Abstract
Cardiac hypertrophy accompanied by maladaptive cardiac remodeling is the uppermost risk factor for the development of heart failure. Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have various biological functions, and their vital role in the regulation of cardiac hypertrophy still needs to be explored. In this study, we demonstrated that lncRNA Plscr4 was upregulated in hypertrophic mice hearts and in angiotensin II (Ang II)–treated cardiomyocytes. Next, we observed that overexpression of Plscr4 attenuated Ang II-induced cardiomyocyte hypertrophy. Conversely, the inhibition of Plscr4 gave rise to cardiomyocyte hypertrophy. Furthermore, overexpression of Plscr4 attenuated TAC (transverse aortic constriction)-induced cardiac hypertrophy. Finally, we demonstrated that Plscr4 acted as an endogenous sponge of miR-214 and forced expression of Plscr4 downregulated miR-214 expression to promote Mfn2 and attenuate hypertrophy. In contrast, knockdown of Plscr4 upregulated miR-214 to induce cardiomyocyte hypertrophy. Additionally, luciferase assay showed that miR-214 was the direct target of Plscr4, and overexpression of miR-214 counteracted the anti-hypertrophy effect of Plscr4. Collectively, these findings identify Plscr4 as a negative regulator of cardiac hypertrophy in vivo and in vitro due to its regulation of the miR-214-Mfn2 axis, suggesting that Plscr4 might act as a therapeutic target for the treatment of cardiac hypertrophy and heart failure.
Keywords: cardiac hypertrophy, lncRNA Plscr4, miR-214, Mfn2
Introduction
Pathological cardiac hypertrophy induced by prolonged hypertrophic stresses, such as hypertension, ischemia, myocarditis, and valvular heart disease, is the uppermost risk factor for the development of heart failure.1, 2 Initially, cardiac hypertrophy is beneficial because it maintains cardiac function. However, sustained hypertrophy leads to such effects as deposition of extracellular collagen, the loss of adrenergic responsivity, and changes in metabolism.3 Together, these changes lead to cell death and irreversible structural cardiac remodeling, ultimately resulting in heart failure and sudden death.3, 4, 5
The cardiomyocytes entirely depend on mitochondrial oxidative phosphorylation to generate ATP for cardiac contraction; thus, mitochondria are crucial organelles for proper heart function.6, 7 In pathological hypertrophy, numerous changes in mitochondrial metabolism are reported.8, 9 The dynamic processes of fusion and fission regulate mitochondrial morphology and function. Alterations in both the fusion and fission processes are associated with pathological cardiac hypertrophy.10, 11 Further studies that focus on looking for molecules that regulate mitochondrial fusion and fission could be of significant therapeutic value for cardiac hypertrophy. Mitofusin 2 (Mfn2), also named the hyperplasia-suppressive gene, is a vital protein located at the mitochondrial outer membrane.12 Mfn2 plays an important role in the maintenance of mitochondrial homeostasis. Recent reports reveal that Mfn2 is a critical negative regulator of cardiac hypertrophy by regulating mitochondrial fusion.10, 13 The expression level of Mfn2 is downregulated in various cardiac hypertrophy models,14, 15 and Mfn2-deficient mice represent modest cardiac hypertrophy accompanied with mild functional reduction.16
The mammalian genome is tremendously large, of which protein-coding genes account for only approximately 1.5%, and the larger portion of the genome remains either untranscribed or transcribed to non-coding RNAs (ncRNAs).17 The ncRNAs play a key role in maintaining the normal physiological functions of the cell.18, 19, 20 MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are two crucial types of ncRNAs. Numerous miRNAs have been demonstrated to influence cardiac hypertrophy, such as miR-1, miR-133, and miR-214.21, 22, 23 In addition, recent reports describe important regulatory roles for several lncRNAs in cardiovascular disease, especially in cardiac hypertrophy.24, 25, 26, 27, 28 However, how lncRNAs are involved in the regulation of cardiac hypertrophy has not been thoroughly elucidated to date.
In this study, we confirmed that overexpression of lncRNA Plscr4 alleviated pressure overload-induced cardiac hypertrophy in mice, and similar results were also demonstrated in cultured neonatal mice cardiomyocytes treated with Ang II. Furthermore, we identified that Plscr4 exerts the anti-hypertrophy effects by regulating the miR-214-Mfn2 pathway, which provides a novel strategy for the treatment of pathological cardiac hypertrophy and heart failure.
Results
Silencing of lncRNA Plscr4 Leads to a Hypertrophic Response in Cultured CMs
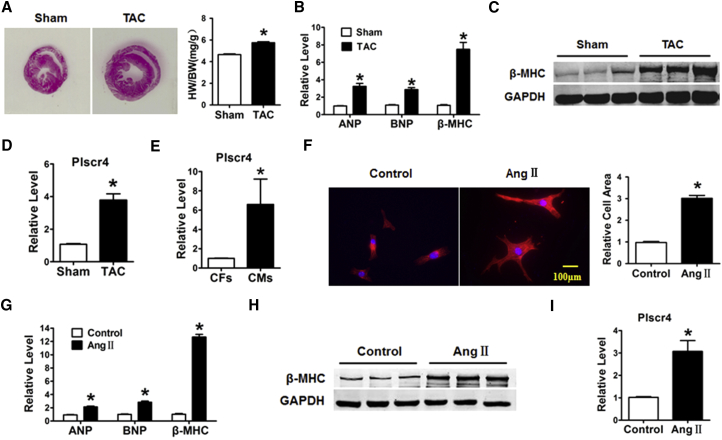
To identify the role of lncRNA Plscr4 in pathological cardiac hypertrophy, C57BL/6 mice underwent transverse aortic constriction (TAC) surgery to induce pathological cardiac hypertrophy or sham operation as a negative control. As shown in Figure 1A, the TAC mice showed a significant cardiac enlargement and an increased heart weight to body weight (HW/BW) ratio compared with the sham group. Moreover, the mRNA levels of the hypertrophic biomarkers atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and β-myosin heavy chain (β-MHC) were upregulated in the TAC mice (Figure 1B). In addition, the β-MHC protein level was also significantly upregulated in the TAC mice compared with the sham mice (Figure 1C). We further performed qRT-PCR to detect alterations in the Plscr4, and we observed that Plscr4 was significantly increased in the TAC mice compared to the sham group (Figure 1D).
Figure 1.
Upregulation of lncRNA Plscr4 in Hypertrophic Mice Hearts
(A) H&E staining for the assessment of gross cardiac enlargement, and the statistical results of the heart weight (HW)/body weight (BW) ratios of mice subjected to TAC or sham surgery for 4 weeks (n = 6 mice per group; *p < 0.05 versus sham group). (B) The relative mRNA level of the hypertrophic markers atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and β-myosin heavy chain (β-MHC) in the hearts of mice subjected to TAC or sham surgery for 4 weeks (n = 6 mice per group; *p < 0.05 versus sham group). (C) Representative western blot bands of β-MHC in the sham and TAC groups. The protein expression was quantified and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (n = 6 per group; *p < 0.05). (D) The relative mRNA level of Plscr4 in the mice 4 weeks after sham or TAC surgery (n = 6 mice per group; *p < 0.05 versus sham group). (E) The relative level of Plscr4 in the cardiac fibroblasts (CFs) and caridiomyocytes (CMs). (F) The immunofluorescence results of the CMs. Cells seeded in 24-well plates were cultured for 48 hr and then treated with PBS or Ang II for 48 hr. Both cell groups were then stained with antibodies directed against α-actinin and with DAPI for nuclear staining. Cell size was measured in 10 fields/well in both groups (n = 4 independent experiments; blue, nuclear; red, α-actinin; scale bars, 100 μm; n > 50 cells per experimental group; *p < 0.05 versus control). (G) The relative mRNA level of the hypertrophic markers ANP, BNP, and β-MHC in the CMs. Cells were treated with PBS or Ang II for 48 hr (n = 6; *p < 0.05 versus control). (H) Representative western blot bands of the β-MHC level in the CMs. Cells were treated with PBS or Ang II for 48 hr (n = 6; *p < 0.05). (I) The relative level of Plscr4 in the CMs after a 48-hr treatment with PBS or Ang II (n = 6; *p < 0.05 versus control). All of the data are presented as the mean ± SEM.
Furthermore, the cellular fractionation experiment revealed that the expression of lncRNA Plscr4 was specifically higher in the cardiomyocytes (CMs) than in the cardiac fibroblasts (CFs) (Figure 1E). We identified CMs by positive staining with α-actinin (Figure S1). These results indicated that Plscr4 was involved in pathological cardiac hypertrophy in vivo. To test the change of Plscr4 in a cellular model, CMs were incubated with 1 μmol/L Ang II for 48 hr or with PBS as a negative control. As shown in Figures 1F–1H, the cell surface area increased along with an increase in the mRNA levels of ANP, BNP, and β-MHC and the protein level of β-MHC. Consistent with the in vivo studies, the expression of Plscr4 was upregulated in the Ang II-treated CMs (Figure 1I).
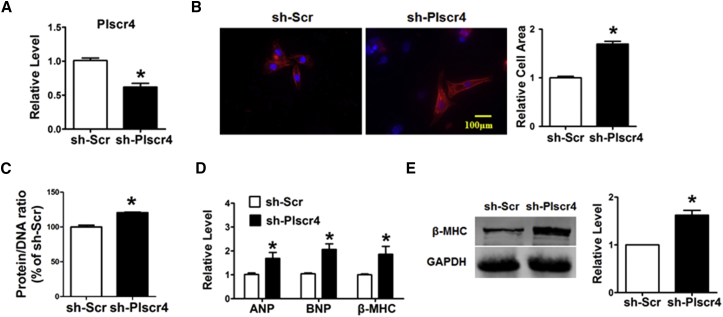
Next, we knocked down the expression of Plscr4 using short hairpin RNA (shRNA) to assess the potential effects of Plscr4 inhibition on cardiac hypertrophy (Figure 2A). As illustrated in Figures 2B–2E, inhibition of Plscr4 resulted in pronounced increases in the cell surface area as well as the protein/DNA ratio, which occurred along with the increased mRNA levels of ANP, BNP, and β-MHC and the protein level of β-MHC compared with the sh-Scramble treatment.
Figure 2.
Inhibition of Plscr4 Induces a Hypertrophic Response in CMs
(A) The successful inhibition of Plscr4 was verified. The CMs were transfected with sh-Scramble or sh-Plscr4 for 48 hr (n = 6; *p < 0.05 versus sh-Scr). (B) Microscopic images of the CMs. Cells seeded in 24-well plates were cultured for 48 hr and then transfected with sh-Scramble or sh-Plscr4 for 48 hr. Both cell groups were then stained with antibodies directed against α-actinin and with DAPI for nuclear staining. Cell size was measured in 10 fields/well in both groups (n = 4 independent experiments; blue, nuclear; red, α-actinin; scale bars, 100 μm; n > 50 cells per experimental group; *p < 0.05 versus sh-Scr). (C) Protein/DNA ratio of the CMs. Cells were transfected with sh-Scramble or sh-Plscr4 for 48 hr. Then cells were lysed with standard sample buffer, protein concentration was determined by the BCA method with BSA as a standard, and DNA concentration was detected by fluorescence assay. The ratio of protein to DNA was then calculated to estimate potential protein synthesis (n = 6; *p < 0.05 versus sh-Scr). (D) The relative mRNA levels of the hypertrophic markers ANP, BNP, and β-MHC in the CMs treated as in (A) (n = 6; *p < 0.05 versus sh-Scr). (E) Representative western blot bands of β-MHC in the CMs that were transfected with sh-Scramble or sh-Plscr4 for 48 hr (n = 6; *p < 0.05 versus sh-Scr). All of the data are presented as the mean ± SEM.
Forced Expression of Plscr4 Attenuates Cardiac Hypertrophy In Vitro and In Vivo
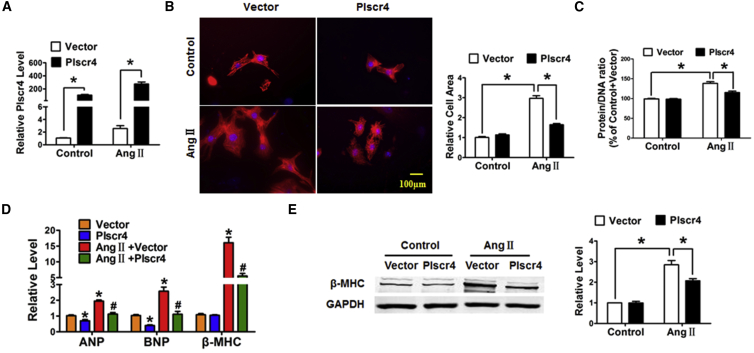
According to the above results, we wanted to determine whether overexpression of Plscr4 displays protective effects on cardiac hypertrophy, and we therefore transfected Plscr4 or a vector control into CMs treated with Ang II (1 μmol/L) for 48 hr. The transfection efficiency was verified by a significant increase in the Plscr4 level in the overexpression group compared with the vector control group in both the Ang II- and PBS-treated cells (Figure 3A). Notably, overexpression of Plscr4 did not exhibit marked abnormalities in CM area, protein/DNA ratio, and the mRNA and protein levels for the hypertrophic biomarker β-MHC, whereas mRNA levels of ANP and BNP revealed a slight decrease under basal conditions (Figures 3B–3E). However, the forced expression of Plscr4 significantly attenuated the increased cell surface area and protein/DNA ratio and inhibited the upregulated mRNA levels of ANP, BNP, β-MHC, and the protein level of β-MHC in the Ang II-treated CMs (Figures 3B–3E).
Figure 3.
Overexpression of lncRNA Plscr4 Attenuates the Hypertrophic Response in Ang II-Treated CMs
(A) The successful transfection of lncRNA-plscr4 was verified. CMs were transfected with lncRNA-plscr4 or the vector control and were subsequently treated with PBS or Ang II (1 μmol/L) for 48 hr (n = 6, *p < 0.05). (B) Microscopic images of the CMs. Cells were seeded in 24-well plates, cultured for 48 hr, and then transfected with lncRNA-plscr4 or the vector control; they were subsequently treated with PBS or Ang II (1 μmol/L) for 48 hr. Both cell groups were then stained with antibodies directed against α-actinin and with DAPI for nuclear staining. Cell size was measured in 10 fields/well in all groups (n = 3 independent experiments; blue, nuclear; red, α-actinin; scale bars, 100 μm; n > 50 cells per experimental group; *p < 0.05). (C) The protein/DNA ratio of the CMs. Cells were seeded in 12-well plates, cultured for 48 hr, and then transfected with lncRNA-plscr4 or the vector control; they were subsequently treated with PBS or Ang II (1 μmol/L) for 48 hr. Then cells were lysed with standard sample buffer, protein concentration was determined by the BCA method with BSA as a standard, and DNA concentration was detected by fluorescence assay. The ratio of protein to DNA was then calculated to estimate potential protein synthesis (n = 6; *p < 0.05). (D) The relative mRNA levels of the hypertrophic markers ANP, BNP, and β-MHC in the CMs as treated in (A) (n = 6; *p < 0.05 versus control, #p < 0.05 versus Ang II + vector). (E) Representative western blot bands of β-MHC in the CMs transfected with lncRNA-plscr4 or the vector control that were subsequently treated with PBS or Ang II (1 μmol/L) for 48 hr (n = 5; *p < 0.05). All of the data are presented as the mean ± SEM.
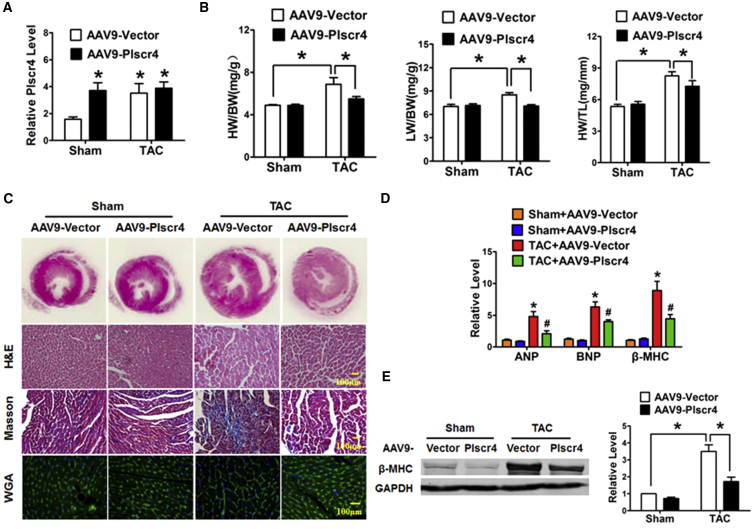
Next, we performed gain-of-function experiments in the mice to determine whether Plscr4 overexpression has a similar protective effect on pressure overload-induced cardiac hypertrophy. The mice were injected through the tail vein with AAV9 viral particles carrying Plscr4 or a vector for 3 weeks, and the mice were later subjected to TAC to induce cardiac hypertrophy. qRT-PCR assays were conducted to confirm overexpression of lncRNA Plscr4 (Figure 4A). We found that overexpression of Plscr4 in mice did not cause detectable changes in cardiac structure or function at baseline. However, after 4 weeks of TAC treatment, the hypertrophic response was blunted in the Plscr4-overexpressing mice, which was indicated by the lower HW/BW, lung weight/BW (LW/BW), and HW/tibia length (HW/TL) ratios and reduced cardiac left ventricular wall thickness in the lncRNA-plscr4 overexpression mice compared with their littermate control mice (Figure 4B; Figure S2). Moreover, an enlargement of the heart and the CMs was identified via H&E or wheat germ agglutinin (WGA), and the Plscr4-overexpressing mice exhibited a smaller heart size and fewer enlarged CMs compared with the vector mice after 4 weeks of TAC surgery. Consistently, the Masson staining exhibited less fibrosis (Figure 4C) and mRNA levels of ANP, BNP, and β-MHC and protein level of β-MHC in the Plscr4-overexpressing mice dramatically decreased when compared to the vector control (Figures 4D and 4E).
Figure 4.
Overexpression of Plscr4 Mitigates Pressure Overload-Induced Cardiac Hypertrophy
(A) The specific overexpression of lncRNA-plscr4 in the mouse heart was verified. The mice were injected through the tail vein with AAV9 viral particles carrying Plscr4 or a vector for 3 weeks, and then they were subjected to TAC or sham operation for 4 weeks (n = 6; *p < 0.05 versus sham). (B) The statistical results for the HW/BW, lung weight (LW)/HW, and HW/tibia length (TL) ratios in the sham or TAC mice infected with the AAV9-Vector or AAV9-Plscr4 (n = 6; *p < 0.05). (C) Representative histological results of the H&E staining, Masson staining to assess fibrosis, and wheat germ agglutinin (WGA) staining of the mouse heart tissues. The AAV9-Vector- and AAV9-Plscr4-overexpressing mice underwent 4 weeks of the TAC or sham operation. Then the mice hearts were collected and fixed in 4% paraformaldehyde for 24 hr followed by embedding in paraffin, and they were cut into 5-μm-thick cross-sectional slices. The slices were stained by H&E, Masson trichrome, and WGA. (D) The relative mRNA levels of the hypertrophic markers ANP, BNP, and β-MHC in the AAV9-Vector- and AAV9-Plscr4-overexpressing mice after 4 weeks of the TAC or sham operation (n = 6; *p < 0.05 versus sham + AAV9-Vector, #p < 0.05 versus TAC + AAV9-Vector). (E) Representative western blot bands of β-MHC in the AAV9-Vector- and AAV9-Plscr4-overexpressing mice after 4 weeks of the TAC or sham operation (n = 6; *p < 0.05).
Plscr4 Inhibits the Expression and Activity of miR-214
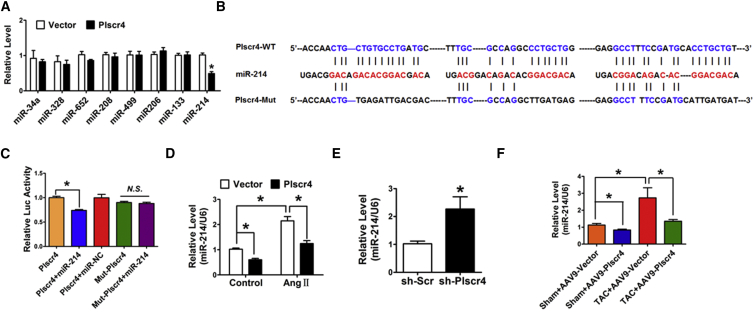
The lncRNAs are reported to act as sponges for miRNAs. To explore whether Plscr4 elicits its effect on cardiac hypertrophy through miRNAs, we tested whether one or more of the miRNAs is the downstream target of Plscr4 by using RegRNA 2.0 software, which is an integrated web-based system that has been developed for comprehensively identifying functional RNA motifs and target sites.29 A range of miRNAs have been predicted to be the potential targets of Plscr4. We selected miRNAs, including miR-214, miR-34a, and miR-328, that have been proven to be involved in the process of cardiac hypertrophy.30, 31, 32, 33 qRT-PCR was performed to detect the differently expressed miRNAs upon overexpression of Plscr4, and miR-214 was substantially reduced (Figure 5A), which is also proven to be an upregulated miRNA in hypertrophy in several miRNA array tests.34, 35 Then we compared the sequences of Plscr4 and miR-214 and noticed that Plscr4 contained three binding sites of miR-214 (Figure 5B). Furthermore, we found that the length of Plscr4 is 1,551 bp, which transcribed and spliced from Chromosome 9: 92,457,394-92,485,332 forward strand. An alignment of the mouse Plscr4 sequence to human expressed sequence tags (ESTs) revealed that the similar sequence is 76% (76% homology). It is conserved between mouse and human in the binding site of miR-214 (Table S1).
Figure 5.
The lncRNA Plscr4 Directly Binds to miR-214
(A) The relative levels of the miRNAs that interact with Plscr4 provided from the RegRNA database prediction tools (n = 6; *p < 0.05 versus vector). (B) The sequence alignment analysis revealed that Plscr4 contains three sites complementary to miR-214, the miR-214-binding site in the Plscr4 wild-type form (Plscr4-WT) and the mutated form (Plscr4-Mut). (C) HEK293 cells were co-transfected with Plscr4-WT or Plscr4-Mut and miR-214 or miR-NC. The luciferase activity was analyzed (n = 6; *p < 0.05). (D) The relative levels of miR-214 in the CMs transfected with Plscr4 or vector control, which were subsequently treated with PBS or Ang II (1 μmol/L) for 48 hr (n = 6; *p < 0.05). (E) The relative level of miR-214 in the CMs transfected with sh-Scramble or sh-Plscr4 (n = 6; *p < 0.05 versus sh-Scr). (F) The relative level of miR-214 in the AAV9-Vector- and the AAV9-Plscr4-overexpressing mice after 4 weeks of the TAC or sham operation (n = 6; *p < 0.05). All of the data are presented as the mean ± SEM.
Meanwhile, our study revealed that the expression of miR-214 was upregulated in the hypertrophic heart and in Ang II-treated CMs (Figure S3), which was in accordance with previous studies.30, 36 Therefore, we presume that there may be particular interaction between Plscr4 and miR-214. To investigate the molecular communications between Plscr4 and miR-214, we performed a luciferase assay using a luciferase construct lncRNA-plscr4 (Plscr4-wild-type [WT]) and a mutated form (Plscr4-Mut) (Figure 5B). The luciferase assay revealed that the transfection of miR-214 suppressed the luciferase activity of Plscr4, but it had less effect on the mutated Plscr4 (Figure 5C). Moreover, overexpression of Plscr4 reduced miR-214 levels both in the Ang II-treated and the normal CMs (Figure 5D). In contrast, the inhibition of Plscr4 induced the upregulation of miR-214 in CMs at baseline (Figure 5E). The inhibitory effect of Plscr4 on miR-214 was further validated in the mouse heart (Figure 5F). Taken together, these results show that Plscr4 may directly regulate miR-214 by these three putative binding sites.
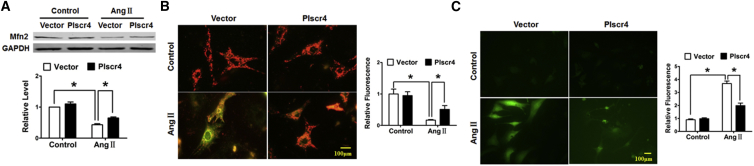
Recent findings suggest that Mfn2, which is located at the mitochondrial outer membrane and plays a critical role in the maintenance of mitochondrial dynamic homeostasis and the normal function of mammalian cells, is the molecular target of miR-214.37 Mitochondria are crucial organelles in CMs, the diminished metabolism of which is reported to be associated with CM hypertrophy.38 Thus, we wanted to explore whether Plscr4 influences mitochondrial homeostasis by regulating Mfn2. The protein level of Mfn2 was downregulated in the TAC mice and the Ang II-treated CMs (Figure S4). Furthermore, overexpression of Plscr4 rescued the decreased Mfn2 under Ang II treatment (Figure 6A). Finally, we examined the integrity and function of the mitochondria. Similar to the protein level of Mfn2, the JC-1 staining and the quantified reactive oxygen species (ROS) production showed that, following Ang II treatment, the mitochondria membrane suffered a tremendous depolarization, as represented by the green staining of the mitochondria, and an increased ROS production, which was reversed by Plscr4 overexpression (Figures 6B and 6C).
Figure 6.
The lncRNA Plscr4 Protected Mitochondrial Function in Response to Hypertrophic Stress
(A) Representative western blot bands of Mitofusin-2 (Mfn2) in the CMs transfected with Plscr4 or the vector control and that were subsequently treated with Ang II (1 μmol/L) for 48 hr (n = 6; *p < 0.05). (B) Representative images of the JC-1 fluorescence. The CMs were transfected with Plscr4 or the vector control and subsequently treated with Ang II (1 μmol/L) for 48 hr, then incubated with JC-1 staining solution. JC-1 exhibited a potential-dependent accumulation in the mitochondria; in healthy cells with a high potential, JC-1 forms complexes emitting a red fluorescence, and in unhealthy cells with a low mitochondrial membrane potential, JC-1 remains in a monomeric form, emitting green fluorescence. The state of the membrane potential was expressed by the ratio of red to green fluorescence (n > 50 cells per experimental group; *p < 0.05). (C) The ROS levels were determined in the CMs that were transfected with Plscr4 or the vector control, subsequently treated with Ang II (1 μmol/L) for 48 hr, and then incubated with the ROS-specific probe DCFH-DA. DCFH-DA could pass through the cell membrane and change into DCFH in the live cells. Intracellular ROS in live cells could interact with DCFH resulting in a fluorometric product DCF, which indicates the amount of ROS present. The cells were imaged with a fluorescence microscope (n > 50 cells per experimental group; *p < 0.05). All of the data are presented as the mean ± SEM.
miR-214 Mediates the Anti-hypertrophy Effects of Plscr4
To confirm whether miR-214 is necessary for Plscr4-regulated cardiac hypertrophy, we transfected Plscr4 or vector alone or in combination with miR-214 into CMs, and we subsequently treated the cells with Ang II for 48 hr. The successful transfection of miR-214 and Plscr4 was confirmed by qRT-PCR (Figure 7A; Figure S5). As depicted in Figures 7B–7E, co-transfection of miR-214 abated the protective effects of Plscr4 on Ang II-induced hypertrophic responses in CMs, which was exhibited by the increased cell size; protein/DNA ratio; and the upregulation of the hypertrophic markers ANP, BNP, and β-MHC. Similarly, co-transfection of miR-214 attenuated the inhibitory effect of Plscr4 on mitochondria membrane depolarization and on ROS production in the Ang II-treated CMs (Figures 7E–7G). However, overexpression of Plscr4 could not affect the expression level of Ezh2, one of the direct targets of miR-214 during cardiac hypertrophy,21, 30 both in vivo and in vitro (Figure S6). Taken together, our results showed that Plscr4 protects against cardiac hypertrophy through the miR-214-Mfn2 axis.
Figure 7.
Plscr4 Regulates Hypertrophy by Targeting miR-214
(A) The successful transfection of miR-214 was verified in the CMs transfected with Plscr4 or the vector alone or in combination with miR-214 or miR-NC followed by 48 hr of Ang II treatment (n = 6; *p < 0.05). (B) The immunofluorescence results of the cell surface area in the CMs transfected with Plscr4 or the vector alone or in combination with miR-214 or miR-NC followed by 48 hr of Ang II treatment. Cells were then stained with antibodies directed against α-actinin and with DAPI for nuclear staining. Cell size was measured in 10 fields/well in both groups (n = 4 independent experiments; blue, nuclear; red, α-actinin; n > 50 cells per experimental group, n = 6; *p < 0.05). (C) The protein/DNA ratio of the CMs. Cells were transfected with Plscr4 or the vector alone or in combination with miR-214 or miR-NC followed by 48 hr of Ang II treatment. Then cells were lysed with standard sample buffer, protein concentration was determined by the BCA method with BSA as a standard, and DNA concentration was detected by fluorescence assay. The ratio of protein to DNA was then calculated to estimate potential protein synthesis (n = 6; *p < 0.05). (D) The relative mRNA levels of the hypertrophic markers ANP, BNP, and β- MHC as treated in (A) (n = 6; *p < 0.05). (E) Representative western blot bands of β- MHC and Mfn2 in the CMs treated as in (A) (n = 6; *p < 0.05). (F) The representative images of the JC-1 fluorescence in the CMs that were transfected with Plscr4 or the vector alone or in combination with miR-214 or miR-NC followed by 48 hr of Ang II treatment, and then incubated with JC-1 staining solution. JC-1 exhibited a potential-dependent accumulation in the mitochondria; in healthy cells with a high potential, JC-1 forms complexes emitting a red fluorescence, and in unhealthy cells with a low mitochondrial membrane potential, JC-1 remains in a monomeric form, emitting green fluorescence. The state of the membrane potential was expressed by the ratio of red to green fluorescence (n > 50 cells per experimental group; *p < 0.05). (G) The ROS levels were determined in the CMs that were transfected with Plscr4 or the vector alone or in combination with miR-214 or miR-NC followed by 48 hr of Ang II treatment, and then incubated with the ROS-specific probe DCFH-DA. DCFH-DA could pass through the cell membrane and change into DCFH in the live cells. Intracellular ROS in live cells could interact with DCFH, resulting in a fluorometric product DCF, which indicates the amount of ROS present. The cells were imaged with a fluorescence microscope (n > 50 cells per experimental group; *p < 0.05).
Discussion
In this study, we demonstrated that lncRNA Plscr4 is an important inhibitor of cardiac hypertrophy. The lncRNA Plscr4-overexpressing mice exhibited a reduced hypertrophic growth in response to the pressure overload, which was accompanied by a preserved cardiac function and decreased cardiac fibrosis. Furthermore, we observed that lncRNA-plscr4 exerts its cardioprotective effect through the lncRNA Plscr4-miR-214-Mfn2 axis. Our study provides a new possible approach for understanding and restraining the pathogenesis of cardiac hypertrophy.
Increasing evidence is pointing toward lncRNAs as regulators of cardiac hypertrophy. Chaer (Cardiac Hypertrophy-Associated Epigenetic Regulator) can directly interact with polycomb repressor complex 2 (PRC2) to inhibit histone H3 lysine 27 methylation at the promoter regions of genes related to cardiac hypertrophy, which thus promotes cardiac hypertrophy.26 Chast (Cardiac Hypertrophy-Associated Transcript) is another pro-hypertrophy lncRNA, which is upregulated in hypertrophic hearts by negatively regulating Plekhm1 to restrain CM autophagy and induce hypertrophy.39 CHRF (Cardiac Hypertrophy-Related Factor) is upregulated in both the hypertrophic mouse heart and human HF samples. CHRF also plays a pro-hypertrophy role by downregulating the miR-489 expression level. miR-489 is an anti-hypertrophy miRNA whose target gene is Myd88, which upregulates the NF-κB (nuclear factor kappaB light-chain enhancer of activated B cells) pathway.28 Interestingly, our study demonstrated that TAC surgery dramatically enhanced Plscr4 expression compared with the sham group after 4 weeks, and we further demonstrated that Plscr4 plays an obvious anti-hypertrophy role. This finding is in accordance with the report of ADAMTS2 (A Disintegrin and Metalloproteinase With Thrombospondin Motifs 2) in cardiac hypertrophy.40 To date, our study first confirmed that, even if an lncRNA is upregulated in response to hypertrophic stress, it may play a protective role.
We demonstrated that Plscr4 exerts its effects on hypertrophy by repressing the pro-hypertrophy gene miRNA-214. Stressed hearts often exhibit alternations of several different molecules and signals concurrently. We speculate that, at early stages, the adaptive response is triggered and leads to the upregulation of Plscr4, which exerts an anti-hypertrophy effect to offset the stress-caused maladaptive changes in CMs. In addition, the pro-hypertrophy signals, such as miR-214, are upregulated. At the early stage, the adaptive activating Plscr4 is sufficiently strong to partially offset the pro-hypertrophy effect of miR-214, resulting in modest increases in hypertrophic growth. However, with long-term stress, a sustained increase of the pro-hypertrophy signal miR-214 beyond a certain threshold is sufficient to counteract the function of Plscr4 activation, leading to the development of cardiac hypertrophy (Figure 8). With the consideration of these, we injected AAV9-Plscr4 through the tail vein to generate mice with Plscr4 overexpression from the initial period of hypertrophy. We wanted to make sure that a continued high expression of Plscr4 existed from the beginning stage to the later period of hypertrophy and could attenuate hypertrophic growth. Finally, our results show that the cardiac-specific overexpression of Plscr4 attenuated hypertrophic growth induced by pressure overload compared with the empty vector group. This finding certified our above theory and might help us to look for a new effective cardiac hypertrophy therapy.
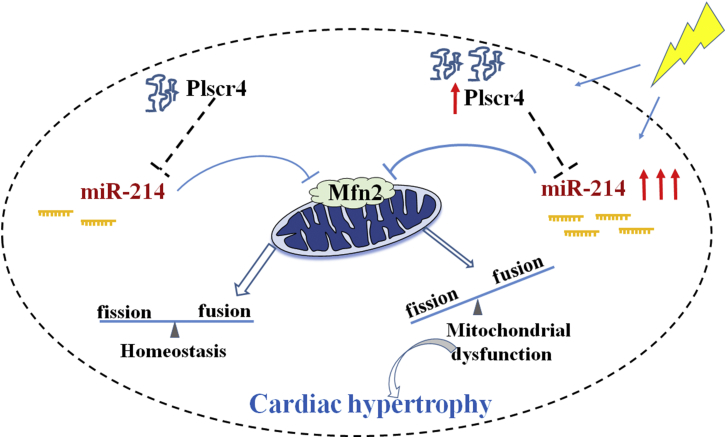
Figure 8.
Schematic Diagram of Molecular Mechanisms Underlying Plscr4-miR-214-Regulated Cardiac Hypertrophy
Under normal conditions, Plscr4 inhibits the expression of miR-214, which is the suppresser of Mfn2, to keep mitochondrial homeostasis. When CMs are exposed to pathological stimuli, the adaptive response leads to the upregulation of Plscr4, which exerts an anti-hypertrophy effect. Meanwhile, the pro-hypertrophy signal miR-214 is also upregulated. However, with prolonged stimulus, when the upregulation of miR-214 is beyond a certain threshold, it is sufficient to counteract the function of Plscr4 activation, leading to a significant decline of Mfn2 with the unbalanced mitochondrial fusion and fission, which promotes the development of cardiac hypertrophy.
More and more studies confirm that lncRNAs act as miRNA sponges to regulate their activity.41, 42 In accordance with these studies, we also demonstrated that Plscr4 exerted its anti-hypertrophy function by binding and sequestering miR-214. miR-214 is verified as a pro-hypertrophy miRNA in previous studies.30 The interaction between Plscr4 and miR-214 attenuates the inhibitory effects of miR-214 on Mfn2, which is demonstrated to be a direct target of miR-214 in the hypertrophic heart.37 Consistent with these reports, our study showed that the Mfn2 expression level decreased in both the TAC-induced hypertrophic hearts and in the Ang II-induced hypertrophic CMs. Previous studies confirmed that the upregulation of Mfn2 inhibits the increase of protein synthesis and hypertrophy in CMs.13, 15 Our study showed that overexpression of Plscr4 rescued the decreased Mfn2 in response to hypertrophic stress and, therefore, resisted mitochondrial dysfunction to alleviate hypertrophic growth. There also may be some other mechanisms to discover. In future work, we will study other potential mechanisms involved in the anti-hypertrophy effects of Plscr4.
In summary, our present study revealed that lncRNA Plscr4 plays a crucial role in anti-hypertrophy and regulates the hypertrophic response in cellular and animal models. Thus, the modulation of Plscr4 may offer novel approaches for the treatment of cardiac hypertrophy. This finding helps to elucidate the functions of lncRNAs and miRNA-controlled cellular events.
Materials and Methods
Animal Experiments
Healthy male C57BL/6 mice (weighing 20–25 g) were purchased from the Experimental Animal Center of the Harbin Medical University. The animals were kept under standard animal room conditions (temperature 21°C ± 1°C and humidity 55%–60%), with free access to food and water. To make the cardiac pressure overload model, the mice underwent a TAC surgery for 4 weeks, as described in a previous study.43 In addition, C57BL/6 mice were injected, through the tail vein, with adeno-associated virus serotype 9 (AAV9) carrying a vector or Plscr4 (Hanbio Biotechnology, Shanghai, China) 3 weeks before further experiments. All the experimental procedures were performed in accordance with and approved by the Institutional Animal Care and Use Committee of the Harbin Medical University.
Echocardiography
Mice were anesthetized with Avertin (160 mg/kg intraperitoneally [i.p.]; Sigma-Aldrich) and were placed on a platform. An echocardiography was performed with the ultrasound machine Vevo2100 high-resolution imaging system (VisualSonics, Toronto, ON, Canada) that was equipped with a 10-MH phased-array transducer with M-mode recordings.
Real-Time qPCR
Total RNA samples were extracted from cultured neonatal mice CMs or the left ventricle of mice hearts using the TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse-transcribed with 5X All-In-One RT MasterMix (abmGood, Vancouver, Canada). Real-time qPCR was carried out on an ABI 7500 fast real time PCR system (Applied Biosystems, Foster City, CA, USA) with the EvaGreen qPCR Mastermix Kit (abmGood, Vancouver, Canada) to detect the mRNA levels of Plscr4, ANP, BNP, and β-MHC, and GAPDH was set as the internal control. The miR-214 level was quantified by the mirVana qRT-PCR miRNA detection kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocols and as previously described,44, 45 and these data were normalized to U6. The data were analyzed using the 2-ΔΔCT method. The sequences of the primers were synthesized by Sangon Biotech (Shanghai, China) and are listed in Table S2.
Western Blot Analysis
The mouse heart tissue and the cultured CMs were lysed with standard sample buffer. The concentration of the proteins was determined with a BCA Protein Assay Kit (Beyotime, Shanghai, China). The samples were subjected to electrophoresis in 10% SDS-PAGE and were later transferred to a nitrocellulose filter membrane. The following primary antibodies were used: an anti-Myosin antibody (1:5,000, mouse monoclonal; Sigma, St. Louis, MO, USA) and an anti-Mitofusin 2 antibody (1:1,000, rabbit polyclonal; Abcam, Cambridge, UK). The western blot bands were analyzed using Odyssey version (v.)1.2 software by measuring the band and normalizing to GAPDH (anti-GAPDH, 1:1,000, mouse polyclonal; Kangcheng, Shanghai, China).
Luciferase Assays
A fragment of Plscr4 containing the miR-214-binding site (Plscr4-WT) was amplified by PCR, and it was, therefore, a fragment with nucleotide replacement mutation (designated Plscr4-Mut). For the luciferase reporter gene assay, the HEK293 cells were cultured in 24-well culture plates, and they were transfected with varying constructs as described in the corresponding figure legends. The firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA).
CM Culture and Treatment
CMs were isolated from C57BL/6 mice (1–2 days) as previously described.46 Briefly, the cardiac tissue was digested by pancreatin (Beyotime, Shanghai, China), and it was maintained in DMEM supplemented with 100 U/mL penicillin and 10% fetal bovine serum (FBS) (Biological Industries [BI]). CMs were purified by differential plating, and 0.1 mmol/L BrdU (5-bromo-2-deoxyuridine) was added to exclude the cardiac fibroblasts. The cells were cultured in a 5% CO2 and 37°C humidified atmosphere for 48 hr after plating. To induce hypertrophy, angiotensin II (Ang II) was added to the CMs at a concentration of 1 μmol/L for 48 hr.
Plasmid and miRNA Transfection
The lncRNA Plscr4 overexpression plasmid and the control vector were synthesized by GeneChem (Shanghai, China). The miR-214 mimic and the negative control miRNA (miR-NC) were synthesized by RIBOBIO (Guangzhou, China). The CMs were transfected with the plasmid and miR-214 or miR-NC using Lipo2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol.
Cell Surface Area Measurement
The cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100 in PBS for 45 min, and stained with α-actinin (Abcam, Cambridge, UK) at 4°C overnight. Next, the cells were incubated with a Daylight 594 goat anti-Mouse antibody for 1 hr at room temperature. The cells were incubated with DAPI for 10 min before immunofluorescence capture. Immunofluorescence was visualized under a fluorescence microscope (Carl Zeiss, 37081); quantification of cell surface area was achieved by measuring 50 random cells from three independent experiments, and the average value was used for analysis. The surface areas were measured using Image-Pro Plus 6.0 software.
Determinations of Protein/DNA Ratio
Total protein and DNA contents were analyzed as previously described.47 Briefly, protein concentration was determined by the BCA method (P0012; Beyotime, Shanghai, China), as described by the manufacturer with BSA as a standard. DNA concentration was detected by fluorescence assay (DNAQF; Sigma, St. Louis, MO, USA), according to the manufacturer’s instructions. The ratio of protein to DNA was then calculated to estimate potential protein synthesis.
Histological Analysis
The mice hearts were collected and fixed in 4% paraformaldehyde for 24 hr followed by embedding in paraffin according to standard histological protocols. Next, the tissue was cut into 5-μm-thick cross-sectional slices. The slices were stained by H&E and Masson trichrome to evaluate histopathology and collagen volume, respectively. The myocyte cross-sectional areas were measured via fluorescein isothiocyanate-conjugated WGA (L4895; Sigma, St. Louis, MO, USA) staining. The fibrotic areas were calculated with image analysis software (Image-Pro Plus 6.0 software).
Mitochondrial Membrane Potential (Δψm) Assessment
JC-1 staining (Beyotime, Shanghai, China) was used to assess the mitochondrial membrane potential of the CMs. Briefly, the CMs were incubated with an equal volume of JC-1 staining solution following the manufacturer’s protocol. JC-1 exhibited a potential-dependent accumulation in the mitochondria, as indicated by a fluorescence emission shift from green 525 ± 10 nm to red 610 ± 10 nm. In healthy cells with a high potential, JC-1 forms complexes emitting a red fluorescence, and in unhealthy cells with a low mitochondrial membrane potential, JC-1 remains in a monomeric form, emitting green fluorescence. The state of the membrane potential was expressed by the ratio of red to green fluorescence.
Measurement of Production of ROS
A hydrogen peroxide assay kit (Solarbio, Beijing, China) was used to determine the H2O2 production. DCFH-DAC is the probe for H2O2. The CMs were washed with PBS and were incubated with DCFH-DA in DMEM for 20 min at 37°C. After removal of the DCFH-DA and washing with PBS, the cells were imaged with a fluorescence microscope (Carl Zeiss, 37081). The same experiment was repeated four times independently.
Data Analysis
The data are presented as the means ± SEM. The statistical analyses were performed using one-way ANOVA and Student’s test. In all cases, p < 0.05 was considered to be statistically significant. The data were analyzed using GraphPad Prism 5.0.
Author Contributions
H.L. and H.S. designed and supervised the experiments. L.L. performed the majority of the experiments and wrote the manuscript. T.L. and X.L. analyzed the majority of experimental data. C.X., Q.L., and H.J. performed the western blot, PCR, and luciferase assays. Y. Li and Y. Liu performed H&E, Masson, and WGA staining. H.Y. and Q.H. cultured cells. Y.Z. and M.Z. analyzed the bioinformation data.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81770284, 81473213, and 81673425) and the Major Program of National Natural Science Foundation of China (81730012).
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.018.
Contributor Information
Hongli Shan, Email: shanhongli@ems.hrbmu.edu.cn.
Haihai Liang, Email: lianghaihai@ems.hrbmu.edu.cn.
Supplemental Information
References
- 1.Veselka J., Anavekar N.S., Charron P. Hypertrophic obstructive cardiomyopathy. Lancet. 2017;389:1253–1267. doi: 10.1016/S0140-6736(16)31321-6. [DOI] [PubMed] [Google Scholar]
- 2.Stanton T., Dunn F.G. Hypertension, Left Ventricular Hypertrophy, and Myocardial Ischemia. Med. Clin. North Am. 2017;101:29–41. doi: 10.1016/j.mcna.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Schirone L., Forte M., Palmerio S., Yee D., Nocella C., Angelini F., Pagano F., Schiavon S., Bordin A., Carrizzo A. A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxid. Med. Cell. Longev. 2017;2017:3920195. doi: 10.1155/2017/3920195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardo B.C., Weeks K.L., Pretorius L., McMullen J.R. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol. Ther. 2010;128:191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Diwan A., Dorn G.W., 2nd Decompensation of cardiac hypertrophy: cellular mechanisms and novel therapeutic targets. Physiology (Bethesda) 2007;22:56–64. doi: 10.1152/physiol.00033.2006. [DOI] [PubMed] [Google Scholar]
- 6.Ong S.B., Subrayan S., Lim S.Y., Yellon D.M., Davidson S.M., Hausenloy D.J. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 7.Kanzaki Y., Terasaki F., Okabe M., Otsuka K., Katashima T., Fujita S., Ito T., Kitaura Y. Giant mitochondria in the myocardium of a patient with mitochondrial cardiomyopathy: transmission and 3-dimensional scanning electron microscopy. Circulation. 2010;121:831–832. doi: 10.1161/CIR.0b013e3181d22e2d. [DOI] [PubMed] [Google Scholar]
- 8.Neubauer S. The failing heart--an engine out of fuel. N. Engl. J. Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 9.Huss J.M., Kelly D.P. Mitochondrial energy metabolism in heart failure: a question of balance. J. Clin. Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan X., Wang L., Liu Z., Guo X., Jiang Y., Lu Y., Peng Y., Liu T., Yang B., Shan H. miR-106a promotes cardiac hypertrophy by targeting mitofusin 2. J. Mol. Cell. Cardiol. 2016;99:207–217. doi: 10.1016/j.yjmcc.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Wüst R.C., de Vries H.J., Wintjes L.T., Rodenburg R.J., Niessen H.W., Stienen G.J. Mitochondrial complex I dysfunction and altered NAD(P)H kinetics in rat myocardium in cardiac right ventricular hypertrophy and failure. Cardiovasc. Res. 2016;111:362–372. doi: 10.1093/cvr/cvw176. [DOI] [PubMed] [Google Scholar]
- 12.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H., Guo Y., Mi L., Wang X., Li L., Gao W. Mitofusin 2 inhibits angiotensin II-induced myocardial hypertrophy. J. Cardiovasc. Pharmacol. Ther. 2011;16:205–211. doi: 10.1177/1074248410385683. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Ponnusamy M., Liu C., Tian J., Dong Y., Gao J., Wang C., Zhang Y., Zhang L., Wang K., Li P. MiR-485-5p modulates mitochondrial fission through targeting mitochondrial anchored protein ligase in cardiac hypertrophy. Biochim. Biophys. Acta. 2017;1863:2871–2881. doi: 10.1016/j.bbadis.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Niu Q., Peng X., Li M., Liu K., Liu Y., Liu J., Jin F., Li X., Wei Y. Candesartan cilexetil attenuated cardiac remodeling by improving expression and function of mitofusin 2 in SHR. Int. J. Cardiol. 2016;214:348–357. doi: 10.1016/j.ijcard.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Papanicolaou K.N., Khairallah R.J., Ngoh G.A., Chikando A., Luptak I., O’Shea K.M., Riley D.D., Lugus J.J., Colucci W.S., Lederer W.J. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol. Cell. Biol. 2011;31:1309–1328. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jandura A., Krause H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Klingenberg M., Matsuda A., Diederichs S., Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 21.Yang T., Gu H., Chen X., Fu S., Wang C., Xu H., Feng Q., Ni Y. Cardiac hypertrophy and dysfunction induced by overexpression of miR-214 in vivo. J. Surg. Res. 2014;192:317–325. doi: 10.1016/j.jss.2014.06.044. [DOI] [PubMed] [Google Scholar]
- 22.Karakikes I., Chaanine A.H., Kang S., Mukete B.N., Jeong D., Zhang S., Hajjar R.J., Lebeche D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 24.Uchida S. Besides Imprinting: Meg3 Regulates Cardiac Remodeling in Cardiac Hypertrophy. Circ. Res. 2017;121:486–487. doi: 10.1161/CIRCRESAHA.117.311542. [DOI] [PubMed] [Google Scholar]
- 25.Micheletti R., Plaisance I., Abraham B.J., Sarre A., Ting C.C., Alexanian M., Maric D., Maison D., Nemir M., Young R.A. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017;9:eaai9118. doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z., Zhang X.J., Ji Y.X., Zhang P., Deng K.Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L., An X., Li Z., Song Y., Li L., Zuo S., Liu N., Yang G., Wang H., Cheng X. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 28.Wang K., Liu F., Zhou L.Y., Long B., Yuan S.M., Wang Y., Liu C.Y., Sun T., Zhang X.J., Li P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 29.Chang T.H., Huang H.Y., Hsu J.B., Weng S.L., Horng J.T., Huang H.D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang T., Zhang G.F., Chen X.F., Gu H.H., Fu S.Z., Xu H.F., Feng Q., Ni Y.M. MicroRNA-214 provokes cardiac hypertrophy via repression of EZH2. Biochem. Biophys. Res. Commun. 2013;436:578–584. doi: 10.1016/j.bbrc.2013.05.079. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Liew O.W., Richards A.M., Chen Y.T. Overview of MicroRNAs in Cardiac Hypertrophy, Fibrosis, and Apoptosis. Int. J. Mol. Sci. 2016;17:E749. doi: 10.3390/ijms17050749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Del Re D.P., Nakano N., Sciarretta S., Zhai P., Park J., Sayed D., Shirakabe A., Matsushima S., Park Y. miR-206 Mediates YAP-Induced Cardiac Hypertrophy and Survival. Circ. Res. 2015;117:891–904. doi: 10.1161/CIRCRESAHA.115.306624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardo B.C., Nguyen S.S., Winbanks C.E., Gao X.M., Boey E.J., Tham Y.K., Kiriazis H., Ooi J.Y., Porrello E.R., Igoor S. Therapeutic silencing of miR-652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J. 2014;28:5097–5110. doi: 10.1096/fj.14-253856. [DOI] [PubMed] [Google Scholar]
- 34.da Costa Martins P.A., Bourajjaj M., Gladka M., Kortland M., van Oort R.J., Pinto Y.M., Molkentin J.D., De Windt L.J. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y., Ji R., Yue J., Yang J., Liu X., Chen H., Dean D.B., Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou Y., Sun Y., Shan H., Li X., Zhang M., Zhou X., Xing S., Sun H., Chu W., Qiao G., Lu Y. β-adrenoceptor regulates miRNA expression in rat heart. Med. Sci. Monit. 2012;18:BR309–BR314. doi: 10.12659/MSM.883263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun M., Yu H., Zhang Y., Li Z., Gao W. MicroRNA-214 Mediates Isoproterenol-induced Proliferation and Collagen Synthesis in Cardiac Fibroblasts. Sci. Rep. 2015;5:18351. doi: 10.1038/srep18351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan J., Zhu W., Rahman M.S., Liu M., Li D., Su S., Zhang N., Hu X., Yu H., Gupta M.P., Wang J. Molecular regulation of mitochondrial dynamics in cardiac disease. Biochim. Biophys. Acta. 2017;1864:1260–1273. doi: 10.1016/j.bbamcr.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., Dittrich M., Maetzig T., Zimmer K., Remke J. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Chen W., Zhang J., Khan A., Li L., Huang F., Qiu Z., Wang L., Chen X. Critical Role of ADAMTS2 (A Disintegrin and Metalloproteinase With Thrombospondin Motifs 2) in Cardiac Hypertrophy Induced by Pressure Overload. Hypertension. 2017;69:1060–1069. doi: 10.1161/HYPERTENSIONAHA.116.08581. [DOI] [PubMed] [Google Scholar]
- 41.Wang K., Liu C.Y., Zhou L.Y., Wang J.X., Wang M., Zhao B., Zhao W.K., Xu S.J., Fan L.H., Zhang X.J. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 42.Wang K., Long B., Zhou L.Y., Liu F., Zhou Q.Y., Liu C.Y., Fan Y.Y., Li P.F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 43.Dong D.L., Chen C., Huo R., Wang N., Li Z., Tu Y.J., Hu J.T., Chu X., Huang W., Yang B.F. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: a novel mechanism for progressive cardiac hypertrophy. Hypertension. 2010;55:946–952. doi: 10.1161/HYPERTENSIONAHA.109.139519. [DOI] [PubMed] [Google Scholar]
- 44.Pan Z., Zhao W., Zhang X., Wang B., Wang J., Sun X., Liu X., Feng S., Yang B., Lu Y. Scutellarin alleviates interstitial fibrosis and cardiac dysfunction of infarct rats by inhibiting TGFβ1 expression and activation of p38-MAPK and ERK1/2. Br. J. Pharmacol. 2011;162:688–700. doi: 10.1111/j.1476-5381.2010.01070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan X., Li J., Wang X., Chen N., Cai B., Wang G., Shan H., Dong D., Liu Y., Li X. Tanshinone IIA protects against cardiac hypertrophy via inhibiting calcineurin/NFATc3 pathway. Int. J. Biol. Sci. 2011;7:383–389. doi: 10.7150/ijbs.7.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C., Li X., Gao X., Zhang R., Zhang Y., Liang H., Xu C., Du W., Zhang Y., Liu X. MicroRNA-328 as a regulator of cardiac hypertrophy. Int. J. Cardiol. 2014;173:268–276. doi: 10.1016/j.ijcard.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 47.Johnson T.B., Kent R.L., Bubolz B.A., McDermott P.J. Electrical stimulation of contractile activity accelerates growth of cultured neonatal cardiocytes. Circ. Res. 1994;74:448–459. doi: 10.1161/01.res.74.3.448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.