Intra-tumor heterogeneity (ITH) measured by MATH independently predicted survival in male CRC patients. Additionally, the association between MATH and related genomic aberration was different based on gender. We further provided potential mechanisms of ITH on RNA and DNA level.
Abstract
Intra-tumor heterogeneity (ITH) is crucial in tumorigenesis and resistance to target therapy. Here, we used mutant-allele tumor heterogeneity (MATH) to measure ITH based on next-generation sequencing data and high MATH was proven as an independent risk prognostic factor in male CRC patients in both a training set of 284 colorectal cancer (CRC) patients with from The Cancer Genome Atlas (TCGA) and a validating set of 187 CRC patients from International Cancer Genome Consortium (ICGC). Further, the genomic pattern according to MATH demonstrated that mutation rates of TP53, IRF5 and KRAS were independently associated with MATH, and the latter two were only significant in male patients. As MATH increased, the fraction of somatic copy number alteration (SCNA) elevated. Moreover, more SCNA events was independently associated with MATH in male than in female. WNT pathway, TGF-β pathway and DNA repair deficiency was enriched in high MATH group and the latter two showed up only in male patients. In summary, we reveal the gender-related prognostic value of MATH and relevant genomic pattern in CRC. Potential mechanisms are provided and it remains to be proven whether they are drivers of subclone formation and ITH. Taking MATH into consideration in clinical trial might contribute to better therapeutic strategies in CRC with researches added on in the future.
Introduction
With the development of precision medicine, the genetic and phenotypic heterogeneity of tumor has been widely noted recently. The heterogeneity, including inter-tumor heterogeneity and intra-tumor heterogeneity (ITH), is significant in tumor progression and clinical choices (1,2). ITH, also known as variation between tumor cells within individual tumor, was attributed to genetic diversity from acquisition of genomic instability and subsequent mutations. The coexistence of multiple subclones and heterogeneous cell phenotypes have impact on clinical diagnosis, therapeutic responses and metastases. Therefore, patients with high level of ITH are less sensitive to single targeted therapy (3). To date, few articles have documented the association between high ITH and poor outcome in cancers (4,5).
ITH was used to be assessed by several methods such as combined analyses of markers associated with cellular differentiation, multiregional biopsies, single cell sequencing and ultra-deep sequencing of mutations (6–8). However, these direct techniques face a great mount of challenges to be clinically applied on large population. Taking analysis of markers as an example, the markers are difficult to be standardized and quantitated. Moreover, application of imaging techniques is required but guidelines for scoring and reporting have not yet been established.
Nowadays, some methods have been proposed to measure intra-tumor genomic heterogeneity with the proliferation of next-generation sequencing (NGS) (9,10). Mutations were found more frequent in the ancestral subclones, while less in other progeny ones. Consequently, more heterogeneous tumors have a broader distribution of mutant-allele fractions among tumor-specific mutated loci (3). Mutant-allele tumor heterogeneity (MATH), calculated by the width of distribution, avoids the practical and theoretical difficulties and measures ITH deriving directly from whole-exome sequencing data (11). MATH is a simple, quantitative and applicable way to demonstrate ITH, since NGS of tumor DNA is expected to be widely applied and generalized in clinic in the future.
ITH assessed by MATH was evaluated as a prognostic biomarker in head and neck squamous cell carcinoma and applied to other cancers (4,12,13). Nonetheless, the prognostic significance of ITH and genomic pattern associated with different levels have not been explored in colorectal cancer (CRC) yet. CRC is a complex disease with considerable divergences in the response to treatment, even in tumors with similar histopathological features. Thus, ITH has been known as one of the most plausible explanation (14). Using published NGS data of a training set of 284 CRC patients from The Cancer Genome Atlas (TCGA) and an independent validating set of 187 CRC patients from International Cancer Genome Consortium (ICGC) data portal, we investigated the connection between not only clinical value but also genomic portrait and intra-tumor genetic heterogeneity measured by MATH.
Materials and methods
Patients and outcome measurement
Clinical characteristics, whole-exome sequencing (~1% of the genome, at 150-fold mean sequence coverage), RNAseq Version 2 RSEM and segmented SCNA data of patients with CRC were obtained from TCGA. We exclude patients without available data of whole-exome sequencing, age, sex, stage, primary site, histology type and follow-up time. 284 patients with CRC diagnosed between 1999 and 2011 were enrolled into the training cohort. Simple somatic mutations of cancer genomes in the validation cohort was from release 23 of ICGC. Finally, the data used for further analysis contained 187 patients with complete information of simple somatic mutation, age, sex, TNM stage, primary site and follow-up time. Outcomes of interest in our study was overall survival. Overall survival was calculated from the date of diagnosis to the date of death. Deaths of any cause were treated as events, and patients were still alive at the time of the last follow-up were treated as censored observations.
Mutant Allele Tumor Heterogeneity
The MATH score is calculated as the percentage ratio of MAD and the median of its mutant-allele fractions at tumor-specific mutated loci, which is based on whole-exome sequencing data of tumor and matched normal DNA: MATH = 100*MAD/median. The tertiles of MATH was used as cutoff points to divide patients into low MATH, intermediate MATH and high MATH groups.
Genomic analysis
Somatic copy number alteration was analyzed by Genomic Identification of Significant Targets in Cancer (GISTIC) 2.0. The amplification and deletion threshold was respectively set at log2 ratio >0.9 and log2 ratio <−0.3. False-discovery rate (FDR) q values obtained for each region reflected and the statistical significance of the aberrations. The fraction of SCNA was obtained from University of California at Santa Cruz Human Genome Browser (http://genome.cse.ucsc.edu/). Gene Set Enrichment Analysis (GSEA) was performed to present the gene set enriching in high MATH group than the low using 13311 gene sets from Molecular Signatures Database (MSigDB).
Statistical analysis
Patient baseline characteristics were compared among the MATH groups by ANOVA and Bonferroni test for pair-wise comparison. Log-rank (Mantel-Cox) test was applied to compare the overall survival between different groups. Univariate and multivariate Cox regression were employed to determine the independently prognostic role of MATH, which was treated as not only a continuous variable but also a categorical variable. The prevalence of non-silent somatic mutations was compared among different MATH groups with Chi-square test and logistic regression model adjusting tumor stage and primary site. Fraction of SCNA were compared by MATH using one-way analysis of variance by ranks (Kruskal–Wallis Test).
Identification of SCNA events and analysis of gene set enrichment were performed by GISTIC 2.0 and GSEA software, respectively. Other statistical methods were all analyzed by R version 3.2.3 (http://cran.r-project.org) and Stata statistical software, version 12.0 (StataCorp, College Station, TX). P values were all two-sided and statistical significance was set at P < 0.05 if not mentioned. All confidence intervals (CIs) were stated at the 95% confidence level.
Results
Baseline characteristics and MATH
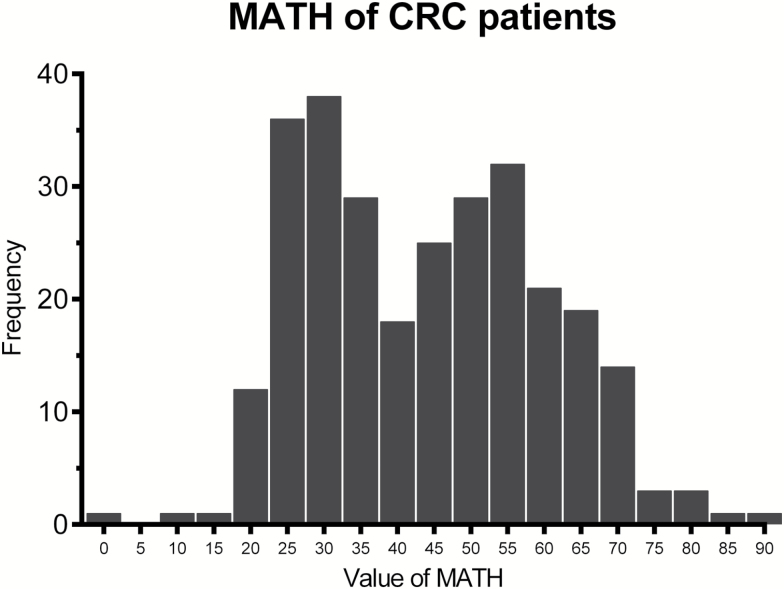
The mean value of MATH was 41.58 in the training cohort and 46.41 in the validating cohort. Among patients in the training cohort, 94, 93 and 93 patients were in low, intermediate and high MATH group, with 24.05, 40.90, and 59.62 as mean value of MATH in each group, respectively. The distribution of MATH was shown in Figure 1. The MATH with different clinicopathological characteristics of tumors was summarized in Table 1. In one-way analysis of variance (ANOVA), we found significant association between the MATH and clinicopathological characteristics including tumor stage, N stage, primary site and MATH groups. Patients with stage II had lower level of MATH score than stage III (P < 0.001) and stage IV (P = 0.003) by Bonferroni test for pair-wise comparison. Similarly, the MATH score of patients with N0 stage was significantly lower than N1 stage (P < 0.001) and N2 stage (P < 0.001). To further explore relationship of clinicopathological characteristics and MATH in gendered subgroup, the same methods mentioned above were also performed on female and male patients.
Figure 1.
Frequency of mutant-allele tumor heterogeneity (MATH) score among 284 patients with colorectal cancer in the training cohort. MATH score is displayed along the horizontal axis and number of patients with MATH within the specific ranges on the vertical axis.
Table 1.
Baseline characteristics and MATH in training cohorta
| Characteristic | All (n = 284) | Female (n = 124) | Male (n = 160) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MATH | N | Per. | P value | MATH | N | Per. | P value | MATH | N | Per. | P value | |
| 41.58 | 284 | 100.00% | 41.05 | 124 | 100.00% | 42.00 | 160 | 100.00% | ||||
| Age | 0.330 | 0.176 | 0.858 | |||||||||
| ≤50 | 40.68 | 45 | 15.85% | 38.06 | 20 | 16.13% | 42.77 | 25 | 15.63% | |||
| 51–70 | 43.10 | 130 | 45.77% | 43.96 | 55 | 44.35% | 42.47 | 75 | 46.88% | |||
| >70 | 40.16 | 109 | 38.38% | 39.00 | 49 | 39.52% | 41.10 | 60 | 37.50% | |||
| Race | 0.867 | 0.632 | 0.994 | |||||||||
| White | 41.39 | 235 | 82.75% | 40.74 | 106 | 85.48% | 41.93 | 129 | 80.63% | |||
| Black | 41.44 | 20 | 7.04% | 41.40 | 8 | 6.45% | 41.48 | 12 | 7.50% | |||
| Asian | 40.34 | 8 | 2.82% | 26.86 | 1 | 0.81% | 42.27 | 7 | 4.38% | |||
| Unknown | 44.36 | 21 | 7.39% | 45.91 | 9 | 7.26% | 43.19 | 12 | 7.50% | |||
| Sex | 0.615 | / | / | |||||||||
| Female | 41.05 | 124 | 43.66% | |||||||||
| Male | 42.00 | 160 | 56.34% | |||||||||
| Stage | <0.001 | 0.018 | 0.004 | |||||||||
| Stage I | 38.74 | 40 | 14.08% | 39.09 | 16 | 12.90% | 38.50 | 24 | 15.00% | |||
| Stage II | 36.96 | 108 | 38.03% | 35.85 | 43 | 34.68% | 37.70 | 65 | 40.63% | |||
| Stage III | 45.73 | 95 | 33.45% | 43.87 | 45 | 36.29% | 47.41 | 50 | 31.25% | |||
| Stage IV | 46.91 | 41 | 14.44% | 47.42 | 20 | 16.13% | 46.43 | 21 | 13.13% | |||
| T | 0.839 | 0.727 | 0.375 | |||||||||
| T1 | 38.58 | 8 | 2.82% | 38.22 | 5 | 4.03% | 39.19 | 3 | 1.88% | |||
| T2 | 40.97 | 41 | 14.44% | 41.73 | 17 | 13.71% | 40.43 | 24 | 15.00% | |||
| T3 | 41.51 | 201 | 70.77% | 41.70 | 89 | 71.77% | 41.35 | 112 | 70.00% | |||
| T4 | 43.50 | 34 | 11.97% | 36.77 | 13 | 10.48% | 47.66 | 21 | 13.13% | |||
| N | <0.001 | 0.018 | <0.001 | |||||||||
| N0 | 37.31 | 153 | 53.87% | 37.05 | 61 | 49.19% | 37.48 | 92 | 57.50% | |||
| N1 | 45.45 | 81 | 28.52% | 44.64 | 38 | 30.65% | 46.17 | 43 | 26.88% | |||
| N2 | 48.39 | 50 | 17.61% | 45.33 | 25 | 20.16% | 51.45 | 25 | 15.63% | |||
| Site | 0.004 | 0.061 | 0.031 | |||||||||
| Colon | 39.96 | 207 | 72.89% | 39.37 | 88 | 70.97% | 40.40 | 119 | 74.38% | |||
| Rectum | 45.94 | 77 | 27.11% | 45.15 | 36 | 29.03% | 46.64 | 41 | 25.63% | |||
| Histology | 0.084 | 0.317 | 0.142 | |||||||||
| Mucinous adenocarcinoma | 37.18 | 34 | 11.97% | 36.59 | 11 | 8.87% | 37.46 | 23 | 14.38% | |||
| Other adenocarcinoma | 42.18 | 250 | 88.03% | 41.48 | 113 | 91.13% | 42.76 | 137 | 85.63% | |||
| MATH | <0.001 | <0.001 | <0.001 | |||||||||
| Low | 24.05 | 94 | 33.10% | 22.94 | 40 | 32.26% | 24.87 | 54 | 33.75% | |||
| Intermediate | 40.90 | 95 | 33.45% | 40.92 | 41 | 33.06% | 40.88 | 54 | 33.75% | |||
| High | 59.62 | 95 | 33.45% | 58.00 | 43 | 34.68% | 60.95 | 52 | 32.50% | |||
aMATH score in this table is the mean of MATH scores in each subgroup.
The mean value of MATH in low, intermediate and high MATH group of validation cohort was 26.91, 47.30 and 46.45, respectively. The MATH according to different clinicopathological characteristics of tumors was summarized in Supplementary Table 1, available at Carcinogenesis Online. In ANOVA, we found age, N stage, M stage and MATH groups were significantly associated with MATH. Further, we performed the same analysis on female and male patients.
Effect of MATH on survival
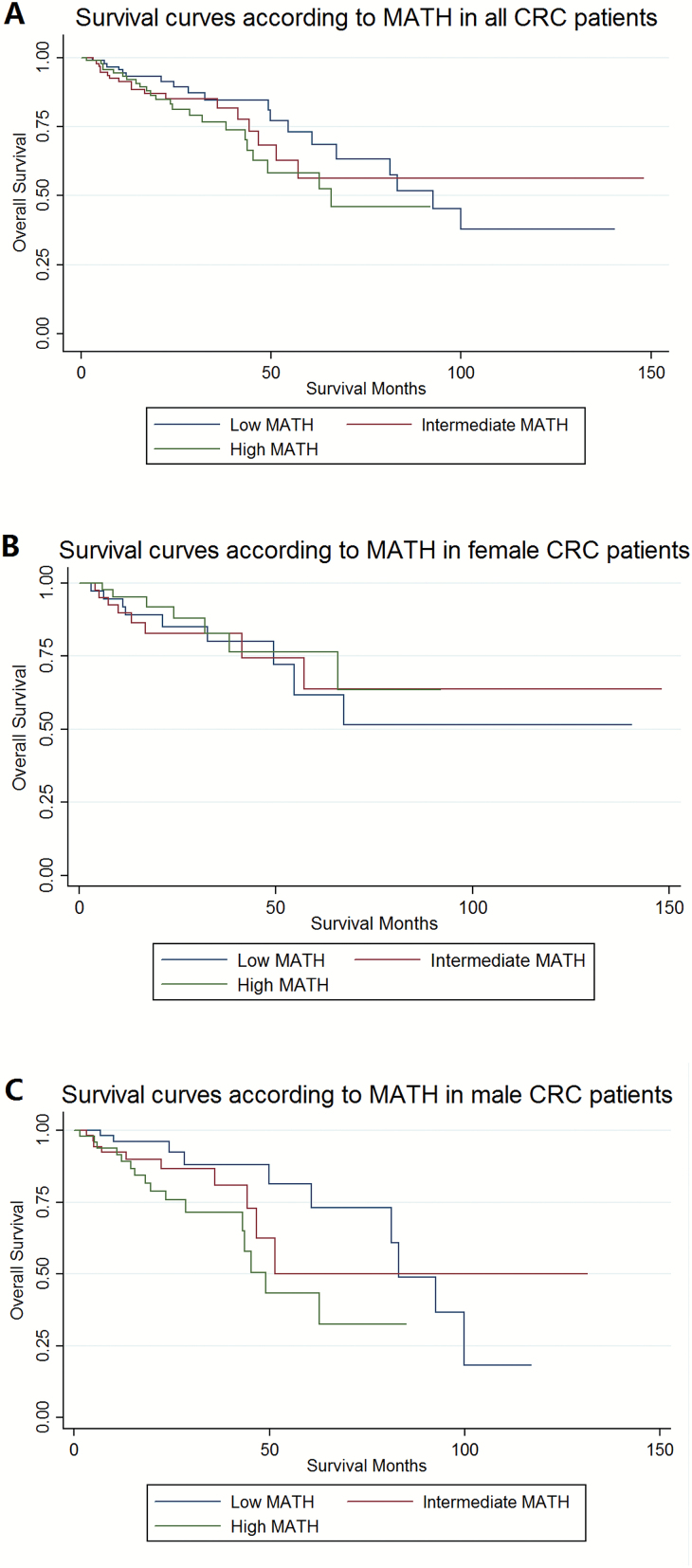
Then we explored the prognostic value of MATH and other variables in all patients and subgroups. In the training cohort, we found that age and tumor stage were significantly associated with OS in univariate Cox regression in all patients, likewise in gender-subgroups including male and female patients. Interestingly, MATH score was a significantly prognostic factor only in male patients by univariate Cox regression analysis, with P = 0.025 as continuous variable and P = 0.019 in high MATH group as categorical variable. The unadjusted OS by MATH was calculated by Kaplan–Meier curves (Figure 2). After adjusting above dependent variables in multivariate Cox regression, they were still independent prognostic factors in all or subgroup patients except tumor stage in male patients. In conclusion, we found that high MATH score was an independent risk factor which predicted the outcome of male patients with CRC, when treated as both continuous variable (HR = 1.025, 95% CI 1.004–1.047, P = 0.017) and categorical variable (Ref: low; high: HR = 3.617, 95% CI 1.438–9.099, P = 0.006), but not a significant predictor for OS in either all or female patients. Detailed information was presented in Table 2.
Figure 2.
Kaplan–Meier survival curves in the training cohort: The overall survival of patients with colorectal cancer according to mutant-allele tumor heterogeneity (MATH) groups. (A) In all patients: χ2 = 1.72, P = 0.424. (B) In female patients: χ2 = 0.45, P = 0.798. (C) In male patients: χ2 = 6.15, P = 0.046.
Table 2.
Univariate and multivariate survival analysis for training cohort
| Characteristic | Univariate analysis | Multivariate analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-year OS | Log rank χ2 | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | ||||||
| All (n = 284) | Age | 16.93 | <0.001 | |||||||||||||
| ≤50 | 92.44% | 1.007 | 0.366 | 2.774 | 0.989 | 0.903 | 0.321 | 2.541 | 0.847 | |||||||
| 51–70 | 88.85% | Ref | Ref | |||||||||||||
| >70 | 69.07% | 2.941 | 1.623 | 5.33 | <0.001 | 3.306 | 1.812 | 6.03 | <0.001 | |||||||
| Race | 1.88 | 0.598 | ||||||||||||||
| White | 80.22% | Ref | ||||||||||||||
| Black | 86.12% | 0.471 | 0.115 | 1.936 | 0.297 | |||||||||||
| Asian | 87.50% | 1.537 | 0.372 | 6.356 | 0.553 | |||||||||||
| Unknown | 82.05% | 0.708 | 0.22 | 2.271 | 0.561 | |||||||||||
| Sex | 1.32 | 0.251 | ||||||||||||||
| Female | 81.70% | 0.738 | 0.438 | 1.242 | 0.253 | |||||||||||
| Male | 80.19% | Ref | ||||||||||||||
| Stage | 12.38 | 0.006 | ||||||||||||||
| Stage I | 87.16% | 0.995 | 0.371 | 2.671 | 0.992 | 1.067 | 0.395 | 2.881 | 0.898 | |||||||
| Stage II | 90.52% | Ref | Ref | |||||||||||||
| Stage III | 72.80% | 1.411 | 0.761 | 2.616 | 0.274 | 1.668 | 0.884 | 3.148 | 0.114 | |||||||
| Stage IV | 62.39% | 3.117 | 1.558 | 6.236 | 0.001 | 4.076 | 2.003 | 8.295 | <0.001 | |||||||
| Site | 0.79 | 0.373 | ||||||||||||||
| Colon | 82.62% | Ref | ||||||||||||||
| Rectum | 76.13% | 1.305 | 0.726 | 2.345 | 0.374 | |||||||||||
| Histology | 0.26 | 0.613 | ||||||||||||||
| Mucinous adenocarcinoma | 89.29% | 1.266 | 0.506 | 3.167 | 0.614 | |||||||||||
| Other adenocarcinoma | 80.18% | Ref | ||||||||||||||
| MATH | 1.72 | 0.424 | ||||||||||||||
| Low | 84.66% | Ref | ||||||||||||||
| Intermediate | 81.72% | 1.143 | 0.599 | 2.183 | 0.685 | |||||||||||
| High | 76.80% | 1.49 | 0.806 | 2.755 | 0.203 | |||||||||||
| MATH (continuous variable) | 1.009 | 0.993 | 1.025 | 0.288 | ||||||||||||
| Female (n = 124) | Characteristic | Univariate analysis | Multivariate analysis | |||||||||||||
| 3-year OS | Log rank χ2 | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | ||||||
| Age | 13.35 | 0.001 | ||||||||||||||
| ≤50 | 94.44% | 0.528 | 0.063 | 4.4 | 0.555 | 0.36 | 0.042 | 3.111 | 0.353 | |||||||
| 51~70 | 92.36% | Ref | Ref | |||||||||||||
| >70 | 64.02% | 3.948 | 1.551 | 10.051 | 0.004 | 5.18 | 1.941 | 13.828 | 0.001 | |||||||
| Race | 2.42 | 0.490 | ||||||||||||||
| White | 80.48% | Ref | ||||||||||||||
| Black | 100.00% | 0 | 0 | / | 1 | |||||||||||
| Female (n = 124) | Characteristic | Univariate analysis | Multivariate analysis | |||||||||||||
| 3-year OS | Log rank χ2 | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | ||||||
| Asian | 100.00% | 0 | 0 | / | 1 | |||||||||||
| Unknown | 75.00% | 1.378 | 0.323 | 5.873 | 0.665 | |||||||||||
| Stage | 6.42 | 0.093 | ||||||||||||||
| Stage I | 80.00% | 0.552 | 0.123 | 2.476 | 0.438 | 0.491 | 0.108 | 2.232 | 0.357 | |||||||
| Stage II | 95.12% | 0.329 | 0.115 | 0.944 | 0.039 | 0.209 | 0.071 | 0.615 | 0.004 | |||||||
| Stage III | 68.63% | Ref | Ref | |||||||||||||
| Stage IV | 77.19% | 1.283 | 0.451 | 3.652 | 0.641 | 1.666 | 0.555 | 5 | 0.363 | |||||||
| Site | 1.74 | 0.187 | ||||||||||||||
| Colon | 85.67% | Ref | ||||||||||||||
| Rectum | 72.85% | 1.747 | 0.755 | 4.045 | 0.193 | |||||||||||
| Histology | 0.06 | 0.814 | ||||||||||||||
| Mucinous adenocarcinoma | 90.91% | 1.191 | 0.278 | 5.108 | 0.814 | |||||||||||
| Other adenocarcinoma | 81.08% | Ref | ||||||||||||||
| MATH | 0.45 | 0.798 | ||||||||||||||
| Low | 80.15% | Ref | ||||||||||||||
| Intermediate | 82.75% | 0.938 | 0.361 | 2.434 | 0.895 | |||||||||||
| High | 82.72% | 0.722 | 0.269 | 1.942 | 0.519 | |||||||||||
| MATH (continuous variable) | 0.988 | 0.963 | 1.013 | 0.338 | ||||||||||||
| Multivariate analysis | ||||||||||||||||
| Male (n = 160) | Univariate analysis | Categorical variable (MATH) | Continuous variable (MATH) | |||||||||||||
| Characteristic | 3-year OS | Log rank χ2 | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | |
| Age | 4.76 | 0.093 | ||||||||||||||
| ≤50 | 90.79% | 1.272 | 0.39 | 4.149 | 0.69 | 1.09 | 0.322 | 3.689 | 0.89 | 1.187 | 0.358 | 3.94 | 0.779 | |||
| 51~70 | 86.24% | Ref | Ref | Ref | ||||||||||||
| >70 | 72.83% | 2.244 | 1.032 | 4.882 | 0.041 | 2.363 | 1.077 | 5.182 | 0.032 | 2.298 | 1.053 | 5.015 | 0.037 | |||
| Race | 1.72 | 0.633 | ||||||||||||||
| White | 79.85% | Ref | ||||||||||||||
| Black | 75.76% | 1.013 | 0.24 | 4.284 | 0.986 | |||||||||||
| Multivariate analysis | ||||||||||||||||
| Univariate analysis | Categorical variable (MATH) | Continuous variable (MATH) | ||||||||||||||
| Male (n = 160) | Characteristic | 3-year OS | Log rank χ2 | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value | HR | 95%CI low | 95%CI high | P value |
| Asian | 85.71% | 1.456 | 0.341 | 6.218 | 0.612 | |||||||||||
| Unknown | 87.50% | 0.322 | 0.044 | 2.37 | 0.266 | |||||||||||
| Stage | 10.3 | 0.016 | ||||||||||||||
| Stage I | 91.30% | 0.755 | 0.215 | 2.644 | 0.66 | 0.613 | 0.174 | 2.164 | 0.447 | 0.634 | 0.18 | 2.239 | 0.479 | |||
| Stage II | 86.57% | Ref | Ref | Ref | ||||||||||||
| Stage III | 76.91% | 0.762 | 0.323 | 1.799 | 0.535 | 0.467 | 0.183 | 1.192 | 0.111 | 0.534 | 0.215 | 1.327 | 0.177 | |||
| Stage IV | 52.98% | 2.88 | 1.229 | 6.748 | 0.015 | 2.361 | 0.966 | 5.769 | 0.06 | 2.189 | 0.894 | 5.355 | 0.086 | |||
| Site | 0.01 | 0.921 | ||||||||||||||
| Colon | 80.25% | Ref | ||||||||||||||
| Rectum | 79.07% | 1.044 | 0.444 | 2.458 | 0.921 | |||||||||||
| Histology | 0.12 | 0.725 | ||||||||||||||
| Mucinous adenocarcinoma | 88.89% | 1.238 | 0.377 | 4.066 | 0.725 | |||||||||||
| Other adenocarcinoma | 79.49% | Ref | ||||||||||||||
| MATH | 6.15 | 0.046 | ||||||||||||||
| Low | 88.04% | Ref | Ref | |||||||||||||
| Intermediate | 80.88% | 1.469 | 0.605 | 3.566 | 0.396 | 1.454 | 0.554 | 3.817 | 0.447 | |||||||
| High | 71.55% | 2.664 | 1.174 | 6.047 | 0.019 | 3.617 | 1.438 | 9.099 | 0.006 | |||||||
| MATH (continuous variable) | 1.023 | 1.003 | 1.043 | 0.025 | 1.025 | 1.004 | 1.047 | 0.017 | ||||||||
Abbreviations: OS: overall survival; CI: confidence interval; HR: hazard ratio
For patients of validation set, N stage and MATH were significantly associated with OS in univariate Cox regression. In subgroup analysis, MATH was a prognostic factor only in male patients as continuous variable or categorical variable. The unadjusted OS by MATH was calculated by Kaplan–Meier curves (Supplementary Figure 1, available at Carcinogenesis Online). After adjusting above dependent variables in multivariate Cox regression, MATH was still an independent prognostic factor in male patients. Detailed information was presented in Supplementary Table 2, available at Carcinogenesis Online.
MATH and somatic mutations
In order to find out the reasons behind the gender-related prognostic value of MATH, the relationship between MATH and genomic pattern in 284 CRC patients from TCGA was further analyzed, especially based on gender. Then we studied the connection between MATH and non-silent somatic mutation rate of distinct genes. The five most prevalent somatic mutations in each MATH group were shown in Supplementary Figure 2, available at Carcinogenesis Online. The mutation rates of APC, TP53, TTN, KRAS and NEFH ranked in the top five in all MATH group for all patients. OBSCN and MUC16 were only shown in the low MATH group on the list of top five, while NEFH and IRF5 were only in the high MATH group. Additionally, KRAS did not make the top five prevalent mutations in high MATH group as in low and intermediate group. In the subgroup analysis, we found that the somatic mutation pattern in different MATH groups was various by gender. For example, female patients had obviously lower mutation rates of APC than male in the low MATH group (55 versus 70.37%), but much higher in the high MATH group (90.7 versus 75%).
To further make research on the independent association between MATH and somatic mutation rate, we chose candidate genes by P < 0.05 with Chi-square test and evaluated independent effect of MATH by logistic regression model adjusting tumor stage, histology and primary site (Supplementary Table 3, available at Carcinogenesis Online). Only genes with a minimum mutation rate of 20% was included. All genes with mutation rate independently increasing or decreasing as MATH elevated was shown in Supplementary Table 4, available at Carcinogenesis Online. Then we explored deeply the impact of MATH score on TP53, IRF5 and KRAS in Supplementary Figure 3, available at Carcinogenesis Online. The mutation of TP53, as the second top prevalent mutations with 62.68% of patients, was independently correlated to higher MATH score in all patients (P < 0.001), as same as female (P = 0.003) and male (P < 0.001) subgroups. The frequency of IRF5 mutation significantly increased as MATH elevated in all (P = 0.017) and male patients (P = 0.005), however, there was no similar tendency in female patients (P = 0.82).The mutation rate of KRAS, which was associated with lower MATH, was revealed to drop independently when MATH increased only in all (P = 0.019) and male patients (P < 0.001), but not in female patients (0.444).
MATH score and somatic copy number alteration
Next, we would like to look for the relationship between MATH score and somatic copy number alteration (SCNA). As shown in Supplementary Table 5, available at Carcinogenesis Online, median fraction of SCNA for in 284 CRC patients from TCGA in low, intermediate and high MATH group was 9.34, 24.03 and 30.49% respectively. The fraction of SCNA was compared by MATH using one-way analysis of variance by ranks (Kruskal–Wallis Test) and P < 0.001. In pairwise comparison, it was validated that the fraction of SCNA significantly elevated along with an increasing in MATH, whether in all, female or male patients.
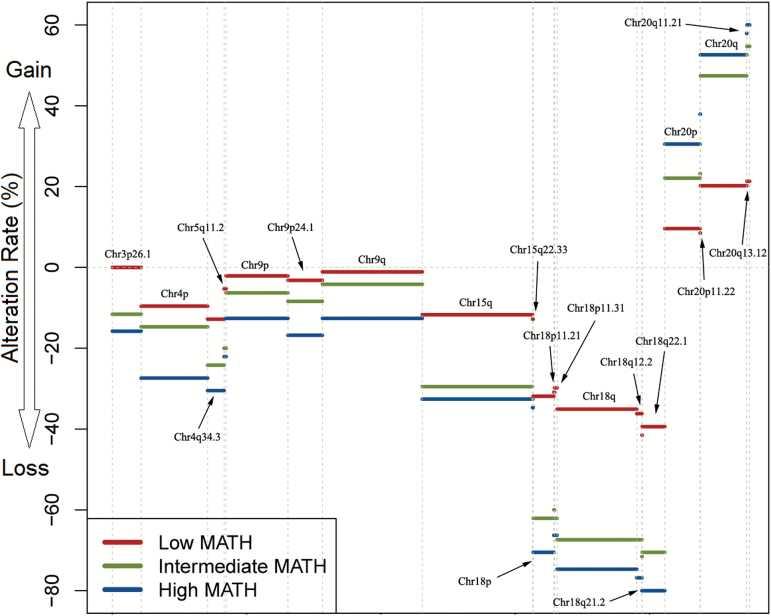
GISTIC 2.0 was utilized to find SCNA events related to MATH. Then logistic regression and Benjamini–Hochberg test was used to identify focal and arm-level SCNA event independently associated with MATH score (Figure 3). In a logistic regression model adjusting for multiple testing using the Benjamini–Hochberg approach, we demonstrated 21 SCNA events to be independently associated with MATH score for all patients, with 3 and 20 for female and male at a P value <0.05 respectively, which were shown in Supplementary Table 6, available at Carcinogenesis Online.
Figure 3.
Somatic copy number alteration events independently associated with mutant-allele tumor heterogeneity (MATH) with P < 0.05 in the logistic regression adjusting for multiple testing by Benjamini–Hochberg approach in the training cohort. Red indicates low MATH group, green indicates intermediate MATH group and blue indicates high MATH group. Alteration rate is displayed along the vertical axis, while the direction of arrow represents SCNA gain (above 0) or loss (below 0).
Gene enrichment in the high MATH groups
Finally, we investigated the association between MATH and gene expression on RNA level in 284 CRC patients from TCGA. GSEA was used to carry out analysis with 13311 gene sets downloaded from MSigDB. Compared to low MATH group, 166, 6 and 166 gene sets were upregulated with P < 0.05 as well as FDR q value <0.5 in high MATH group for all, female, and male patients, respectively. All these significant gene sets were summarized in Supplementary Tables 7–9, available at Carcinogenesis Online. The results presented higher expression of gene sets related to WNT pathway, TGF-β pathway and DNA repair deficiency arising at higher MATH score, while the latter two appeared only in male patients (Table 3). Heat maps for gene expression profile in these pathways of CRC patients with high and low MATH score (pink: high expression and blue: low expression) were show in Supplementary Figures 4–6, available at Carcinogenesis Online.
Table 3.
Hallmark gene sets that are highly expressed in high MATH group
| Gene set | ||
|---|---|---|
| ALL (n = 284) | NOM P-value | FDR q-value |
| HALLMARK_WNT_BETA_ CATENIN_SIGNALING | 0.002 | 0.330 |
| Male (n = 160) | ||
| HALLMARK_TGF_BETA_SIGNALING | 0.037 | 0.260 |
| HALLMARK_UV_RESPONSE_DN | 0.006 | 0.367 |
Discussion
ITH is recognized as a vital barrier to treatment of cancer because of resistance to targeted therapy resulting from heterogeneous subclones (15). Hence, high genetic heterogeneity was supposed to provide a risk for poor survival in patients with cancers. As a heterogeneous disease, ITH in CRC mainly derived from three general ways of genetic alteration: chromosomal instability (CIN), microsatellite instability and CpG island methylator phenotype (CIMP) (16,17).
As a novel method to measure ITH, MATH has been reported as biomarker of outcome of patients with head and neck squamous cell carcinoma in several previous studies (4,5). However, the differences according to ITH evaluated by MATH have not been thoroughly explored at genomic level so far. As far as we know, this is the first study focusing the prognostic value of MATH and further investigating the pattern of somatic mutations and SCNA and gene set enrichment according to MATH in CRC patients.
With published NGS data of a training set of 284 CRC patients from TCGA and an independent validation set of 187 CRC patients from ICGC, we found that higher ITH measured by MATH was an independent risk factor in male patients with CRC. Interestingly, the prognostic value of MATH is gender-dependent. Even though there is no obvious difference in MATH between female and male, our study reveal that genomic aberrations according to MATH in different sex groups are quite different, which may account for the gender-related prognostic value. It will be discussed in depth later.
Our study also indicates that MATH is associated with unique biological pattern on DNA and RNA level, such as somatic mutations, copy number alteration and expression of gene sets. Factors related to higher MATH score include advanced tumor stage, more lymph nodes involved, primary site in rectum, TP53 mutation and higher fraction of SCNA. As a well-known tumor suppressor, TP53 plays an important role in DNA repair and apoptosis regulation (18,19). The finding of more TP53 mutations in patients with higher MATH is in line with previous reports that TP53 involved in maintaining genetic stability (20,21). Additionally, it remains to be further studied why some mutations, such as KRAS, have significant lower frequency as MATH elevated.
Considering unique genomic pattern associated with MATH in male may contribute to its gender-related prognostic value, analyses were performed on the distinct genetic pattern related to MATH in gender subgroups. Evidences suggest that the impact of MATH on somatic mutations was different based on gender. IRF5, also known as the transcription factor interferon regulatory factor 5, has been reported as a p53 target gene and is required for DNA damage-induced apoptosis (22). It is interesting to note that IRF5 mutation increased as MATH elevated only in male patients. Therefore, we suggest male patients with higher MATH are more likely to be defective in DNA repair resulting from mutation of TP53 and IRF5, which would contribute to poor outcome. SCNA events are regarded to be relevant to tumor behavior (23,24). Some of SCNA events associated with MATH score in this study were reported as indicators of poor prognosis across human cancer (25–27). For example, we found copy number deletion of Chr18q21.2 was an independent indicator of elevated MATH only in male subgroup, with SMAD4 in peak region. SMAD4, a known CRC predisposition gene, has a key role in transforming growth factor-β (TGF-β) signaling pathway, and decreased SMAD4 mRNA levels appear to be associated with a worse prognosis and poorer response to 5-FU (28,29). It is yet to be further determine whether these SCNA events defined by GISTIC 2.0 are driver events of elevated ITH. Moreover, male patients have more SCNA events independently related to MATH, which might explain the gender-related prognostic value of MATH.
Another analysis on genomic aberration according to MATH was gene enrichment analysis by GSEA. We found that WNT associated gene sets were related to high MATH. Subgroup analysis in male patients demonstrated that gene sets related to TGF-βpathway and DNA repair deficiency were significantly unregulated in high MATH group. WNT pathway has a crucial role in oncogenesis. Mutation and altered expression of components of the canonical WNT pathway are linked to cancer as well as other human diseases. As a negative regulator of the WNT pathway, the mutation of tumor suppressor gene APC ranked the first in all MATH group for all CRC patients as presented above. TGF-β signaling pathway is involved in a wide range of cellular process and SMAD4 participates in composition of a heterotrimeric complex, which translocates into the nucleus and acts as a TGF-β-induced transcription factor for various genes. Considering the fact that deletion of Chr18q21.2 was linked to high MATH only for male patients, SMAD4 loss may account for altering TGF-β signaling. Numbers of studies have reported that loss of SMAD4 protein expression or SMAD4 copy number involves the generation of genomic instability, although the underlying mechanism remains to be further investigated (30–32). The UV response pathway, which is downregulated in high MATH group for the male, is a vital repair mechanisms of DNA damage-related injury (33). Generally, the ability of response to DNA damage is crucial to the maintenance of genomic stability and the prevention of cancers (34,35). The overexpression of gene sets related to DNA repair deficiency in the high MATH group within male patients corresponded to higher mutation of TP53 and IRF5 in male, which were both required for DNA repair (36). These results shed light on the deep mechanisms of how genomic aberration on genetic level predicted ITH. We supposed that the strategies targeting WNT signaling, TGF-β signaling and DNA repair deficiency, such as IWR-1 targeting APC–Axin–GSK3β complex, restoration of SMAD4 and DNA repair enzyme, may have potential clinical value in CRC patients with higher ITH (37–39).
Inevitably, our study had several limitations. Information of therapies was incomplete in the TCGA and ICGC data portal, which could act as confounding factors in the effect of MATH on survival and genetic pattern. Moreover, more detailed and direct evidences are needed to validate mechanisms of ITH with multiple methods in further researches. Nonetheless, we reveal for the first time that higher ITH measured by MATH in male patients with CRC was an independent risk factor for shorter overall survival. According evidences above, we hypothesize this finding resulted from biological distinction at different MATH score in male. Mutation of IRF5 and TP53, SCNA events independently related to MATH including deletion of Chr18q21.2, as well as TGF-β signaling and DNA repair deficiency predicted higher genetic heterogeneity measured by MATH and probably account for the gender-related prognostic value of MATH, since they were only observed in male patients.
Previous studies reported that genomic aberrations involving CIN and microsatellite instability and CIMP status in CRC are gender-associated (40–42). Even though CIN, microsatellite instability and CIMP reflect one aspect of ITH, they had little influence on MATH calculation. Median absolute deviation (MAD) and the median of its mutant-allele fractions at tumor-specific mutated loci are used to calculate MATH and minimize the influence of outlier loci. Although CIN is not reported in this article, it could be inferred from the data of SCNA events. It is evident from Supplementary Table 3, available at Carcinogenesis Online, that the difference of the proportion of SCNA between the two gender groups is little. However, we found that the SCNA events associated with MATH in male were obviously more than in female. So, we suggest that the association between MATH and SCNA might be different in the two gender groups, in other words, the association between MATH and CIN might be different in the two gender groups. Still, our study didn’t reveal the reason for this difference, but it might contribute to the gender-related prognostic value.
Notwithstanding, it remains to be studied in the future whether these molecular characteristics are driver of subclone formation and why they only show up in male. At present, the failure to target mutations driving resistance to chemotherapy in subclones imposes a restriction on predicting the response to treatment (15). Novel therapeutic approaches targeting potential mechanisms of ITH as well as therapeutic strategies according to specific MATH score are expected to be studied in further researches. Considering the promising role of NGS in future clinical practice, MATH, as a simple way to measure ITH, will be a significant biomarker in therapy selection and survival prediction for male patients with CRC.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This study was supported by the grant from the Hospital Foundation of Fudan University Shanghai Cancer Center (YJ201504).
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- CIN
chromosomal instability
- CRC
colorectal cancer
- ICGC
International Cancer Genome Consortium
- ITH
intra-tumor heterogeneity
- MAD
median absolute deviation
- MATH
mutant-allele tumor heterogeneity
- NGS
next-generation sequencing
- SCNA
somatic copy number alteration
- TCGA
The Cancer Genome Atlas
- TGF-β
transforming growth factor-β
References
- 1. Burrell R.A., et al. (2013) The causes and consequences of genetic heterogeneity in cancer evolution. Nature, 501, 338–345. [DOI] [PubMed] [Google Scholar]
- 2. Marusyk A., et al. (2010) Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta, 1805, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marusyk A., et al. (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer, 12, 323–334. [DOI] [PubMed] [Google Scholar]
- 4. Mroz E.A., et al. (2013) High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer, 119, 3034–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mroz E.A., et al. (2015) Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med., 12, e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Navin N., et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature, 472, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shah S.P., et al. (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature, 486, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J., et al. (2014) Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science, 346, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andor N., et al. (2016) Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med., 22, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roth A., et al. (2014) PyClone: statistical inference of clonal population structure in cancer. Nat. Methods, 11, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mroz E.A., et al. (2013) MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol., 49, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keenan T., et al. (2015) Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J. Clin. Oncol., 33, 3621–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rocco J.W. (2015) Mutant allele tumor heterogeneity (MATH) and head and neck squamous cell carcinoma. Head Neck Pathol., 9, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blanco-Calvo M., et al. (2015) Colorectal cancer classification and cell heterogeneity: a systems oncology approach. Int. J. Mol. Sci., 16, 13610–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turner N.C., et al. (2012) Genetic heterogeneity and cancer drug resistance. Lancet. Oncol., 13, e178–e185. [DOI] [PubMed] [Google Scholar]
- 16. Goel A., et al. (2003) Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res., 63, 1608–1614. [PubMed] [Google Scholar]
- 17. Goel A., et al. (2007) The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology, 132, 127–138. [DOI] [PubMed] [Google Scholar]
- 18. Fei P., et al. (2003) P53 and radiation responses. Oncogene, 22, 5774–5783. [DOI] [PubMed] [Google Scholar]
- 19. Marión R.M., et al. (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature, 460, 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Negrini S., et al. (2010) Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol., 11, 220–228. [DOI] [PubMed] [Google Scholar]
- 21. Wahl G.M., et al. (1997) Maintaining genetic stability through TP53 mediated checkpoint control. Cancer Surv., 29, 183–219. [PubMed] [Google Scholar]
- 22. Hu G., et al. (2005) Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res., 65, 7403–7412. [DOI] [PubMed] [Google Scholar]
- 23. Muzny D.M., et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sebat J., et al. (2004) Large-scale copy number polymorphism in the human genome. Science, 305, 525–528. [DOI] [PubMed] [Google Scholar]
- 25. Carethers J.M., et al. (1998) Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology, 114, 1188–1195. [DOI] [PubMed] [Google Scholar]
- 26. Douglas E.J., et al. (2004) Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res., 64, 4817–4825. [DOI] [PubMed] [Google Scholar]
- 27. Tuupanen S., et al. (2009) The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet., 41, 885–890. [DOI] [PubMed] [Google Scholar]
- 28. Alhopuro P., et al. (2005) SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin. Cancer Res., 11, 6311–6316. [DOI] [PubMed] [Google Scholar]
- 29. Boulay J.L., et al. (2002) SMAD4 is a predictive marker for 5-fluorouracil-based chemotherapy in patients with colorectal cancer. Br. J. Cancer, 87, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bornstein S., et al. (2009) Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J. Clin. Invest., 119, 3408–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salovaara R., et al. (2002) Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut, 51, 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan P., et al. (2016) Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin. Cancer Res., 22, 3037–3047. [DOI] [PubMed] [Google Scholar]
- 33. Sinha R.P., et al. (2002) UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci., 1, 225–236. [DOI] [PubMed] [Google Scholar]
- 34. Blokhina O., et al. (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot., 91 Spec No, 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park J.M., et al. (2014) Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PLoS One, 9, e100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith M.L., et al. (1995) Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene, 10, 1053–1059. [PubMed] [Google Scholar]
- 37. Benjdia A., et al. (2012) Structural insights into recognition and repair of UV-DNA damage by spore photoproduct lyase, a radical SAM enzyme. Nucleic Acids Res., 40, 9308–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwarte-Waldhoff I., et al. (2002) Smad4 transcriptional pathways and angiogenesis. Int. J. Gastrointest. Cancer, 31, 47–59. [DOI] [PubMed] [Google Scholar]
- 39. Takebe N., et al. (2015) Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol., 12, 445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ali R.H., et al. (2014) Gender-associated genomic differences in colorectal cancer: clinical insight from feminization of male cancer cells. Int. J. Mol. Sci., 15, 17344–17365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brim H., et al. (2012) Genomic aberrations in an African American colorectal cancer cohort reveals a MSI-specific profile and chromosome X amplification in male patients. PLoS One, 7, e40392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ogino S., et al. (2006) CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J. Mol. Diagn., 8, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.