581–6588. doi: 10.1111/febs.12561. [DOI] [PubMed] [Google Scholar]
- 19.Schwienhorst A. Direct thrombin inhibitors—a survey of recent developments. Cell. Mol. Life Sci. 2006;63:2773–2791. doi: 10.1007/s00018-006-6219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendelboum Raviv S., Horváth A., Aradi J., Bagoly Z., Fazakas F., Batta Z., Muszbek L., Hársfalvi J. 4-Thio-deoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J. Thromb. Haemost. 2008;6:1764–1771. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 21.Bonifacio L., Church F.C., Jarstfer M.B. Effect of locked-nucleic acid on a biologically active g-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasternak A., Hernandez F.J., Rasmussen L.M., Vester B., Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen T.B., Henriksen J.R., Rasmussen B.E., Rasmussen L.M., Andresen T.L., Wengel J., Pasternak A. Thermodynamic and biological evaluation of a thrombin binding aptamer modified with several unlocked nucleic acid (UNA) monomers and a 2′-C-piperazino-UNA monomer. Bioorg. Med. Chem. 2011;19:4739–4745. doi: 10.1016/j.bmc.2011.06.087. [DOI] [PubMed] [Google Scholar]
- 24.Nallagatla S.R., Heuberger B., Haque A., Switzer C. Combinatorial synthesis of thrombin-binding aptamers containing iso-guanine. J. Comb. Chem. 2009;11:364–369. doi: 10.1021/cc800178m. [DOI] [PubMed] [Google Scholar]
- 25.Saccà B., Lacroix L., Mergny J.L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaitseva M., Kaluzhny D., Shchyolkina A., Borisova O., Smirnov I., Pozmogova G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys. Chem. 2010;146:1–6. doi: 10.1016/j.bpc.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Martino L., Virno A., Randazzo A., Virgilio A., Esposito V., Giancola C., Bucci M., Cirino G., Mayol L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagano B., Martino L., Randazzo A., Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esposito V., Scuotto M., Capuozzo A., Santamaria R., Varra M., Mayol L., Virgilio A., Galeone A. A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance. Org. Biomol. Chem. 2014;12:8840–8843. doi: 10.1039/c4ob01475h. [DOI] [PubMed] [Google Scholar]
- 30.Smirnov I., Shafer R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 31.Scuotto M., Rivieccio E., Varone A., Corda D., Bucci M., Vellecco V., Cirino G., Virgilio A., Esposito V., Galeone A. Site specific replacements of a single loop nucleoside with a dibenzyl linker may switch the activity of TBA from anticoagulant to antiproliferative. Nucleic Acids Res. 2015;43:7702–7716. doi: 10.1093/nar/gkv789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin S.R., Schilstra M.J. Circular dichroism and its application to the study of biomolecules. Methods Cell Biol. 2008;84:263–293. doi: 10.1016/S0091-679X(07)84010-6. [DOI] [PubMed] [Google Scholar]
- 33.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang C.F., Shafer R.H. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mergny J.L., Li J., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly J.A., Feigon J., Yeates T.O. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 37.Coppola T., Varra M., Oliviero G., Galeone A., D’Isa G., Mayol L., Morelli E., Bucci M.R., Vellecco V., Cirino G., Borbone N. Synthesis, structural studies and biological properties of new TBA analogues containing an acyclic nucleotide. Bioorg. Med. Chem. 2008;16:8244–8253. doi: 10.1016/j.bmc.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 38.Borbone N., Bucci M., Oliviero G., Morelli E., Amato J., D’Atri V., D’Errico S., Vellecco V., Cirino G., Piccialli G. Investigating the role of T7 and T12 residues on the biological properties of thrombin-binding aptamer: enhancement of anticoagulant activity by a single nucleobase modification. J. Med. Chem. 2012;55:10716–10728. doi: 10.1021/jm301414f. [DOI] [PubMed] [Google Scholar]
- 39.Padmanabhan K., Padmanabhan K.P., Ferrara J.D., Sadler J.E., Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 40.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 41.Tarkanyi I., Horváth A., Szatmari I., Eizert H., Vámosi G., Damjanovich S., Ségal-Bendirdjian E., Aradi J. Inhibition of human telomerase by oligonucleotide chimeras, composed of an antisense moiety and a chemically modified homo-oligonucleotide. FEBS Lett. 2005;579:1411–1416. doi: 10.1016/j.febslet.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 42.Horváth A., Tokés S., Hartman T., Watson K., Turpin J.A., Buckheit R.W., Jr., Sebestyén Z., Szöllosi J., Benko I., Bardos T.J. Potent inhibition of HIV-1 entry by (s4dU)35. Virology. 2005;334:214–223. doi: 10.1016/j.virol.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 43.Huntington J.A. Structural insights into the life history of thrombin. In: Tanaka K., Davie E.W., Ikeda Y., Iwanaga S., Saito H., Sueishi K., editors. Recent Advances in Thrombosis and Hemostasis. Springer Japan; 2008. pp. 80–106. [Google Scholar]
- 44.Coppens M., Eikelboom J.W., Gustafsson D., Weitz J.I., Hirsh J. Translational success stories: development of direct thrombin inhibitors. Circ. Res. 2012;111:920–929. doi: 10.1161/CIRCRESAHA.112.264903. [DOI] [PubMed] [Google Scholar]
- 45.Crawley J.T., Zanardelli S., Chion C.K., Lane D.A. The central role of thrombin in hemostasis. J. Thromb. Haemost. 2007;5(Suppl 1):95–101. doi: 10.1111/j.1538-7836.2007.02500.x. [DOI] [PubMed] [Google Scholar]
- 46.Patching S.G. Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta. 2014;1838(1 Pt A):43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi E.W., Nayak L.V., Bates P.J. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38:1623–1635. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varizhuk A.M., Tsvetkov V.B., Tatarinova O.N., Kaluzhny D.N., Florentiev V.L., Timofeev E.N., Shchyolkina A.K., Borisova O.F., Smirnov I.P., Grokhovsky S.L. Synthesis, characterization and in vitro activity of thrombin-binding DNA aptamers with triazole internucleotide linkages. Eur. J. Med. Chem. 2013;67:90–97. doi: 10.1016/j.ejmech.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 50.Agarwal T., Kumar S., Maiti S. Unlocking G-quadruplex: effect of unlocked nucleic acid on G-quadruplex stability. Biochimie. 2011;93:1694–1700. doi: 10.1016/j.biochi.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Fennewald S., Luxon B.A., Aronson J., Herzog N.K., Gorenstein D.G. Aptamers containing thymidine 3′-O-phosphorodithioates: synthesis and binding to nuclear factor-kappaB. Bioorg. Med. Chem. Lett. 1999;9:3357–3362. doi: 10.1016/s0960-894x(99)00600-9. [DOI] [PubMed] [Google Scholar]
- 52.Zandarashvili L., Nguyen D., Anderson K.M., White M.A., Gorenstein D.G., Iwahara J. Entropic enhancement of protein-DNA affinity by oxygen-to-sulfur substitution in DNA phosphate. Biophys. J. 2015;109:1026–1037. doi: 10.1016/j.bpj.2015.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langkjaer N., Pasternak A., Wengel J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 54.McBride L.J., Caruthers M.H. An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 1983;24:245–248. [Google Scholar]
- 55.Kotkowiak W., Kotkowiak M., Kierzek R., Pasternak A. Unlocked nucleic acids: implications of increased conformational flexibility for RNA/DNA triplex formation. Biochem. J. 2014;464:203–211. doi: 10.1042/BJ20141023. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Gomollon S., Nicolas F.E. Purification of DNA oligos by denaturing polyacrylamide gel electrophoresis (PAGE) Methods Enzymol. 2013;529:65–83. doi: 10.1016/B978-0-12-418687-3.00006-9. [DOI] [PubMed] [Google Scholar]
- 57.Borer P.N. Optical properties of nucleic acids, absorption and circular dichroism spectra. In: Fasman G.D., editor. Handbook of Biochemistry and Molecular Biology: Nucleic Acids. Third Edition. CRC Press; 1975. pp. 589–595. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.