The relationship between EMT-related genes and cancer susceptibility is largely unknown. We conduct a genome-wide association study and found that genetic variants in EMT-related genes are associated with NSCLC risk, suggesting that these genes may be involved in tumorigenesis.
Abstract
The epithelial–mesenchymal transition (EMT) process is a crucial step for tumor invasion and metastasis. Previous research investigating EMT has mostly focused on its role in cancer progression. Recent studies showed that EMT and EMT-driving transcription factor (EMT-TF) expression are early events in lung cancer pathogenesis, implying a potential association between EMT and lung cancer risk. In this study, we examined whether genetic variants in EMT-related genes are associated with risk of non-small cell lung cancer (NSCLC). We used data from a genome-wide association study of 1482 NSCLC cases and 1544 healthy controls as the discovery phase, in which we analyzed 1602 single-nucleotide polymorphisms (SNPs) within 159 EMT-related genes. We then validated the significant SNPs in another 5699 cases and 5815 controls from the National Cancer Institute lung cancer genome-wide association study. Cumulative effects were evaluated for validated SNPs, and a gene-based test was performed to explore gene-level association with disease risk. In the discovery phase, 174 SNPs demonstrated significant associations with NSCLC risk. In the validation phase, seven SNPs mapped to EGFR, NOTCH3, ADGRF1 and SMAD3 were confirmed. Cumulative effect analysis of the significant SNPs demonstrated increasing risk with the number of unfavorable genotypes in the discovery and validation datasets. Gene-based analysis implicated ADGRF1, NOTCH3 and CDH1 as significant for NSCLC risk. Functional prediction revealed several potential mechanisms underlying these associations. Our results suggest that EMT-related gene variants may be involved in susceptibility to NSCLC; if confirmed, they might help identify higher-risk individuals.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide (1). In the United States, an estimated 224390 new cases and approximately 158080 deaths are expected to occur in 2016 (2). Although it is primarily caused by environmental exposure, growing evidence has implied genetic factors in susceptibility to this disease. Recent large-scale genome-wide association studies have extensively evaluated the association between genetic variants and lung cancer risk, with multiple significant cancer-risk loci being identified (3–11). Despite these discoveries, additional loci that do not exceed the commonly used genome-wide significance threshold (P < 5 × 10−8) may still exist and remain to be identified (12).
Epithelial–mesenchymal transition (EMT) is essential during embryonic development, when polarized immotile epithelial cells give rise to motile mesenchymal cells, allowing them to adopt a migratory and invasive behavior (13). EMT is driven by some transcription factors (e.g. Snail, Zeb and Twist), together with epigenetic and post-translational regulators, and is characterized by the loss of cell adhesion, downregulation of epithelial markers (E-cadherin, occludins and claudins) and upregulation of mesenchymal markers (vimentin, fibronectin and N-cadherin) (14). The abnormal induction of EMT in cancer cells has been linked to their disassociation from the primary tumor and initiation of metastasis (14,15). It has been well established that the EMT process is a crucial step in the invasion–metastatic cascade, linked with immunosuppression and with chemotherapy and immunotherapy resistance (16). However, the observations that EMT-TFs are expressed in non-invasive tumors and that EMT-TFs have antiapoptotic effects under stress conditions suggest that EMT can be initiated in the early stages of tumorigenesis, long before the completion of the trans-differentiation process and initiation of tumor metastasis (17). For lung cancer, recent studies also showed that EMT and EMT-TF expression are early events in lung cancer pathogenesis, supporting a biologic basis for a relationship between the EMT process and lung cancer susceptibility (18,19).
Given the important role of EMT in lung cancer pathogenesis, we examined whether genetic variants in EMT-related genes are associated with non-small cell lung cancer (NSCLC) risk using a large two-phase genetic association study that analyzed 1602 single-nucleotide polymorphisms (SNPs) in 159 EMT-related genes.
Materials and methods
Study population and data collection
Study participants signed the informed consent and the study was approved by the Institutional Review Board of MD Anderson Cancer Center. The study design and participant recruitment for the discovery phase were described previously (20,21). Briefly, cases were identified from an ongoing lung cancer case–control study at MD Anderson. All patients were newly diagnosed and histologically confirmed to have NSCLC from 1995 through 2008. There were no age, sex, ethnicity or disease stage restrictions on case recruitment. The controls were healthy individuals with no prior history of any type of cancer (except for non-melanoma skin cancer) who were recruited from a Kelsey Seybold Clinic located in the Houston metropolitan area. A structured questionnaire was used to collect epidemiological data for all participants. A total of 1482 cases and 1544 controls were included in the discovery stage.
In the validation phase, the genotype data for cases and controls were obtained from the genome-wide association study lung cancer dataset from the Database of Genotypes and Phenotypes (dbGAP) (dbGAP Study Accession number phs000336.v1.p1) (6,22). This dataset consists of 5699 NSCLC and 5815 controls.
Genes and SNP selection
Genes whose expression can predict that NSCLC had undergone EMT and genes involved in the EMT pathway were selected (15,17,23). Overall, 159 genes were identified based on an extensive survey of the literature on EMT in cancer and NSCLC pathogenesis (Supplementary Table 1 is available at Carcinogenesis Online). The UCSC Genome Browser was used to obtain the chromosome positions of the start and end of each gene. We used data from the International HapMap Project for tagging SNPs identification. For each gene, tag SNPs located within 10 kb of the transcribed intervals were selected. The Tagger pairwise method (Broad Institute, Cambridge, MA) was used for tagging SNPs selection with r2 of 0.8 or higher and allele frequency of at least 0.05 in Caucasians. In addition, potential functional SNPs in the coding region, 5′-untranslated region, 3′-untranslated region, promoter region or splice sites were also included. A total of 1602 SNPs were identified for genotyping analysis.
Genotyping
Genomic DNA had been isolated from peripheral blood using the QIAamp DNA extraction kit (QIAGEN, Valencia, CA). Genotyping and quality control for the cohort in the discovery phase have been previously described (7,24). Briefly, genotyping was carried out using Illumina HumanHap 317k, 610k and 660k BeadChips. Quality control filters included samples or SNPs with a call rate of at least 95% and minor allele frequencies of at least 0.01. The same approach was used for the validation phase.
Statistical analysis
All statistical analyses were two-sided. Statistical analysis in the study was performed using Plink and Intercooled Stata 10.0 statistical software package (StataCorp LP, College Station, TX) (25). Deviations from the Hardy–Weinberg equilibrium were evaluated by calculating and then comparing the observed and expected frequencies of genotypes using the χ2 test. Differences between the case and control subjects were compared by the χ2 test or Fisher’s exact test for categorical variables and by Student’s t-test for continuous variables. Univariate and multivariate logistic regressions were applied in both the discovery and validation phases to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for each variant while adjusting for age, sex and smoking status in discovery phase and for age and sex in validation phase. Risk associations between genotypes and NSCLC susceptibility were estimated by OR using three different genetic models (dominant, additive and recessive models) to define the best-fitting model with the most significant P value, and only the best-fitting models were reported. Results of discovery and validation studies were polled by meta-analysis. The fixed-effect model is selected if the result of Cochran’s Q test for heterogeneity is 0.05 or higher; otherwise, a random-effect model would be adopted.
Cumulative analysis was performed to evaluate the cumulative effect of multiple variants on NSCLC risk. Unfavorable genotypes were divided into three groups according to the number of unfavorable genotypes in subjects (low risk: fewer than three unfavorable genotypes, median risk: three or four unfavorable genotypes and high risk: five to seven unfavorable genotypes). The group with the lowest risk was used as the reference group.
For gene-based testing, we used Versatile Gene-based Association Study (VEGAS) to summarize the effect of individual variants within each specific gene (26). For a defined gene, an extended range of 10 kb up- and downstream was used. SNP IDs and their corresponding P-values were inputted in the offline version of the program, which then produced the gene-based P-values for each gene.
For in silico functional assessment, online database HaploReg v4.1 was used for functional annotation and to identify proxy variants in high linkage disequilibrium (r2 > 0.8) with candidate SNPs (27). Another variant-centered interactive tool, SNiPA, was applied to predict the expression quantitative trait locus (eQTL) effects for each variant we examined (28).
Results
Characteristics of the study population
The characteristics of our discovery phase population are presented in Table 1. There were 1482 cases and 1544 controls in the discovery set, with no significant difference in the mean age between the two groups. Compared with the cases, the control group contained a significantly larger proportion of male participants. There was a statistically significant difference with regard to smoking status between the cases and controls (P < 0.001); specifically, more cases than controls were current smokers (37.2 versus 31.3%, respectively). There were 5699 cases and 5815 controls in the validation set; as in the discovery set, the age distributions for the two groups were similar and there were significantly more male participants in the control group (6).
Table 1.
Host characteristics of cases and controls in the discovery dataset
| Number of patients (%) | |||
|---|---|---|---|
| Variable | Cases, n = 1482 | Controls, n = 1544 | P-value |
| Age, mean (SD) | 61.56 (9.92) | 61.98 (11.36) | 0.274 |
| Sex | |||
| Male | 762 (51.42) | 940 (60.88) | <0.001 |
| Female | 720 (48.58) | 604 (39.12) | |
| Smoking status | |||
| Never | 328 (22.13) | 407 (26.36) | 0.001 |
| Former | 603 (40.69) | 654 (42.36) | |
| Current | 551 (37.18) | 483 (31.28) | |
| Pack-years, mean (SD)a | 44.58 (30.16) | 51.49 (31.41) | <0.001 |
SD, standard deviation.
aIn smokers only.
Risk associated with individual EMT-related gene SNPs
In the discovery phase, we screened 1602 SNPs within 159 EMT-related genes. None of these SNPs were deviated from Hardy–Weinberg equilibrium after adjustment for multiple testing (data not shown). We identified 174 SNPs with P values < 0.05 that were significantly associated with NSCLC risk (Supplementary Table 2 is available at Carcinogenesis Online); 10 SNPs remained significant after validation in the dbGAP dataset. We selected tagged SNPs at an r2 threshold of ≥0.8 to filter out highly correlated SNPs; 7 of the 10 SNPs met this criterion. These tagged SNPs were mapped to EGFR, ADGRF1 (GPR110), NOTCH3 and SMAD3 (Table 2). Among these SNPs, the SNPs from EGFR [rs884904 (AA+AG) and rs7809332 (CC+CT)] and from NOTCH3 [rs3815188 (AA+AG) and rs2238643 (TT+TC)] were associated with decreased risk of NSCLC, whereas the SNPs from ADGRF1 [rs6941183 (TT+TC) and rs1226500 (AA+AC)] and the SNP from SMAD3 [rs2118610 (TT+TC)] were shown to be associated with increased risk of NSCLC.
Table 2.
SNPs associated with NSCLC risk in both the discovery and validation populations
| Discovery | Validation | Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | Gene | SNP | Model | OR (95% CI)a | P-value | OR (95% CI)b | P-value | OR (95% CI) | P-value | P-het |
| chr7:55049691 | EGFR | rs884904 | Add | 0.83 (0.69–1.00) | 0.046 | 0.90 (0.82–0.99) | 0.027 | 0.89 (0.82–0.96) | 4.36 × 10–3 | 0.433 |
| chr7:55067570 | EGFR | rs7809332 | Add | 0.86 (0.76–0.97) | 0.014 | 0.91 (0.86–0.97) | 5.76 × 10–3 | 0.90 (0.85–0.95) | 3.54 × 10–4 | 0.397 |
| chr6:47069911 | ADGRF1 | rs1226500 | Add | 1.17 (1.05–1.31) | 5.53 × 10–3 | 1.06 (1.00–1.12) | 0.049 | 1.08 (1.03–1.13) | 2.77 × 10–3 | 0.108 |
| chr6:47105069 | ADGRF1 | rs6941183 | Dom | 1.23 (1.06–1.42) | 5.37 × 10–3 | 1.09 (1.01–1.18) | 0.021 | 1.12 (1.05–1.20) | 8.97 × 10–4 | 0.155 |
| chr19:15164225 | NOTCH3 | rs3815188 | Dom | 0.85 (0.72–0.99) | 0.042 | 0.91 (0.84–1.00) | 0.045 | 0.90 (0.83–0.97) | 6.62 × 10–3 | 0.423 |
| chr19:15160648 | NOTCH3 | rs2238643 | Add | 0.88 (0.78–0.99) | 0.027 | 0.94 (0.89–1.00) | 0.048 | 0.93 (0.88–0.98) | 6.80 × 10–3 | 0.318 |
| chr15:65215388 | SMAD3 | rs2118610 | Dom | 1.26 (1.08–1.47) | 3.98 × 10–3 | 1.12 (1.04–1.21) | 4.23 × 10–3 | 1.15 (1.07–1.23) | 1.02 × 10–4 | 0.190 |
Add, additive model; Dom, dominant model; P-het, P for heterogeneity test.
aAdjusted for age, sex and smoking status.
bAdjusted for age and sex.
Cumulative effects of genetic variants on NSCLC risk
To determine the combined effect of multiple genetic variants, the cumulative effect of the seven SNPs was assessed. We observed a dose effect in both the discovery and validation phases, with P-values of 3.84 × 10–6 and 6.37 × 10–4, respectively. In the discovery phase, compared with patients with fewer than three unfavorable genotypes, patients carrying three to four unfavorable genotypes had a 1.34-fold increase in risk (95% CI, 1.04–1.71; P = 0.02), and patients carrying five to seven unfavorable genotypes had a 1.69-fold increase in risk (95% CI, 1.33–2.15, P = 1.83 × 10–5). In the validation population, we did not observe a difference in the risk of developing NSCLC in patients with three to four risk genotypes compared with those with fewer than three risk genotypes (P = 0.61), but patients with five to seven unfavorable genotypes had a significantly higher NSCLC risk (OR, 1.18; 95% CI, 1.03–1.33; P = 0.012). To illustrate the effect per increase in 1 unfavorable genotype, we calculate the OR for trends, this yielded an estimated OR of 1.12 and 1.06 in the discovery and validation population, respectively. This means that, compared with those subjects with no unfavorable genotype, the OR for NSCLC risk in individuals with 5 unfavorable genotypes would be 1.76 (1.125) in the discovery and 1.34 (1.065) in the validation phases. For those carrying 6 unfavorable genotypes, the OR will increase to 1.97 and 1.42 for discovery and validation phase, respectively (Table 3). We also examined the cumulative effects stratified by smoking status in the discovery population and did not find any significant difference between ever-smokers and never-smokers (Supplementary Table 3 is available at Carcinogenesis Online).
Table 3.
Cumulative effect of unfavorable genotypes in NSCLC
| Group | Number of unfavorable genotypes | Cases, n (%) | Controls, n (%) | Adjusted OR | P-value |
|---|---|---|---|---|---|
| Discovery | |||||
| 0 | 0–2 | 138 (39.54) | 211 (60.46) | 1 (reference) | |
| 1 | 3–4 | 518 (46.46) | 597 (53.54) | 1.34 (1.04–1.71)a | 0.02 |
| 2 | 5–7 | 762 (52.62) | 686 (47.38) | 1.69 (1.33–2.15)a | 1.83 × 10–5 |
| OR per unfavorable genotype increase | 1.12 (1.07–1.17)a | 3.84 × 10–6 | |||
| Validation | |||||
| 0 | 0–2 | 568 (46.75) | 647 (53.25) | 1 (reference) | |
| 1 | 3–4 | 2034 (47.88) | 2214 (52.12) | 1.03 (0.90–1.17)b | 0.61 |
| 2 | 5–7 | 2999 (51.43) | 2832 (48.57) | 1.18 (1.03–1.33)b | 0.012 |
| OR per unfavorable genotype increase | 1.06 (1.03–1.08)b | 7.79 × 10–4 | |||
aAdjusted by age, sex and smoking status.
bAdjusted by age and sex.
Versatile Gene-based Association Study
To identify candidate genes influencing NSCLC risk, we conducted a gene-based analysis using the SNPs from the discovery and validation datasets. For the discovery phase, SNPs in 39 candidate genes were significantly (P < 0.05) associated with the risk of NSCLC (Supplementary Table 4 is available at Carcinogenesis Online). The results for three candidate genes, ADGRF1, NOTCH3 and CDH1, were confirmed in the validation dataset (Table 4). Interestingly, none of the individual SNPs genotyped in CDH1 were validated to be significantly associated with NSCLC risk.
Table 4.
Gene-based analysis for EMT-related genes and risk of NSCLC, presented as P-values
| Gene | CHR | Number of SNPs | P-value | |
|---|---|---|---|---|
| Discovery | Validation | |||
| ADGRF1 | 6 | 6 | 0.0014 | 0.0168 |
| NOTCH3 | 19 | 8 | 0.0010 | 0.0447 |
| CHD1 | 16 | 8 | 0.0010 | 0.0448 |
In silico functional prediction
Online databases, including HaploReg v4.1, SNiPA and GTEx, were applied to predict the potential mechanisms underlying the identified associations and determine proxy variants of the identified variants. HaploReg identified one SNP (rs2118610) that was located in promoter histone marks and five SNPs (rs884904, rs1226500, rs3815188, rs2238643 and rs2118610) that were located in enhancer histone marks. All seven SNPs were predicted to alter regulatory binding motifs, and three SNPs (rs884904, rs1226500 and rs2118610) were located in DNase hypersensitive sites (Table 5).
Table 5.
SNP function prediction
| SNP | Gene | Position | Promoter histone marks | Enhancer histone marks | DNase | Motifs changed | eQTL |
|---|---|---|---|---|---|---|---|
| rs884904 | EGFR | 3′ near gene | GI | GI | 6 altered motifs | ||
| rs7809332 | EGFR | 3′ near gene | 8 altered motifs | ||||
| rs6941183 | ADGRF1 | Intronic | 10 altered motifs | ADGRF1 | |||
| rs1226500 | ADGRF1 | 3′ near gene | 8 tissues | 8 tissues | 7 altered motifs | ||
| rs3815188 | NOTCH3 | Synonymous | IPSC, MUS | Hic1 | ILVBL | ||
| rs2238643 | NOTCH3 | Intronic | IPSC | Ik-3, NRSF, SMC3 | ILVBL | ||
| rs2118610 | SMAD3 | Intronic | LNG | 19 tissues | 6 tissues | Foxp1 | SMAD3 |
We found that SMAD3: rs2118610, an intronic variant, have a functional effect via its predicted location in promoter histone marks, and we identified it as a direct eQTL regulating the expression of SMAD3. Several other potential causal variants within SMAD3 that we identified in the HaploReg database included rs7178117, rs1545161 and rs6494633. These intronic variants were also predicted to have direct regulatory effects on SMAD3 through eQTL with SMAD3. NOTCH3: rs3815188, a synonymous variant, was identified as a potential cis-eQTL with one gene: ILVBL, located approximately 70 kb upstream from NOTCH3. The intronic variant TT+TC genotype of ADGRF1: rs6941183 was associated with increased risk of NSCLC. Although we did not find any significant association of this SNP with ADGRF1 mRNA expression levels in the databases, we did find that a proxy SNP (rs16875384) showing high linkage disequilibrium with this SNP has been reported to have direct eQTL effects on ADGRF1 (P < 1.00 × 10–16) in lung tissue (29). We also conducted eQTL analysis using TCGA data to examine whether the seven SNPs were associated with altered gene expression. For SNPs without genotyping data in TCGA, the linked SNPs (r2 > 0.80) identified from HaploReg (v4.1) were used for analysis. As shown in Supplementary Table 5, which is available at Carcinogenesis Online, we found significant associations of rs7245563 and rs7178117 with NOTCH3 and SMAD3 mRNA expression levels, respectively. A borderline significance (P = 0.073) was also noted for rs1552633 and ADGRF1.
Discussion
Previous studies on EMT-related genes have mostly been limited to their expression and cancer invasion and metastasis. Recent study by Amankwah et al. (30) investigated EMT-related gene variants and susceptibility of epithelial ovarian cancer, and did not observe significant association. We hypothesized that genes implicated in the EMT process may influence the risk of NSCLC. The results of this two-stage study support our hypothesis; we identified seven SNPs of EMT-related genes that were associated with NSCLC risk.
Two of the identified SNPs are within the EGFR gene. EGFR is a member of a family consisting of four related members of transmembrane receptor tyrosine kinases (31). It is important for cell proliferation, differentiation, migration and resistance to apoptosis (32). EGFR is frequently overexpressed or mutated in NSCLC and is of prognostic significance (33). Studies have demonstrated that the activation of the EGFR pathway induces EMT in NSCLC (34). Furthermore, the pathway is also involved in tumorigenesis; EGFR ligands are expressed in NSCLCs and may activate EGFR by autocrine loops, subsequently leading to the stimulation of downstream signaling pathways, driving the malignant phenotype (35). Previous studies have reported several SNPs within EGFR to be associated with lung cancer risk (36–39). In our study, EGFR: rs884904 and rs7809332 were validated to be associated with decreased risks of developing NSCLC. Interestingly, rs7809332 (CC + CT) was previously identified as a protective genotype in a study conducted in Asian never-smoking women (39). Further functional studies are warranted to address the mechanism underlying this association.
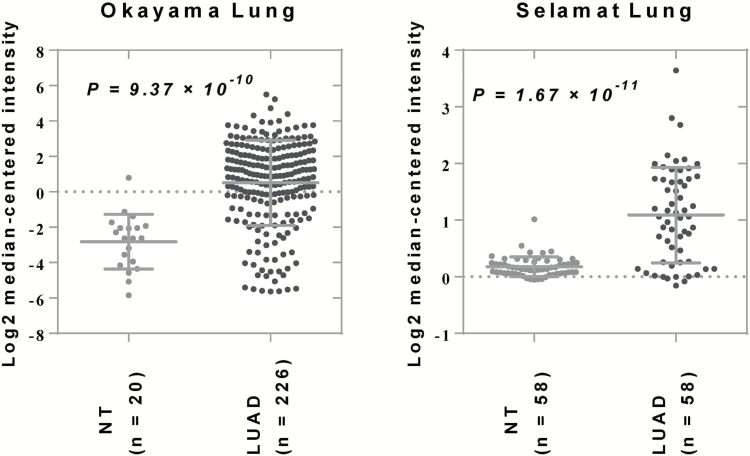
Both the individual SNP analysis and the gene-level test implicated the ADGRF1 gene as a predictor of NSCLC risk. ADGRF1 is an orphan G-protein-coupled receptor—a receptor with limited known function. Previous studies reported this cell surface protein to be an oncoprotein that is overexpressed in lung cancer (40,41). A later study found that ADGRF1 mRNA expression was positively correlated with E-cadherin (CDH1) and negatively correlated with vimentin and N-cadherin (CDH2), suggesting a potential role of ADGRF1 in the EMT process (23). We identified ADGRF1: rs6941183, an intronic variant, as a significant variant associated with NSCLC risk in both the discovery and validation populations. Our functional prediction found that one 3′-untranslated region variant, rs16875384, was in high linkage disequilibrium with this SNP, which has been described as a strong eQTL for ADGRF1 in human lung tissue (29). Although the association between ADGRF1 and lung cancer remains largely unknown, gene expression analysis based on Oncomine and public gene expression (GEO33479) datasets showed overexpression of ADGRF1 in lung squamous dysplasia or cancer specimens, suggesting there might be a role of this gene in lung carcinogenesis (Figure 1 and Supplementary Figure 1 is available at Carcinogenesis Online).
Figure 1.
Scatter plots showing the mRNA expression of ADGRF1 in two Oncomine datasets, Okayama Lung and Selamat Lung. LUAD, lung adenocarcinoma; NT, normal tissue.
Notch signaling plays a crucial role in the development and homeostasis of most tissues. Dysregulation of Notch signaling has been reported in various types of diseases, including lung carcinogenesis (42,43). As one of the candidate genes was significant in our gene-based analysis, NOTCH3 is expressed in NSCLC and plays a tumor-promoting role in the context of cell adhesion and EMT (44). Using a public gene expression data, we observed an upregulated expression of this gene in multiple steps of lung carcinogenesis (Supplementary Figure 2 is available at Carcinogenesis Online). Apart from its expression in cancer cells, we found in Protein Atlas that Notch3 was also expressed in immune cells in multiple cancer sites including lung cancer. Interestingly, previous studies indicated that high level of Notch3 might result in reduced T-cell activation (45). Later study demonstrated that Notch3 overexpression could trigger the trans-activation of Foxp3 promoter and positively regulate the expression of Foxp3, which is a marker of regulatory T cells (46). These studies suggested that Notch3 might be associated with an inhibitory immune microenvironment. Analysis of the discovery and validation samples supported association of two genetic variants genotyped in NOTCH3, rs3815188 and rs2238643 with NSCLC risk. The risk alleles were associated with reduced NSCLC risks in our study. Rs3815188 is a synonymous SNP that does not alter the resultant protein sequence. However, the literature has shown that synonymous SNPs may directly alter miRNA binding or protein folding or affect mRNA expression (47). Alternatively, this SNP may represent a tagging SNP that tags other functional SNP(s), which warrants further investigation. The other identified SNP, rs2238643, is located in the intronic region. Interestingly, we identified a potential cis-eQTL for these two variants with ILVBL, a gene that is approximately 70 kb upstream from NOTCH3.
Smad3 is a critical transcriptional factor that, through transcriptional regulation, controls the expression of transforming growth factor-β1 and its target genes (48). Compelling evidence supports a central role of Smad3 in transforming growth factor-β-dependent EMT associated with tumor progression and metastasis. Lin et al. (49) reported that genetic variants in SMAD3 were potential predictors of overall survival in NSCLC patients treated with chemotherapy. Previous studies also demonstrated that interference with endogenous Smad2/3 signaling enhanced the malignancy of xenografted tumors of premalignant and well-differentiated tumor cells, suggesting a relationship between this gene and tumorigenesis (50,51). Our results indicated that SMAD3: rs21188610, an intronic variant, was associated with NSCLC risk. Functional prediction implicated this variant as a direct eQTL regulating the expression of SMAD3, suggesting that it is located within a region that directly affects SMAD3 expression, potentially through the modulation of the promoter flanking region (52).
Although our results indicated only a moderate effect of individual SNPs on NSCLC risk, we found that an increasing number of unfavorable genotypes significantly increased risk of NSCLC in the cumulative analysis. It may be possible to use the combined information from the seven SNPs to assess an individual patient’s risk of NSCLC, but more validation must precede any such risk assessments.
While our study has relatively large sample size and a two-stage study design that included a total of 7181 cases and 7359 controls, there is also limitation. Our findings may not be generalized to other ethnicities as our population are mainly limited to non-Hispanic whites. However, our study may also benefit from this as the homogenous population may reduce the effects of population heterogeneity. Additional studies are needed to examine the association of the validated SNPs with NSCLC risk in other racial/ethnic groups.
In summary, the present study suggests that multiple common germline SNPs in EMT-related genes play a significant role in susceptibility to NSCLC. Future studies are needed to confirm these findings and elucidate how these SNPs are involved in NSCLC etiology.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This work was supported, in part, by grants from the NIH (P50 CA070907, R01 CA176568) (to X.W.), Cancer Prevention and Research Institute of Texas (RP130502) (to X.W.), and The University of Texas MD Anderson Cancer Center institutional support for the Center for Translational and Public Health Genomics.
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- CI
confidence interval
- EMT
epithelial–mesenchymal transition
- EMT-TF
EMT-driving transcription factor
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- SNP
single-nucleotide polymorphism
- VEGAS
Versatile Gene-based Association Study
References
- 1. Torre LA, et al. (2012) Global cancer statistics. CA. Cancer. J. Clin., 2015;65, 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R.L., et al. (2016) Cancer statistics, 2016. CA. Cancer J. Clin., 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 3. Broderick P., et al. (2009) Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer. Res., 69, 6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thorgeirsson T.E., et al. (2008) A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature, 452, 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., et al. (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Landi M.T., et al. (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amos C.I., et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu P., et al. (2008) Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J. Natl. Cancer Inst., 100, 1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hung R.J., et al. (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., et al. (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet., 46, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Truong T., et al. (2010) Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl. Cancer Inst., 102, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zuk O., et al. (2012) The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA, 109, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derynck R., et al. (2014) Signaling pathway cooperation in TGF-beta-induced epithelial-mesenchymal transition. Curr. Opin. Cell Biol., 31, 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nieto M.A., et al. (2016) EMT: 2016. Cell. 166, 21–45. [DOI] [PubMed] [Google Scholar]
- 15. De Craene B., et al. (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer., 13, 97–110. [DOI] [PubMed] [Google Scholar]
- 16. Ye L.Y., et al. (2016) Hypoxia-induced epithelial-to-mesenchymal transition in hepatocellular carcinoma induces an immunosuppressive tumor microenvironment to promote metastasis. Cancer Res., 76, 818–830. [DOI] [PubMed] [Google Scholar]
- 17. Puisieux A., et al. (2014) Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol., 16, 488–494. [DOI] [PubMed] [Google Scholar]
- 18. Larsen J.E., et al. (2016) ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Invest., 126, 3219–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei Q., et al. (2016) LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene., 35, 2655–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin J., et al. (2012) Systematic evaluation of apoptotic pathway gene polymorphisms and lung cancer risk. Carcinogenesis, 33, 1699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pu X., et al. (2012) Predictors of survival in never-smokers with non-small cell lung cancer: a large-scale, two-phase genetic study. Clin. Cancer. Res., 18, 5983–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tryka K.A., et al. (2014) NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic. Acids. Res., 42(Database issue):D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Byers L.A., et al. (2013) An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer. Res., 19, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y., et al. (2010) Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet. Oncol., 11, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purcell S., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mishra A., et al. (2015) VEGAS2: software for more flexible gene-based testing. Twin. Res. Hum. Genet., 18, 86–91. [DOI] [PubMed] [Google Scholar]
- 27. Ward L.D., et al. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants . Nucleic. Acids. Res., 40(Database issue):D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnold M. et al. (2015) SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics (Oxford, England)., 31, 1334–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hao K., et al. (2012) Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet., 8, e1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amankwah E.K., et al. (2015) Epithelial–mesenchymal transition (EMT) gene variants and epithelial ovarian cancer (EOC) risk. Genet. Epidemiol., 39, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hynes N.E., et al. (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer., 5, 341–354. [DOI] [PubMed] [Google Scholar]
- 32. Olayioye M.A., et al. (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J., 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gazdar A.F. (2010) Epidermal growth factor receptor inhibition in lung cancer: the evolving role of individualized therapy. Cancer Metastasis Rev., 29, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravi J., et al. (2016) Cannabinoid receptor-2 agonist inhibits macrophage induced EMT in non-small cell lung cancer by downregulation of EGFR pathway. Mol. Carcinog., 55, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laurie S.A., et al. (2013) Role of epidermal growth factor receptor inhibitors in epidermal growth factor receptor wild-type non-small-cell lung cancer. J. Clin. Oncol., 31, 1061–1069. [DOI] [PubMed] [Google Scholar]
- 36. Choi J.E., et al. (2007) Polymorphisms in the epidermal growth factor receptor gene and the risk of primary lung cancer: a case-control study. BMC Cancer., 7, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang W., et al. (2006) Mutation and polymorphism in the EGFR-TK domain associated with lung cancer. J. Thorac. Oncol., 1, 635–647. [PubMed] [Google Scholar]
- 38. Jou Y.S., et al. (2009) Association of an EGFR intron 1 SNP with never-smoking female lung adenocarcinoma patients. Lung Cancer (Amsterdam, Netherlands)., 64, 251–256. [DOI] [PubMed] [Google Scholar]
- 39. Chen KY, et al. (2013) EGFR polymorphisms, hormone replacement therapy and lung adenocarcinoma risk: analysis from a genome-wide association study in never-smoking women. Carcinogenesis., 34, 612–619. [DOI] [PubMed] [Google Scholar]
- 40. Lum A.M., et al. (2010) Orphan receptor GPR110, an oncogene overexpressed in lung and prostate cancer. BMC Cancer., 10, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hasan A.N., et al. (2015) An in silico analytical study of lung cancer and smokers datasets from Gene Expression Omnibus (GEO) for prediction of differentially expressed genes. Bioinformation., 11, 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen T.D., et al. (2011) Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer. Res., 71, 6010–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin L., et al. (2010) Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer Res., 70, 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hassan W.A., et al. (2016) Evaluation of role of Notch3 signaling pathway in human lung cancer cells. J. Cancer Res. Clin. Oncol., 142, 981–993. [DOI] [PubMed] [Google Scholar]
- 45. Maekawa Y., et al. (2003) Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity, 19, 549–559. [DOI] [PubMed] [Google Scholar]
- 46. Barbarulo A., et al. (2011) Notch3 and canonical NF-kappaB signaling pathways cooperatively regulate Foxp3 transcription. J. Immunol., 186, 6199–6206. [DOI] [PubMed] [Google Scholar]
- 47. Ho P.A., et al. (2011) WT1 synonymous single nucleotide polymorphism rs16754 correlates with higher mRNA expression and predicts significantly improved outcome in favorable-risk pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. J. Clin. Oncol., 29, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ju W., et al. (2006) Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. J. Mol. Cell. Biol., 26, 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin M., et al. (2011) Genetic variations in the transforming growth factor-beta pathway as predictors of survival in advanced non-small cell lung cancer. Carcinogenesis., 32, 1050–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian F., et al. (2003) Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res., 63, 8284–8292. [PubMed] [Google Scholar]
- 51. Zavadil J., et al. (2005) TGF-beta and epithelial-to-mesenchymal transitions. Oncogene., 24, 5764–5774. [DOI] [PubMed] [Google Scholar]
- 52. Grundberg E., et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet., 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.